Structural Insights into Plant Viruses Revealed by Small-Angle X-ray Scattering and Atomic Force Microscopy
Abstract
1. Introduction
2. Structural Investigation of Helical Plant Viruses by Small-Angle X-ray Scattering
2.1. Methods of SAXS Measurements, Data Analysis and Interpretation in Relation to Structural Study of Helical Plant Viruses
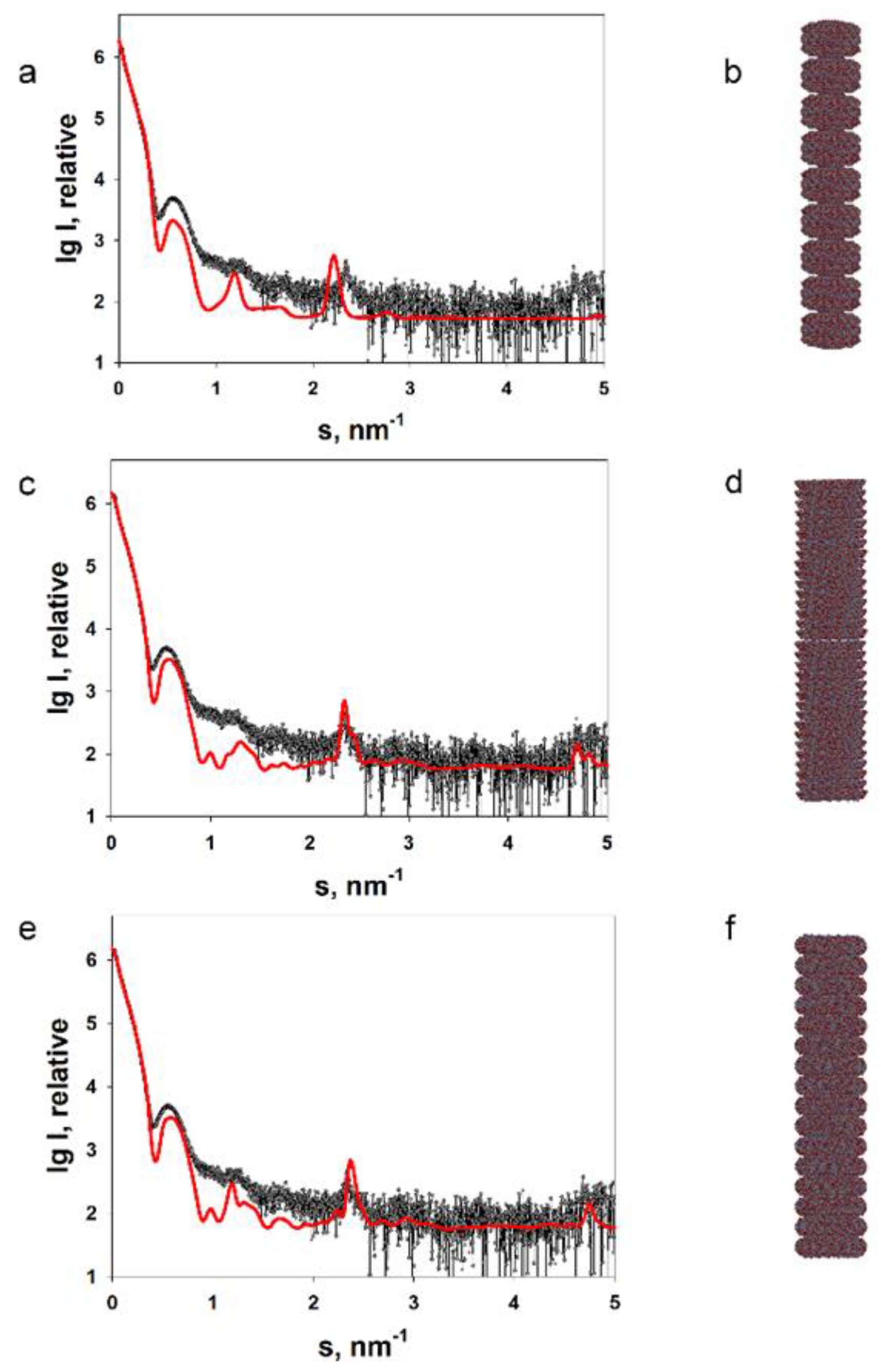
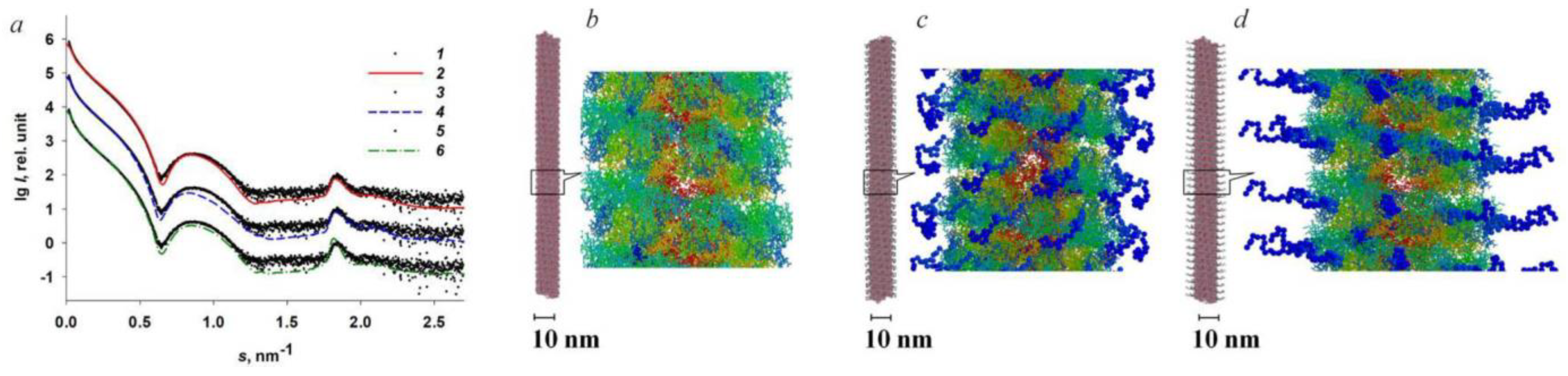
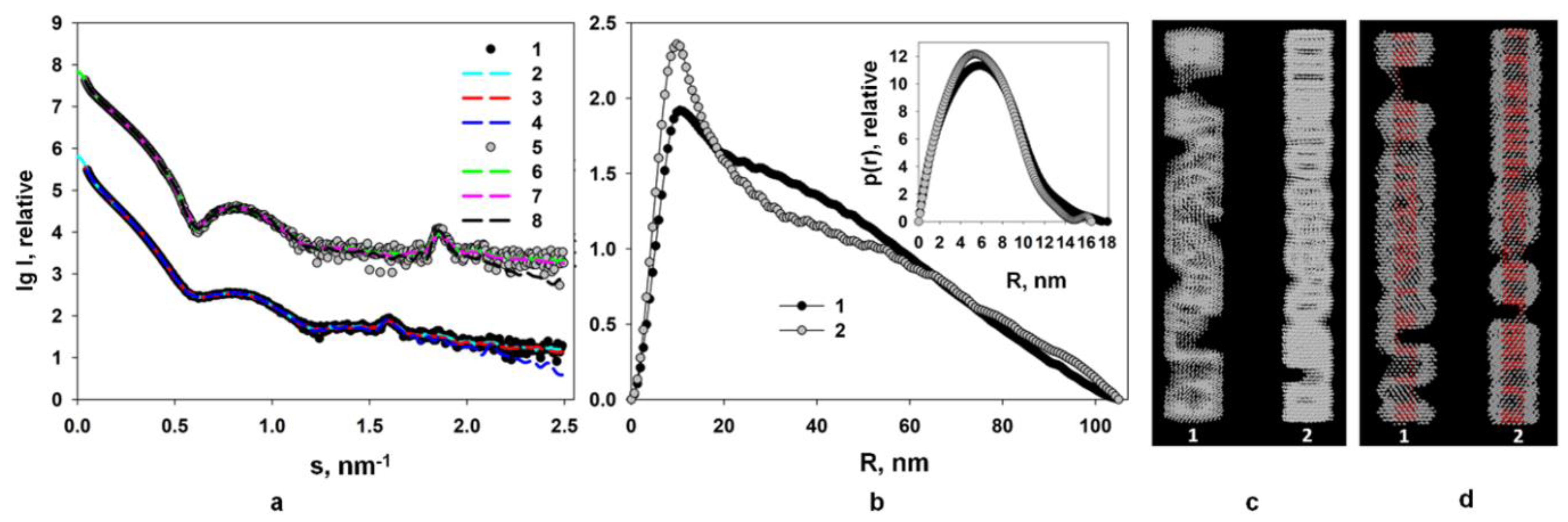
2.2. Structural Study of Helical Plant Viruses Based on SAXS Measurements
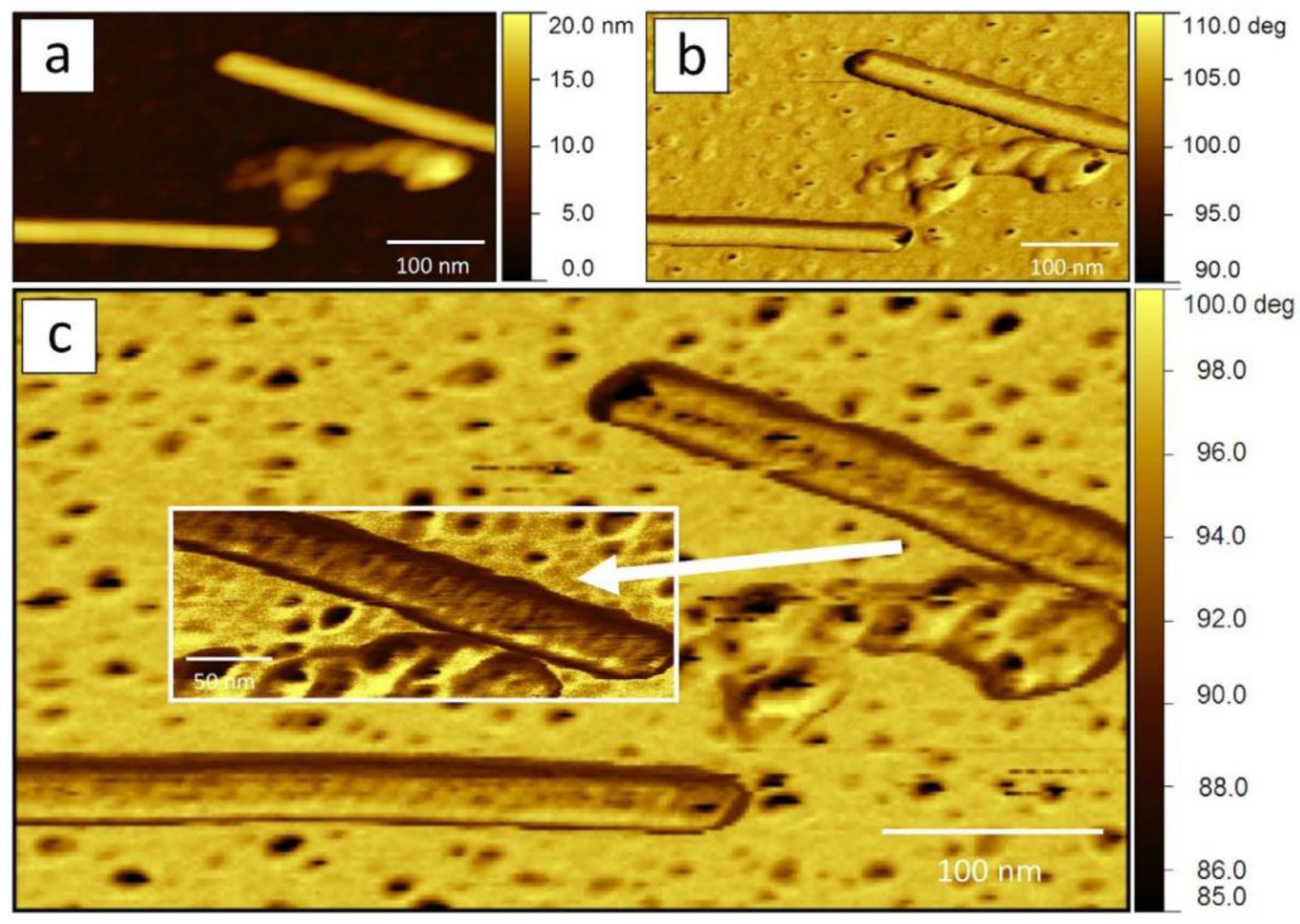
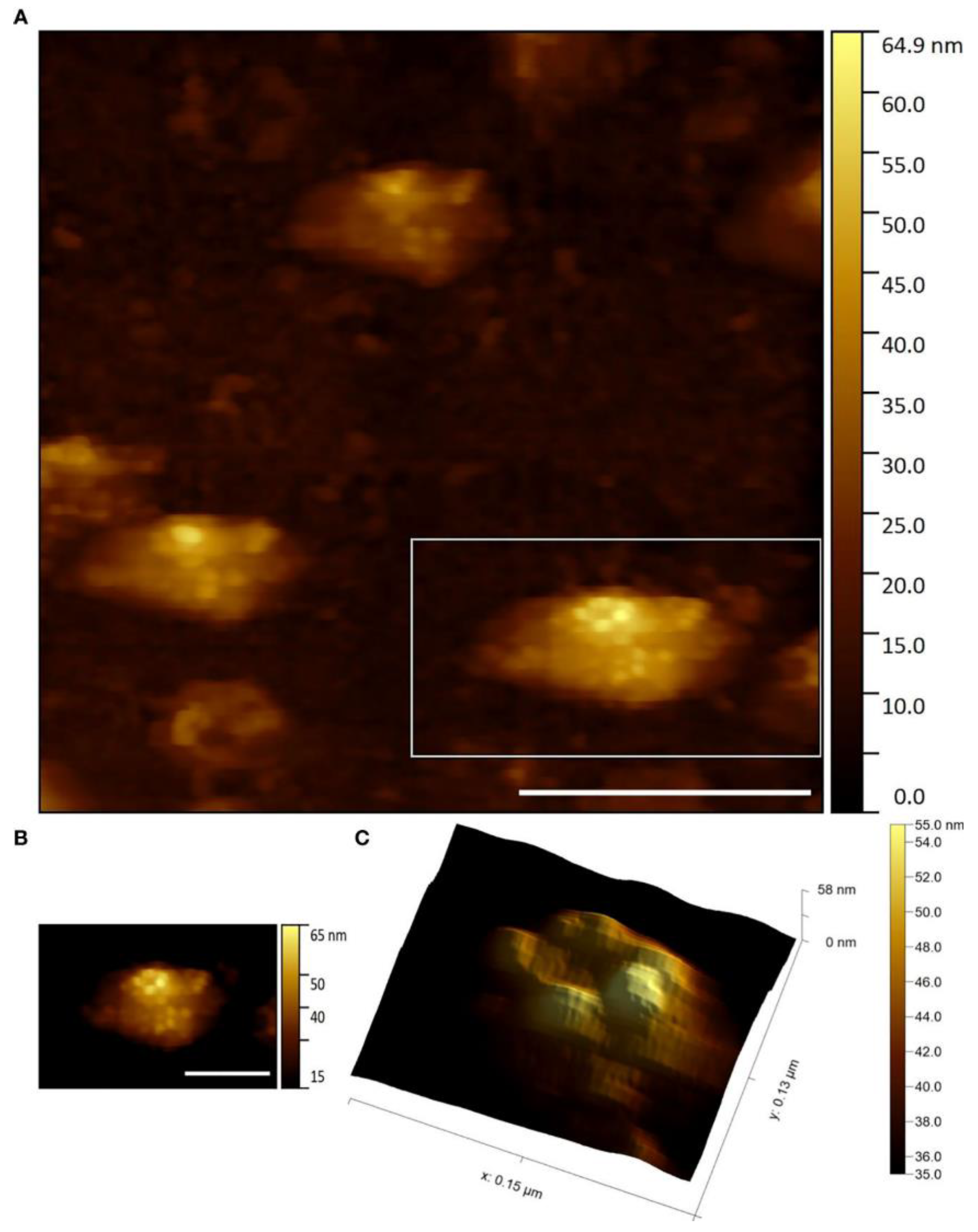
3. Atomic Force Microscopy Investigation of Plant Viruses
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murphy, F.A.; Fauquet, C.M.; Bishop, D.H.L.; Ghabrial, S.A.; Jarvis, A.W.; Martelli, G.P.; Mayo, M.A.; Summers, M.D. (Eds.) Virus Taxonomy; Springer: Vienna, Austria, 1995; ISBN 978-3-211-82594-5. [Google Scholar]
- Carstens, E.B. Ratification Vote on Taxonomic Proposals to the International Committee on Taxonomy of Viruses (2009). Arch. Virol. 2010, 155, 133–146. [Google Scholar] [CrossRef]
- Hefferon, K. Plant-derived Pharmaceuticals for the Developing World. Biotechnol. J. 2013, 8, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Steele, J.F.C.; Peyret, H.; Saunders, K.; Castells-Graells, R.; Marsian, J.; Meshcheriakova, Y.; Lomonossoff, G.P. Synthetic Plant Virology for Nanobiotechnology and Nanomedicine. WIREs Nanomed. Nanobiotechnol. 2017, 9, e1447. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, N.F.; Calder, G.; Lomonossoff, G.P.; Evans, D.J. Plant Viral Capsids as Nanobuilding Blocks: Construction of Arrays on Solid Supports. Langmuir 2006, 22, 10032–10037. [Google Scholar] [CrossRef]
- Lomonossoff, G.P.; Wege, C. TMV Particles: The Journey from Fundamental Studies to Bionanotechnology Applications. In Advances in Virus Research; Elsevier: Berlin/Heidelberg, Germany, 2018; Volume 102, pp. 149–176. ISBN 978-0-12-815194-5. [Google Scholar]
- Love, A.J.; Makarov, V.; Yaminsky, I.; Kalinina, N.O.; Taliansky, M.E. The Use of Tobacco Mosaic Virus and Cowpea Mosaic Virus for the Production of Novel Metal Nanomaterials. Virology 2014, 449, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Rahimian, N.; Miraei, H.R.; Amiri, A.; Ebrahimi, M.S.; Nahand, J.S.; Tarrahimofrad, H.; Hamblin, M.R.; Khan, H.; Mirzaei, H. Plant-Based Vaccines and Cancer Therapy: Where Are We Now and Where Are We Going? Pharmacol. Res. 2021, 169, 105655. [Google Scholar] [CrossRef]
- Solovyev, A.G.; Makarov, V.V. Helical Capsids of Plant Viruses: Architecture with Structural Lability. J. Gen. Virol. 2016, 97, 1739–1754. [Google Scholar] [CrossRef]
- Stubbs, G.; Kendall, A. Helical Viruses. In Viral Molecular Machines; Rossmann, M.G., Rao, V.B., Eds.; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 2012; Volume 726, pp. 631–658. ISBN 978-1-4614-0979-3. [Google Scholar]
- Kendall, A.; McDonald, M.; Bian, W.; Bowles, T.; Baumgarten, S.C.; Shi, J.; Stewart, P.L.; Bullitt, E.; Gore, D.; Irving, T.C.; et al. Structure of Flexible Filamentous Plant Viruses. J. Virol. 2008, 82, 9546–9554. [Google Scholar] [CrossRef]
- Valle, M. Structural Homology Between Nucleoproteins of ssRNA Viruses. In Virus Protein and Nucleoprotein Complexes; Harris, J.R., Bhella, D., Eds.; Subcellular Biochemistry; Springer: Singapore, 2018; Volume 88, pp. 129–145. ISBN 978-981-10-8455-3. [Google Scholar]
- Grinzato, A.; Kandiah, E.; Lico, C.; Betti, C.; Baschieri, S.; Zanotti, G. Atomic Structure of Potato Virus X, the Prototype of the Alphaflexiviridae Family. Nat. Chem. Biol. 2020, 16, 564–569. [Google Scholar] [CrossRef]
- Kežar, A.; Kavčič, L.; Polák, M.; Nováček, J.; Gutiérrez-Aguirre, I.; Žnidarič, M.T.; Coll, A.; Stare, K.; Gruden, K.; Ravnikar, M.; et al. Structural Basis for the Multitasking Nature of the Potato Virus Y Coat Protein. Sci. Adv. 2019, 5, eaaw3808. [Google Scholar] [CrossRef]
- Charon, J.; Theil, S.; Nicaise, V.; Michon, T. Protein Intrinsic Disorder within the Potyvirus Genus: From Proteome-Wide Analysis to Functional Annotation. Mol. BioSyst. 2016, 12, 634–652. [Google Scholar] [CrossRef]
- Ksenofontov, A.L.; Paalme, V.; Arutyunyan, A.M.; Semenyuk, P.I.; Fedorova, N.V.; Rumvolt, R.; Baratova, L.A.; Järvekülg, L.; Dobrov, E.N. Partially Disordered Structure in Intravirus Coat Protein of Potyvirus Potato Virus A. PLoS ONE 2013, 8, e67830. [Google Scholar] [CrossRef]
- Namba, K.; Pattanayek, R.; Stubbs, G. Visualization of Protein-Nucleic Acid Interactions in a Virus. J. Mol. Biol. 1989, 208, 307–325. [Google Scholar] [CrossRef]
- Bloomer, A.C.; Champness, J.N.; Bricogne, G.; Staden, R.; Klug, A. Protein Disk of Tobacco Mosaic Virus at 2.8 Å Resolution Showing the Interactions within and between Subunits. Nature 1978, 276, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G. Structure of Viruses: A Short History. Quart. Rev. Biophys. 2013, 46, 133–180. [Google Scholar] [CrossRef] [PubMed]
- Verdaguer, N.; Garriga, D.; Fita, I. X-Ray Crystallography of Viruses. In Structure and Physics of Viruses; Mateu, M.G., Ed.; Subcellular Biochemistry; Springer: Dordrecht, The Netherlands, 2013; Volume 68, pp. 117–144. ISBN 978-94-007-6551-1. [Google Scholar]
- Castón, J.R. Conventional Electron Microscopy, Cryo-Electron Microscopy and Cryo-Electron Tomography of Viruses. In Structure and Physics of Viruses; Mateu, M.G., Ed.; Subcellular Biochemistry; Springer: Dordrecht, The Netherlands, 2013; pp. 79–115. ISBN 978-94-007-6551-1. [Google Scholar]
- Adams, M.C.; Schiltz, C.J.; Heck, M.L.; Chappie, J.S. Crystal Structure of the Potato Leafroll Virus Coat Protein and Implications for Viral Assembly. J. Struct. Biol. 2022, 214, 107811. [Google Scholar] [CrossRef] [PubMed]
- Ruszkowski, M.; Strugala, A.; Indyka, P.; Tresset, G.; Figlerowicz, M.; Urbanowicz, A. Cryo-EM Reconstructions of BMV-Derived Virus-like Particles Reveal Assembly Defects in the Icosahedral Lattice Structure. Nanoscale 2022, 14, 3224–3233. [Google Scholar] [CrossRef] [PubMed]
- Harder, O.F.; Barrass, S.V.; Drabbels, M.; Lorenz, U.J. Fast Viral Dynamics Revealed by Microsecond Time-Resolved Cryo-EM. Nat. Commun. 2023, 14, 5649. [Google Scholar] [CrossRef] [PubMed]
- Clare, D.K.; Pechnikova, E.V.; Skurat, E.V.; Makarov, V.V.; Sokolova, O.S.; Solovyev, A.G.; Orlova, E.V. Novel Inter-Subunit Contacts in Barley Stripe Mosaic Virus Revealed by Cryo-Electron Microscopy. Structure 2015, 23, 1815–1826. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, T.; Bohon, J.; Gagné, M.-È.L.; Bolduc, M.; Leclerc, D.; Li, H. Crystal Structure of the Coat Protein of the Flexible Filamentous Papaya Mosaic Virus. J. Mol. Biol. 2012, 422, 263–273. [Google Scholar] [CrossRef]
- DiMaio, F.; Chen, C.-C.; Yu, X.; Frenz, B.; Hsu, Y.-H.; Lin, N.-S.; Egelman, E.H. The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses. Nat. Struct. Mol. Biol. 2015, 22, 642–644. [Google Scholar] [CrossRef] [PubMed]
- Agirrezabala, X.; Méndez-López, E.; Lasso, G.; Sánchez-Pina, M.A.; Aranda, M.; Valle, M. The Near-Atomic cryoEM Structure of a Flexible Filamentous Plant Virus Shows Homology of Its Coat Protein with Nucleoproteins of Animal Viruses. eLife 2015, 4, e11795. [Google Scholar] [CrossRef] [PubMed]
- Zamora, M.; Méndez-López, E.; Agirrezabala, X.; Cuesta, R.; Lavín, J.L.; Sánchez-Pina, M.A.; Aranda, M.A.; Valle, M. Potyvirus Virion Structure Shows Conserved Protein Fold and RNA Binding Site in ssRNA Viruses. Sci. Adv. 2017, 3, eaao2182. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, R.; Yuste-Calvo, C.; Gil-Cartón, D.; Sánchez, F.; Ponz, F.; Valle, M. Structure of Turnip Mosaic Virus and Its Viral-like Particles. Sci. Rep. 2019, 9, 15396. [Google Scholar] [CrossRef] [PubMed]
- Egelman, E.H. A Robust Algorithm for the Reconstruction of Helical Filaments Using Single-Particle Methods. Ultramicroscopy 2000, 85, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Chase, O.; Javed, A.; Byrne, M.J.; Thuenemann, E.C.; Lomonossoff, G.P.; Ranson, N.A.; López-Moya, J.J. CryoEM and Stability Analysis of Virus-like Particles of Potyvirus and Ipomovirus Infecting a Common Host. Commun. Biol. 2023, 6, 433. [Google Scholar] [CrossRef]
- Glatter, O.; Kratky, O. (Eds.) Small Angle X-ray Scattering; Academic Press: London, UK; New York, NY, USA, 1982; ISBN 978-0-12-286280-9. [Google Scholar]
- Feigin, L.A.; Svergun, D.I. Structure Analysis by Small-Angle X-ray and Neutron Scattering; Taylor, G.W., Ed.; Springer: Boston, MA, USA, 1987; ISBN 978-1-4757-6626-4. [Google Scholar]
- Svergun, D.I.; Koch, M.H.J.; Timmins, P.A.; May, R.P. Small Angle X-ray and Neutron Scattering from Solutions of Biological Macromolecules; Oxford University Press: Oxford, UK, 2013; ISBN 978-0-19-963953-3. [Google Scholar]
- De Pablo, P.J. The Application of Atomic Force Microscopy for Viruses and Protein Shells: Imaging and Spectroscopy. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2019; Volume 105, pp. 161–187. ISBN 978-0-12-818456-1. [Google Scholar]
- Roos, W.H.; Wuite, G.J.L. Nanoindentation Studies Reveal Material Properties of Viruses. Adv. Mater. 2009, 21, 1187–1192. [Google Scholar] [CrossRef]
- Manalastas-Cantos, K.; Konarev, P.V.; Hajizadeh, N.R.; Kikhney, A.G.; Petoukhov, M.V.; Molodenskiy, D.S.; Panjkovich, A.; Mertens, H.D.T.; Gruzinov, A.; Borges, C.; et al. ATSAS 3.0: Expanded Functionality and New Tools for Small-Angle Scattering Data Analysis. J. Appl. Crystallogr. 2021, 54, 343–355. [Google Scholar] [CrossRef]
- Hopkins, J.B.; Gillilan, R.E.; Skou, S. BioXTAS RAW: Improvements to a Free Open-Source Program for Small-Angle X-Ray Scattering Data Reduction and Analysis. J. Appl. Crystallogr. 2017, 50, 1545–1553. [Google Scholar] [CrossRef]
- Liu, H.; Hexemer, A.; Zwart, P.H. The Small Angle Scattering ToolBox (SASTBX): An Open-Source Software for Biomolecular Small-Angle Scattering. J. Appl. Crystallogr. 2012, 45, 587–593. [Google Scholar] [CrossRef]
- Doucet, M.; Cho, J.H.; Alina, G.; Bakker, J.; Bouwman, W.; Butler, P.; Campbell, K.; Gonzales, M.; Heenan, R.; Jackson, A.; et al. SasView Version 4.2.2. 2019. Available online: https://www.sasview.org (accessed on 7 March 2024).
- Schneidman-Duhovny, D.; Hammel, M.; Tainer, J.A.; Sali, A. FoXS, FoXSDock and MultiFoXS: Single-State and Multi-State Structural Modeling of Proteins and Their Complexes Based on SAXS Profiles. Nucleic Acids Res. 2016, 44, W424–W429. [Google Scholar] [CrossRef] [PubMed]
- Breßler, I.; Kohlbrecher, J.; Thünemann, A.F. SASfit: A Tool for Small-Angle Scattering Data Analysis Using a Library of Analytical Expressions. J. Appl. Crystallogr. 2015, 48, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- Brookes, E.; Rocco, M. Recent Advances in the UltraScan SOlution MOdeller (US-SOMO) Hydrodynamic and Small-Angle Scattering Data Analysis and Simulation Suite. Eur. Biophys. J. 2018, 47, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Spinozzi, F.; Ferrero, C.; Ortore, M.G.; De Maria Antolinos, A.; Mariani, P. GENFIT: Software for the Analysis of Small-Angle X-Ray and Neutron Scattering Data of Macromolecules in Solution. J. Appl. Crystallogr. 2014, 47, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Grant, T.D. Ab Initio Electron Density Determination Directly from Solution Scattering Data. Nat. Methods 2018, 15, 191–193. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-Based System for Small-Angle Scattering Data Analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Svergun, D.I. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.T.; Konarev, P.V.; Svergun, D.I. New Developments in the ATSAS Program Package for Small-Angle Scattering Data Analysis. J. Appl. Crystallogr. 2012, 45, 342–350. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Svergun, D.I. Ambiguity Assessment of Small-Angle Scattering Curves from Monodisperse Systems. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1051–1058. [Google Scholar] [CrossRef]
- Svergun, D.I. Restoring Low Resolution Structure of Biological Macromolecules from Solution Scattering Using Simulated Annealing. Biophys. J. 1999, 76, 2879–2886. [Google Scholar] [CrossRef]
- Konarev, P.V.; Petoukhov, M.V.; Svergun, D.I. MASSHA—A Graphics System for Rigid-Body Modelling of Macromolecular Complexes against Solution Scattering Data. J. Appl. Crystallogr. 2001, 34, 527–532. [Google Scholar] [CrossRef]
- Svergun, D.; Barberato, C.; Koch, M.H.J. CRYSOL—A Program to Evaluate X-Ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Costa, L.; Andriatis, A.; Brennich, M.; Teulon, J.-M.; Chen, S.W.; Pellequer, J.-L.; Round, A. Combined Small Angle X-Ray Solution Scattering with Atomic Force Microscopy for Characterizing Radiation Damage on Biological Macromolecules. BMC Struct. Biol. 2016, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Hiragi, Y.; Inoue, H.; Sano, Y.; Kajiwara, K.; Ueki, T.; Kataoka, M.; Tagawa, H.; Izumi, Y.; Muroga, Y.; Amemiya, Y. Temperature Dependence of the Structure of Aggregates of Tobacco Mosaic Virus Protein at pH 7.2. J. Mol. Biol. 1988, 204, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Sano, Y.; Inoue, H.; Hiragi, Y. Differences of Reconstitution Process between Tobacco Mosaic Virus and Cucumber Green Mottle Mosaic Virus by Synchrotron Small Angle X-ray Scattering Using Low-Temperature Quenching. J. Protein Chem. 1999, 18, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Potschka, M.; Koch, M.H.J.; Adams, M.L.; Schuster, T.M. Time-Resolved Solution x-Ray Scattering of Tobacco Mosaic Virus Coat Protein: Kinetics and Structure of Intermediates. Biochemistry 1988, 27, 8481–8491. [Google Scholar] [CrossRef] [PubMed]
- Ksenofontov, A.L.; Petoukhov, M.V.; Prusov, A.N.; Fedorova, N.V.; Shtykova, E.V. Characterization of Tobacco Mosaic Virus Virions and Repolymerized Coat Protein Aggregates in Solution by Small-Angle X-Ray Scattering. Biochem. Mosc. 2020, 85, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Ksenofontov, A.L.; Petoukhov, M.V.; Matveev, V.V.; Fedorova, N.V.; Semenyuk, P.I.; Arutyunyan, A.M.; Manukhova, T.I.; Evtushenko, E.A.; Nikitin, N.A.; Karpova, O.V.; et al. Effect of the Coat Protein N-Terminal Domain Structure on the Structure and Physicochemical Properties of Virions of Potato Virus X and Alternanthera Mosaic Virus. Biochem. Mosc. 2023, 88, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Shtykova, E.V.; Petoukhov, M.V.; Fedorova, N.V.; Arutyunyan, A.M.; Skurat, E.V.; Kordyukova, L.V.; Moiseenko, A.V.; Ksenofontov, A.L. The Structure of the Potato Virus A Particles Elucidated by Small Angle X-Ray Scattering and Complementary Techniques. Biochem. Mosc. 2021, 86, 230–240. [Google Scholar] [CrossRef]
- Ksenofontov, A.L.; Baratova, L.A.; Semenyuk, P.I.; Fedorova, N.V.; Badun, G.A. Changes in the Structure of Potato Virus A Virions after Limited in Situ Proteolysis According to Tritium Labeling Data and Computer Simulation. Biochem. Mosc. 2023, 88, 2146–2156. [Google Scholar] [CrossRef]
- Ksenofontov, A.L.; Dobrov, E.N.; Fedorova, N.V.; Serebryakova, M.V.; Prusov, A.N.; Baratova, L.A.; Paalme, V.; Järvekülg, L.; Shtykova, E.V. Isolated Potato Virus A Coat Protein Possesses Unusual Properties and Forms Different Short Virus-like Particles. J. Biomol. Struct. Dyn. 2018, 36, 1728–1738. [Google Scholar] [CrossRef]
- Ksenofontov, A.L.; Dobrov, E.N.; Fedorova, N.V.; Arutyunyan, A.M.; Golanikov, A.E.; Järvekülg, L.; Shtykova, E.V. Structure of Potato Virus A Coat Protein Particles and Their Dissociation. Mol. Biol. 2018, 52, 913–921. [Google Scholar] [CrossRef]
- Karpova, O.V.; Arkhipenko, M.V.; Zayakina, O.V.; Nikitin, N.A.; Kiselyova, O.I.; Kozlovsky, S.V.; Rodionova, N.P.; Atabekov, J.G. Regulation of RNA Translation in Potato Virus X RNA-Coat Protein Complexes: The Key Role of the N-Terminal Segment of the Protein. Mol. Biol. 2006, 40, 628–634. [Google Scholar] [CrossRef]
- Dubrovin, E.V.; Kirikova, M.N.; Novikov, V.K.; Drygin, Y.F.; Yaminsky, I.V. Study of the Peculiarities of Adhesion of Tobacco Mosaic Virus by Atomic Force Microscopy. Colloid. J. 2004, 66, 673–678. [Google Scholar] [CrossRef]
- McPherson, A.; Malkin, A.J.; Kuznetsov, Y.G. Atomic Force Microscopy in the Study of Macromolecular Crystal Growth. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 361–410. [Google Scholar] [CrossRef]
- Michel, J.P.; Ivanovska, I.L.; Gibbons, M.M.; Klug, W.S.; Knobler, C.M.; Wuite, G.J.L.; Schmidt, C.F. Nanoindentation Studies of Full and Empty Viral Capsids and the Effects of Capsid Protein Mutations on Elasticity and Strength. Proc. Natl. Acad. Sci. USA 2006, 103, 6184–6189. [Google Scholar] [CrossRef]
- Malkin, A.J.; Land, T.A.; Kuznetsov, Y.G.; McPherson, A.; DeYoreo, J.J. Investigation of Virus Crystal Growth Mechanisms by In Situ Atomic Force Microscopy. Phys. Rev. Lett. 1995, 75, 2778–2781. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Z.; Li, S.; Wang, Q.; Li, X. Nanomechanical Characterization of Polyaniline Coated Tobacco Mosaic Virus Nanotubes. J. Biomed. Mater. Res. 2008, 87A, 8–14. [Google Scholar] [CrossRef]
- García, R. Dynamic Atomic Force Microscopy Methods. Surf. Sci. Rep. 2002, 47, 197–301. [Google Scholar] [CrossRef]
- Dubrovin, E.V.; Drygin, Y.-F.; Novikov, V.K.; Yaminsky, I.V. Atomic Force Microscopy as a Tool of Inspection of Viral Infection. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 128–131. [Google Scholar] [CrossRef]
- Calò, A.; Eleta-Lopez, A.; Stoliar, P.; De Sancho, D.; Santos, S.; Verdaguer, A.; Bittner, A.M. Multifrequency Force Microscopy of Helical Protein Assembly on a Virus. Sci. Rep. 2016, 6, 21899. [Google Scholar] [CrossRef]
- Calò, A.; Eleta-Lopez, A.; Ondarçuhu, T.; Verdaguer, A.; Bittner, A.M. Nanoscale Wetting of Single Viruses. Molecules 2021, 26, 5184. [Google Scholar] [CrossRef]
- Xu, K.; Sun, W.; Shao, Y.; Wei, F.; Zhang, X.; Wang, W.; Li, P. Recent Development of PeakForce Tapping Mode Atomic Force Microscopy and Its Applications on Nanoscience. Nanotechnol. Rev. 2018, 7, 605–621. [Google Scholar] [CrossRef]
- Müller, D.J.; Janovjak, H.; Lehto, T.; Kuerschner, L.; Anderson, K. Observing Structure, Function and Assembly of Single Proteins by AFM. Prog. Biophys. Mol. Biol. 2002, 79, 1–43. [Google Scholar] [CrossRef]
- Kiselyova, O.I.; Yaminsky, I.V.; Karpova, O.V.; Rodionova, N.P.; Kozlovsky, S.V.; Arkhipenko, M.V.; Atabekov, J.G. AFM Study of Potato Virus X Disassembly Induced by Movement Protein. J. Mol. Biol. 2003, 332, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Obraztsova, E.A.; Kalinina, N.O.; Taliansky, M.E.; Gabrenaite-Verkhovskaya, R.; Makinen, K.; Yaminsky, I.V. Atomic Force Microscopy of Potato Virus A. Colloid. J. 2008, 70, 199–201. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; Malkin, A.J.; Lucas, R.W.; Plomp, M.; McPherson, A. Imaging of Viruses by Atomic Force Microscopy. J. Gen. Virol. 2001, 82, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Gurnon, J.R.; Van Etten, J.L.; McPherson, A. Atomic Force Microscopy Investigation of a Chlorella Virus, PBCV-1. J. Struct. Biol. 2005, 149, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; McPherson, A. Atomic Force Microscopy in Imaging of Viruses and Virus-Infected Cells. Microbiol. Mol. Biol. Rev. 2011, 75, 268–285. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.T.; Payton, O.; Picco, L.; Allen, M.J. Visualisation of Microalgal-Viral Interactions by High-Speed Atomic Force Microscopy. Front. Virol. 2023, 3, 1111335. [Google Scholar] [CrossRef]
- Arkhipenko, M.V.; Petrova, E.K.; Nikitin, N.A.; Protopopova, A.D.; Dubrovin, E.V.; Yaminskii, I.V.; Rodionova, N.P.; Karpova, O.V.; Atabekov, J.G. Characteristics of Artificial Virus-like Particles Assembled in Vitro from Potato Virus X Coat Protein and Foreign Viral RNAs. Acta Naturae 2011, 3, 40–46. [Google Scholar] [CrossRef]
- Krieg, M.; Fläschner, G.; Alsteens, D.; Gaub, B.M.; Roos, W.H.; Wuite, G.J.L.; Gaub, H.E.; Gerber, C.; Dufrêne, Y.F.; Müller, D.J. Atomic Force Microscopy-Based Mechanobiology. Nat. Rev. Phys. 2018, 1, 41–57. [Google Scholar] [CrossRef]
- Wilts, B.D.; Schaap, I.A.T.; Schmidt, C.F. Swelling and Softening of the Cowpea Chlorotic Mottle Virus in Response to pH Shifts. Biophys. J. 2015, 108, 2541–2549. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Pérez, M.; Zeng, C.; Miguel, M.C.; Dragnea, B. Intermittency of Deformation and the Elastic Limit of an Icosahedral Virus under Compression. ACS Nano 2019, 13, 7842–7849. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ge, Z.; Fang, J. Elastic Modulus of Viral Nanotubes. Phys. Rev. E 2008, 78, 031914. [Google Scholar] [CrossRef]
- Shtykova, E.V.; Dadinova, L.A.; Fedorova, N.V.; Golanikov, A.E.; Bogacheva, E.N.; Ksenofontov, A.L.; Baratova, L.A.; Shilova, L.A.; Tashkin, V.Y.; Galimzyanov, T.R.; et al. Influenza Virus Matrix Protein M1 Preserves Its Conformation with pH, Changing Multimerization State at the Priming Stage Due to Electrostatics. Sci. Rep. 2017, 7, 16793. [Google Scholar] [CrossRef] [PubMed]
- Shtykova, E.V.; Petoukhov, M.V.; Dadinova, L.A.; Fedorova, N.V.; Tashkin, V.Y.; Timofeeva, T.A.; Ksenofontov, A.L.; Loshkarev, N.A.; Baratova, L.A.; Jeffries, C.M.; et al. Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH. J. Virol. 2019, 93, e01450-18. [Google Scholar] [CrossRef] [PubMed]
- Shtykova, E.V.; Baratova, L.A.; Fedorova, N.V.; Radyukhin, V.A.; Ksenofontov, A.L.; Volkov, V.V.; Shishkov, A.V.; Dolgov, A.A.; Shilova, L.A.; Batishchev, O.V.; et al. Structural Analysis of Influenza A Virus Matrix Protein M1 and Its Self-Assemblies at Low pH. PLoS ONE 2013, 8, e82431. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Makarov, V.V.; Kalinina, N.O. Structure and Noncanonical Activities of Coat Proteins of Helical Plant Viruses. Biochem. Mosc. 2016, 81, 1–18. [Google Scholar] [CrossRef]

| Program Package | Short Description |
|---|---|
| ATSAS [38] https://www.embl-hamburg.de/biosaxs/software.html (accessed on 7 March 2024) | Primary data processing; Ab initio modelling; Atomic structure based (molecular) modelling; Analysis of mixtures and flexible systems. |
| BioXTAS RAW [39] https://bioxtas-raw.readthedocs.io/en/latest/ (accessed on 7 March 2024) | Primary data processing; Utilization of ATSAS modules. |
| SASTBX [40] https://bio.tools/sastbx (accessed on 7 March 2024) | Building molecular shapes; Refinement of existing atomic models. |
| SASview [41] https://www.sasview.org (accessed on 7 March 2024) | Multipurpose data analysis package; Model fitting; Distance distribution function evaluation; Invariant analysis; Correlation Function Analysis. |
| ScÅtter https://bl1231.als.lbl.gov/scatter/ (accessed on 7 March 2024) | Primary data processing. |
| FoXS/FoXSDock/MultiFoXS [42] http://modbase.compbio.ucsf.edu/foxs (accessed on 7 March 2024) | Calculation of SAXS data from atomic coordinates, combined with docking or flexibility modelling. |
| SASfit [43] https://sasfit.org/ (accessed on 7 March 2024) | Primary analysis of SAS data; Integral structural parameters calculation; Size distribution evaluation. |
| US-SOMO [44] https://somo.aucsolutions.com/ (accessed on 7 March 2024) | Evaluation of hydrodynamic parameters from SAS-based models. |
| GENFIT [45] https://sites.google.com/site/genfitweb/ (accessed on 7 March 2024) | SAS data fitting by a set of more than 30 model types. |
| DENSS [46] https://tdgrant.com (accessed on 7 March 2024) | Ab initio modelling; Scattering calculation from atomic models. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shtykova, E.V.; Dubrovin, E.V.; Ksenofontov, A.L.; Gifer, P.K.; Petoukhov, M.V.; Tokhtar, V.K.; Sapozhnikova, I.M.; Stavrianidi, A.N.; Kordyukova, L.V.; Batishchev, O.V. Structural Insights into Plant Viruses Revealed by Small-Angle X-ray Scattering and Atomic Force Microscopy. Viruses 2024, 16, 427. https://doi.org/10.3390/v16030427
Shtykova EV, Dubrovin EV, Ksenofontov AL, Gifer PK, Petoukhov MV, Tokhtar VK, Sapozhnikova IM, Stavrianidi AN, Kordyukova LV, Batishchev OV. Structural Insights into Plant Viruses Revealed by Small-Angle X-ray Scattering and Atomic Force Microscopy. Viruses. 2024; 16(3):427. https://doi.org/10.3390/v16030427
Chicago/Turabian StyleShtykova, Eleonora V., Evgeniy V. Dubrovin, Alexander L. Ksenofontov, Polina K. Gifer, Maxim V. Petoukhov, Valeriy K. Tokhtar, Irina M. Sapozhnikova, Andrey N. Stavrianidi, Larisa V. Kordyukova, and Oleg V. Batishchev. 2024. "Structural Insights into Plant Viruses Revealed by Small-Angle X-ray Scattering and Atomic Force Microscopy" Viruses 16, no. 3: 427. https://doi.org/10.3390/v16030427
APA StyleShtykova, E. V., Dubrovin, E. V., Ksenofontov, A. L., Gifer, P. K., Petoukhov, M. V., Tokhtar, V. K., Sapozhnikova, I. M., Stavrianidi, A. N., Kordyukova, L. V., & Batishchev, O. V. (2024). Structural Insights into Plant Viruses Revealed by Small-Angle X-ray Scattering and Atomic Force Microscopy. Viruses, 16(3), 427. https://doi.org/10.3390/v16030427






