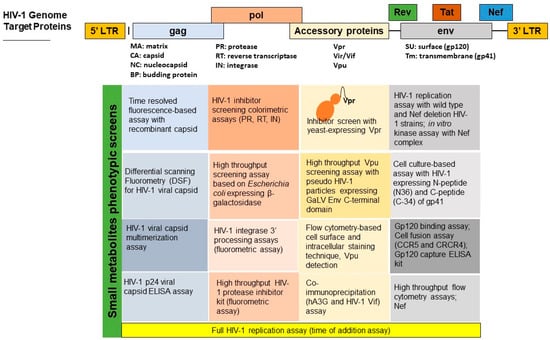
Abstract
The emergence of drug-resistant Human Immunodeficiency Virus-1 strains against anti-HIV therapies in the clinical pipeline, and the persistence of HIV in cellular reservoirs remains a significant concern. Therefore, there is a continuous need to discover and develop new, safer, and effective drugs targeting novel sites to combat HIV-1. The fungal species are gaining increasing attention as alternative sources of anti-HIV compounds or immunomodulators that can escape the current barriers to cure. Despite the potential of the fungal kingdom as a source for diverse chemistries that can yield novel HIV therapies, there are few comprehensive reports on the progress made thus far in the search for fungal species with the capacity to produce anti-HIV compounds. This review provides insights into the recent research developments on natural products produced by fungal species, particularly fungal endophytes exhibiting immunomodulatory or anti-HIV activities. In this study, we first explore currently existing therapies for various HIV-1 target sites. Then we assess the various activity assays developed for gauging antiviral activity production from microbial sources since they are crucial in the early screening phases for discovering novel anti-HIV compounds. Finally, we explore fungal secondary metabolites compounds that have been characterized at the structural level and demonstrate their potential as inhibitors of various HIV-1 target sites.
1. Introduction
The increasing number of people living with Human Immunodeficiency Virus (HIV) remains a significant global public health concern. According to UNAIDS report, more than 38 million people worldwide were living with HIV in the year 2021. About 20.6 million of these people living with HIV were in the Eastern and Southern Africa regions where HIV-1 is the predominant strain [1]. Approximately, 75.5% people infected with HIV globally are receiving antiretroviral therapy which indicate a remarkable progress [1]. The introduction of combination antiretroviral (cARV) therapies has reduced HIV-1 from a fatal disease to a manageable chronic condition owing to reduced probabilities of the disease progression to acquired immunodeficiency disease syndrome (AIDS) and thus resulting in a significant reduction to HIV-1-related morbidity and mortality [2]. Despite these gains, about 650,000 people in 2021 succumbed to AIDS-related illnesses. Although a large extent of these deaths could be a result of limited access to antiretroviral therapy in some regions, some other factors includes the limited therapeutic effect since these drugs do not cure HIV due to failure to eliminate latently integrated HIV proviral DNA in the host genome, which is capable of initiating replication to detectable levels once ARV treatment is interrupted [3]. Virological failure is reported in at least 20% of individuals receiving the first line of ARVs in low- and middle-income countries [4]. The success story of the existing antiretroviral regimens is hindered by the emergence of antiviral-resistant HIV-1 strains and adverse side effects leading to non-adherence [4,5]. For example, long-term use of ARTs comes with the risk of myopathy, lymphadepathy, cardiotoxic effects, and other hematologic disorders [6,7,8]. The repeated use of ARTs may cause the evolution of viral mutations in infected individuals leading to suppression of the bone marrow and high mitochondrial toxicity [9]. Moreover, current antiviral drugs cannot inhibit the virus from replicating in viral reservoirs. Human immunodeficiency virus continues to persist within latent cellular reservoirs, which can become reactivated at any time to produce an infectious virus [10,11]. New therapies are therefore required not only for HIV suppression but also for containing or eliminating HIV reservoirs.
One of the long-term research approaches that is gaining impetus is directed towards implementing small molecule screens of naturally derived compounds, specifically to identify compounds that are effective for provirus reactivation in combination with previously Food and Drugs Administration (FDA) approved drugs. Fungal species, especially endophytic fungi associated with medicinal plants are rich sources of novel and bioactive small molecular (secondary metabolites) compounds [12,13,14,15]. Although this review will focus on all fungal metabolites, endophytic fungi are of particular interest since they represent a largely unexplored niche. To date, no fungal metabolites have been approved as treatment for HIV-1, although there are several early screening studies suggesting fungal derived natural products to hold a high potential as anti-HIV therapies. Some of these studies have been extensively reviewed by Linnakoski et al. [15] and Roy [16]. This review intends to highlight structurally elucidated compounds derived from fungi/endophytic fungi as alternative HIV treatment strategies that have the potential to expand HIV-1 treatment options and escape existing treatment barriers. This review will explore the advances in developing antiviral assays, which are vital to uncovering antiviral agents from microbial sources. Furthermore, we propose comprehensive, whole replication antiviral screening assays as the most informative and unbiased approach for early drug development initiatives that will assist in charting a future direction in the search for antiviral compounds from microbial sources. Finally, we examine the potential of secondary metabolites derived from endophytic/fungi as future antiviral agents that can be used as potentiators, inhibitors, or latency-reversing agents to combat HIV.
2. The Biology of HIV and Currently Available Therapies for HIV Treatment
Human immunodeficiency virus infection begins when the viral glycoprotein gp120 binds to CD4+ receptors on the T helper cell [17]. Once attached, the CD4+ T cell and the HIV capsid fuse, allowing the viral RNA, reverse transcriptase, integrase, and protease to enter the cell [18,19]. Inside the host cell’s cytoplasm, the HIV RNA is converted into HIV DNA, which is facilitated by the reverse transcriptase enzyme [20]. The reverse transcriptase achieves this by transcribing viral RNA into dsDNA [17,21,22]. The viral DNA is transported into the host cell nucleus, which is then incorporated into the host’s genome with the help of the integrase enzyme [19].
Upon integrating the viral DNA into the host genome, the viral DNA can remain latent for many years [17]. In the absence of virus latency, the virus undergoes replication, utilizing the host machinery to produce long chains of viral proteins. The HIV proteins and the viral RNA are transported to the host cell’s surface, where they assemble into non-infectious particles [22,23]. After budding, the viral protease, mediates a proteolytic cleavage of the viral capsid, resulting in a mature infectious virus particle (virion) [22,24,25]. Current treatment requires that people living with HIV stay on antiretroviral therapy for life; otherwise, hidden HIV proviral DNA in cellular reservoirs will reactivate when treatment is interrupted.
There have been significant advances in the treatment, control, and prevention of HIV since its discovery in the 1980s [26]. In 1987, the Food and Drug Administration (FDA) approved its first drug, zidovudine (AZT), which initially failed in cancer screens, to treat HIV [27]. The synthesis of this drug was inspired by a naturally discovered thymidine from a marine sponge, Tectitethya crypta, known as sponge thymidine [28]. The discovery of AZT has paved the way for several classes of antiretroviral drugs currently on the market. The FDA classifies antiretroviral drugs for HIV infection into the following categories: nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), fusion inhibitors, entry inhibitors—CCR5 coreceptor antagonist and HIV integrase strand transfer inhibitors [29,30,31].
Highly active antiretroviral therapy (HAART) has significantly reduced the morbidity and mortality of HIV/AIDS. Antiretroviral therapy is currently recommended for all adults infected with HIV, and it has also reduced mother-to-child transmission to less than 1% [32]. Bhatti et al. [18] showed that taking anti-HIV medication alone is less effective than a combined ARV treatment regimen. A single-drug treatment allows the virus to mutate over time and build resistance to the medication. In combination therapy, more than one antiretroviral drug is administered concurrently to minimize the chances of resistance development. Recommendations for initial regimens include two nucleoside reverse transcriptase inhibitors, such as abacavir with lamivudine or tenofovir disoproxil fumarate, in combination with a third active ARV drug. The third drug can be from one of the three classes integrase strand transfer inhibitor (INSTI), a non-nucleoside reverse transcriptase inhibitor, or protease inhibitor (NNRTI) with a pharmacokinetic enhancer [33]. The first-line regimen currently in use is the combination of dolutegravir, lamivudine and tenofovir disoproxil fumarates. This is the combination of integrase and reverse transcriptase inhibitors. Despite the combination therapy, there is still a problem of HIV-1 drug resistance and the combination is also not effective in eradicating the virus in the latent reservoirs [34]. In fact, since the launch of potent ART, treatment recommendations have continued to prescribe regimens that include two NRTIs plus a third drug as preferred options [35]. The improved therapeutic and safety profile of treating all HIV-positive individuals as soon as a diagnosis is confirmed is paramount as it decreases mortality and increases life expectancy and the standard of treatment in antiretroviral therapy patients living with HIV [36].
The development of drug resistance to currently available antiretroviral drugs is a significant cause of treatment failure in HIV patients. The current antiretroviral therapies are also associated with adverse side effects and cell toxicity which often leads to non-adherence [37,38]. Resistance to NRTIs occurs through two mechanisms; (1) mutations resulting in reduced incorporation of the NRTI into the growing DNA chain and (2) enhanced removal of a drug from its attachment site at the end of the DNA chain [39]. These RT mutations allow ATP or pyrophosphate to bind adjacent to the bound nucleoside analogue at the active site. High-energy ATP or pyrophosphate can attack the bond that binds the drug to DNA, resulting in the drug’s liberation and termination of its effects. To combat antiviral resistance, there is an urgent need for the development of new anti-HIV drugs since approved ARVs are associated with various shortfalls such as toxicity and rapid emergence of resistant strains.
To further optimize therapeutic options and ensure a sustainable flow of new drugs with novel mechanisms of action, there is a need to ramp up early drug development initiatives. New antiviral drugs should meet specific criteria, such as not being susceptible to current resistance mechanisms. This can be achieved by targeting sites independent of the viral mutation development, such as those that target protein–protein interactions and host-mediated immune response to infection. In addition, current antiretroviral drugs only target the structural proteins of HIV. Thus, there is a need to develop novel antiretroviral therapies that will also target accessory proteins to achieve a comprehensive therapeutic approach. Advances have been made with small molecule metabolites, especially from microorganisms such as fungi [12,40]. Although these studies are in the early discovery phases, there are promising developments that call for more focus on optimizing antiviral screening assays and investment in developing hit compounds to lead compounds and eventually, clinical trials.
4. Advances in Developing Fungal-Derived Small Molecular Inhibitors for Major HIV-1 Target Sites
The fungal kingdom presents a great source of small therapeutic compounds with antibacterial, antifungal, and antiviral properties [77]. Unlike plant-derived secondary metabolites, fungal secondary metabolites can be scaled-up at a reasonable cost and time due to the availability of well-established industrial production parameters using fungal species [16]. Fungi from unique ecological niches, such as those found in the inner tissues of healthy plants, known as endophytic fungi, have been of great interest over the last two decades given their potential for use in drug development. The endophytes often reside within their host plants without causing any harm to the host plant [78]. In addition, secondary metabolites from microbial sources in novel niches, such as endophytic fungi, present compounds with structural complexities that cannot be matched by synthetic molecules, making them excellent candidates for new drug development. Fungal secondary metabolites often exhibit novel mechanisms of action that can escape already established mechanisms of drug resistance by microbial pathogens [79,80]. While the exploration of fungal compounds exhibiting antiviral activities is an emerging field, there have been several compounds revealed to possess high potential as viral inhibitors in several early screening programs as reviewed by Roy et al. [16] and Linnakoski et al. [15]. At this stage, there are currently no anti-HIV compounds derived from fungal sources in the clinical pipeline and almost all compounds are at an early discovery stage (hit identification). Although fungi have been widely explored as a source of active secondary metabolites, recent genome data suggest that what is currently known represents only a small perceptible part of a much larger reservoir. There is more biosynthetic potential that has not been explored [81,82,83,84]. Thus, there is plenty of scope for finding many potential anti-HIV drugs by exploring novel microbial species such as endophytic fungi.
5. Fungal Compounds as HIV-1 Cell-Surface Receptor Attachment Inhibitors
The fungal kingdom has been shown as a potential source of several bioactive compounds targeting the HIV-1 cell surface receptor attachment proteins. For instance, two novel chemokine receptor (CCR-5) inhibitors, Sch 210971 and Sch 210972, were isolated from an endophytic fungus, Chaetomium globosum Kunze 1705 with a potent in vitro activity of 79 nM [83]. It is well established that the chemokine receptors CCR5 and CXCR4 are required as coreceptors for binding gp120 and CD4+ T cells [84,85,86]. Inhibition of such binding can prevent viral entry into the host cell and prevent replication, representing a highly effective alternative target for HIV therapy. Several other anti-HIV-1 compounds derived from fungi targeting the gp120–CD4+ binding have been discovered, including isochromophilones I and II obtained from Penicillium multicolor FO-2338. These compounds disrupt gp120 and CD4+ interaction at 6.6 and 3.9 µM in in vitro assays, respectively, and prevent HIV-1 entry into the host cell [85,86,87]. Other isochromophilones A–F isolated from the Marine Mangrove endophytic fungi Diaporthe sp. SCSIO 41011 were reported to be cytotoxic and induced apoptosis in three different cell lines [88]. This finding emphasizes the importance of cytotoxicity screening of the fungal secondary metabolites before antiviral testing. Bioassay-guided isolation of the fungus, Emericella aurantiobrunnea led to the discovery and subsequent purification of a compound known as variecolin. Variecolin was observed to compete with macrophage inflammatory protein (MIP)-1R for binding into human CCR5 with an IC50 value of 9 µM [87]. These results on hit identification provide evidence of the potential of fungal-derived secondary metabolites in inhibiting HIV entry and replication. However, further studies of these hit compounds would offer more insight into their mechanisms and potential for further development into potential therapeutic agents.
6. Fungal Compounds as Reverse Transcriptase Inhibitors
Fungal natural products have also been explored as a source of reverse transcriptase inhibitors. The reverse transcriptase enzyme in HIV-1 infection plays a crucial role in viral replication by generating complementary DNA from an RNA template early in the HIV-1 life cycle. The role of reverse transcriptase makes it one of the most crucial targets for potential anti-HIV therapy [89]. Several other fungal species produce secondary metabolites that inhibit viral reverse transcription. This includes a study by Bashyal et al. [90], altertoxins extracted from a fungus, Alternaria tenuissima were found to inhibit the reverse transcriptase enzyme from transcribing HIV-1 RNA into DNA. Moreover, Melappa et al. [91] reported total coumarins from the crude extract of an endophytic fungus, Alternaria species to inhibit reverse transcriptase enzyme in a single protein in vitro colorimetric assay. The limitations of this study included that they only reported total coumarins and no purification of the active compounds was reported. However, the results of these studies provide some indication on the potential of fungi as a source of reverse transcriptase inhibitors. Additional research efforts are needed to validate this assertion.
7. Fungal Compounds as Integrase Inhibitors
The other important target in the HIV life cycle is the integration of the HIV viral DNA into the nucleus of the human genome facilitated by the enzyme integrase. Briefly, integration occurs via two catalytic actions, i.e., 3′-end processing and strand transfer [92]. The challenge with the current antiretroviral drugs is that HIV-1 has developed resistance to approved protease inhibitors primarily due to amino acid mutations within or proximal to the drug’s catalytic binding site [92].
The initiatives to discover naturally derived integrase inhibitors have started to indicate some successes. For instance, Singh et al. [93] showed that Penicillium chrysogenum produced xanthoviridicatins E and F, showing potent inhibition of HIV-1 integrase enzyme at highly effective concentrations of 6 and 5 µM, respectively. Both compounds inhibited the cleavage process of HIV-1 integrase. In the same year, a novel polyketide, cytosporic acid extracted from the endophytic fungus, Cytospora species, showed an in vitro HIV-1 strand transfer inhibition with IC50 of 20 μM [94]. Two other structurally similar fungal compounds, australifungin and australifungol, were evaluated, and only the former exhibited integrase inhibition activity with the same activity as cytosporic acid [94]. Australifungin was first discovered in fungal cultures of Sporormiella australis as an inhibitor of sphinganine N-acyl transferase, a mechanism common to mycotoxins such as fumonisin B [95]. The activity of both cytosporic acid and australifungin was attributed to the presence of a β-keto aldehyde group and a carboxyl group, respectively. Since both these compounds exhibited weak activities, their activities could further be improved by derivatization of the active pharmacophore to develop drug-like compounds [96]. Other success indicators for fungal-derived HIV-1 integrase inhibitor include the discovery of a phenalenone (atrovenetinone methyl acetal) from Penicillium species [97]. The authors compared this compound to phenalenones that they had previously isolated from fungal cultures, erabulenol B and funalenone, where the latter exhibited an intense integrase inhibition activity at 1.7 μM. Melappa et al. [91] also reported that the endophytic fungi, Trichoderma harzanium and Alternaria species isolated from a plant, Calophyllum inophyllum produced coumarins that showed inhibition of HIV-1 integrase. This study did not identify the coumarin derivative that was responsible for HIV-1 integrase inhibition. Coumestan-type isolated from the Calophyllum inophylum demonstrated anti-HIV-1 activity against integrase [98,99].
8. Fungal Compounds as Protease Inhibitors
Fungal protease inhibitors also reveal strong potential as future candidates in developing antiviral drugs. Singh et al. [100] reported hinnuliquinone as a potent inhibitor of the HIV-1 protease from an unidentified endophytic fungus inhabiting the leaves of Oak trees (Quercus coccifera) [100]. Another study used novel triterpenoids, gadoteridol F, 20-hydroxlucidenic acid N, ganoderic acid GC-2 and 20(21)-dehydrolucidenic acid N which were isolated from fungus, Ganordema sinense exhibited HIV protease-inhibitory activity at IC50 22, 25—30 and 48 μM, respectively [101]. Recently, Vora et al. [56] performed a targeted investigation of mulberroside C based on in silico observations that showed high-affinity binding to various target sites in the HIV cycle, especially as a protease inhibitor. Furthermore, out of the studied endophytic isolates, the extracts of MaF04C identified as Phoma species showed the most significant viral inhibition with IC50 of 8.19 ng/mL and less than 0.001 μg/mL in a TZM-bl cell-based β-glucosidase assay and HIV-1 p24 luciferase-based assay, respectively [56]. They further determined that MaF04C and MaF01SG (identified as Chaetomium species) had the highest relative protease inhibition profile, thus confirming the suggested protease inhibitory properties observed in the in silico studies [56]. These bioactive compounds have not proceeded to clinical trials or been validated beyond the early screening stages. Therefore, laboratory assays and in vivo tests are needed to fully understand the bioactivity, toxicity, and pharmacokinetic profile of the viral protease inhibitors produced by these endophytic fungi.
Ryang et al. [99] reported that paclitaxel could inhibit HIV-1 protease activity like the positive control, pepstatin A, in their in vitro experiment. A combination of paclitaxel and one already existing protease inhibitor (indinavir, nelfinavir, or combinations of these agents) at recommended dosages and schedules have been used to treat patients with HIV-associated Kaposi’s Sarcoma without enhancing toxicity [100,101,102,103,104]. However, patient conditions for paclitaxel applications should be considered because of its side effects on bone marrow suppression. Ryang et al. [99] indicate that fungal-derived compounds can also be used to potentiate existing HIV inhibitors in combination therapy which is a proven strategy for escaping resistance development.
9. Advances in Developing Fungal-Derived Small Molecular Inhibitors for New HIV-1 Target Sites
Despite the cART being potent and life-prolonging, a significant limitation is that it is not curative and does not eradicate HIV-1 infection since treatment interruption inevitably results in a rapid rebound of viremia [105]. The rebound of HIV is due to the presence of HIV reservoirs, mainly in the latently infected resting CD4+ memory T cells and myeloid cells such as macrophages and microglia that are difficult to target by cART or immune effector mechanisms [106,107,108,109]. HIV latency is defined as a reversibly, non-productive state of infection of individual cells that retain the capacity to produce infectious virus particles but allow the virus to evade the host’s immune response [110]. Latently infected cells contain stably integrated, replication-competent proviruses repressed at transcriptional and post-transcriptional levels by many silencing mechanisms [111,112].
Advances have been made in the discovery of fungal latency reversing agents (LRAs) with promising latency-reversing activities. Niu et al. [107] identified 12 eutypellazine (A–L) compounds with antiviral activity from a fungus, Eutypella sp. MCCC 3A00281. These compounds inhibited viral replication in the 293T HIV cell model with low toxicity. The compound eutypellazine J (80 μM) showed latency reactivation of HIV-1 in J-latA2 cells in a dose-dependent manner. The observed latency-reversing activity was comparable to that of the positive controls, prostratin (5 μM) and SAHA (suberoylanilide hydroxamic acid, 2.5 μM) [111]. Stoszko et al. [108] recently isolated gliotoxin from a fungal secondary metabolite library and showed this compound to exhibit latency-reversing properties. The compound was derived from Aspergillus fumigatus CBS 100074, and this was shown to disrupt 7SK small nuclear ribonucleoprotein (snRNP), resulting in the release of the positive transcription elongation factor B (P-TEFb) and subsequent reversal of HIV-1 latency [108]. The latency-reversing mechanism shown by gliotoxin is like that of hexamethylene bis acetamide (HMBA) and disulfiram compounds [113,114,115,116]. This also is an exciting target site since the P-TEFb catalyzes the phosphorylation of several transcriptional regulators at the HIV promoter site, which supports transcriptional initiation and elongation. Gliotoxin was previous reported as a virulent factor in Aspergillus fumigatus and showed immunosuppression properties [117]. It is a mycotoxin produced by species of Aspergillus, Trichoderma and Penicillium [118]. Furthermore, gliotoxin can be toxic when swallowed or inhaled and can cause skin and eye irritation when exposed. The oral dose of gliotoxin to be taken is 67 mg/kg [119]. These findings indicate fungal species as an untapped source of therapeutic agents that could expand treatment options for viral infections such as HIV-1. After the LRAs reactivate latent viruses, the immune system can eradicate the virus-producing cells. However, recent data indicated that CD8+ T cells in HIV-1-infected individuals on cART could not eliminate latently infected cells even after successful reactivation.
The currently available antiretroviral drugs target structural HIV-1 proteins but not HIV-1 accessory proteins (Nef, Vif, Vpu and Vpr). Accessory proteins in HIV-1 act as virulence factors mediating the severity of viral infection, replication, and disease progression. Although these proteins do not show any enzymatic activity, they act mainly with the host factors through protein–protein interactions [112]. Identifying natural products from microbial sources such as fungi could expand HIV-1 treatment options. Some advances have been made with fungal secondary metabolites inhibiting the HIV-1 viral protein R (Vpr). Fumagillin isolated from Aspergillus fumigatus fresenius was reported to inhibit the cell cycle arrest activity of Vpr in yeast and mammalian expression cells [115]. Fumagillin is a known mycotoxin that has been used as treatment for microsporidia fungal infection. It has been researched for cancer treatment by employing its angiogenesis inhibitory properties [115,116,117]. Kamata et al. [116] also showed that damnacanthal, a noni component, inhibits Vpr-associated cell death with no effect on the cell cycle. The advances in high-throughput screening platforms for antiviral compounds have the potential to accelerate the discovery of small molecular inhibitors that will expand to other accessory proteins.
While a few promising and successful efforts have been made in the early screening of fungi for anti-HIV-1 bioactive compounds, the full potential of the fungal kingdom has not been extensively explored to unearth its antiviral activities. Secondary metabolites isolated from Alternaria alternata have revealed the potential to inhibit HIV-1 at distinct stages of its life cycle; however, the compound responsible for the antiviral activity was not reported [49]. However, there are few reports regarding the mechanisms of action of these compounds. Therefore, further research is necessary to identify the mechanisms of these beneficial compounds isolated from endophytic/fungi with low cytotoxicity. More anti-HIV agents have been discovered from fungal species, placing the fungal kingdom as an exciting source for anti-HIV compounds. Table 1 summarizes fungal secondary metabolites that exhibit anti-HIV activities, inhibiting different stages of the HIV life cycle. Considering that natural products are structurally complex and may present with toxicity that could prevent their further development into viable drugs, the structural information of bioactive compounds could serve as a starting point in synthesis or semi-synthesis programs for the generation of analogues. This strategy has been successful in providing a sustainable supply of drugs inspired by naturally occurring bioactive structures and ensured that they are available in sufficient quantities [118,120].
Screening small compounds from natural sources, such as fungi, for antiviral (anti-HIV) bioactivity is still in its infancy. Nevertheless, the promising discoveries made thus far reveal the potential for further development of these bioactive compounds as lead compounds for HIV treatment. However, there are several limitations to the small molecular screens for anti-HIV compounds [48,121]. These obstacles include the need for highly specialized containment laboratories with highly trained personnel and a lack of innovative antiviral screening tools [48].

Table 1.
Anti-HIV-1 compounds isolated from fungal species.
Table 1.
Anti-HIV-1 compounds isolated from fungal species.
| HIV-1 Life Cycle Stages | HIV-1 Agent/Compound | Structure | Fungal Species | Mechanism of Action | Activity Concentration | Reference |
|---|---|---|---|---|---|---|
| Fusion or entry inhibitors | Isochromophilones I C23H25O5Cl Mw 416.9 | 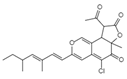 | Penicillium multicolor FO-2338 | Inhibited the gp120, CD4 binding | 6.6 μM | [85,86,122] |
| Isochromophilones II C22H27O4Cl Mw 390.9 | 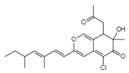 | 3.9 µM | ||||
| Variecolin C25H36O2l Mw 368.55 | 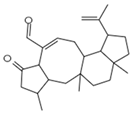 | Emericella aurantiobrumea | Competed with macrophage inflammatory protein (MIP)-1 for binding to CCR5 | 9 µM | [87,122,123] | |
| Novel Sch 210972 C25H35NO6 Mw 445.55 | 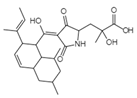 | Chaetomium globosum Kunze SCF 1705 | Strongly inhibited CCR-5 inhibitory activity | 79 nM | [83,120,122,123] | |
| Penicillixanthone A C32H30O14 Mw 638.578 | 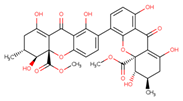 | Aspergillus fumigatus | Inhibited CCR5-tropic HIV-1 SF162 | 0.36 μM | [123,124] | |
| Inhibited CXCR4-tropic HIV-1 NL4-3 | 0.26 μM | |||||
| Reverse Transcription | Stachybotrin D C26H35NO5 Mw 441.56 | 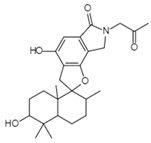 | Stachybotrys chartarum | Inhibited the RT RNA-dependent DNA polymerase activity in a dose-dependent manner | 8.4 µM | [122,125] |
| Helotialins A C23H30O6 Mw 402.48 | 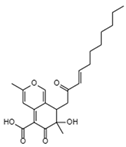 | Helotialean Ascomycete | Not identified | 8.01 nM | [122,126] | |
| Helotialins B C22H30O4 Mw 358.47 | 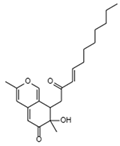 | 27.9 nM | ||||
| Altertoxin Ⅴ C20H14O6 Mw 350.32 | 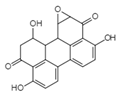 | Alternaria tenuissima | Not identified | ≤2.20 µM | [90,122] | |
| Integration | Xanthoviridicatins E and F C27H20O9 Mw 488.44 | 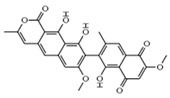 | Penicillium chrysogenum | They inhibited the cleavage reaction of HIV-1 integrase. | 6 and 5 μM | [93,122] |
| Aquastatin A C36H52O12 MW 676.79 | 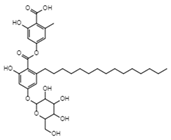 | Fusarium aquaeductuum | Moderately inhibited the strand transfer reaction of HIV-1 integrase | 50 µM | [122,127] | |
| Integracins B C35H54O7 Mw 586.8 | 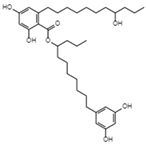 | Cytonaema sp. | Inhibited coupled reaction of recombinant HIV-1 integrase | 6.1 µM | [122,128] | |
| Integric acid C25H52O6 Mw 430.53 | 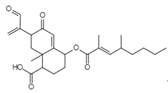 | Xylaria feejeensis | Inhibited the 3′ end processing, strand transfer and disintegration | 5–10 μM | [122,129] | |
| Atrovenetinone methyl acetal C20H20O7 Mw 372.37 | 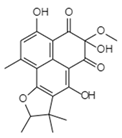 | Penicillium sp. FKI-1463 | Not identified | 19 µM | [97,122] | |
| Translation or budding | Hinnuliquinone C32H30N2O4 Mw 506.59 | 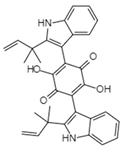 | Unidentified fungus | Not identified | 2.5 µM. | [100,122] |
| Mulberroside C C24H26O9 Mw 458.46 | 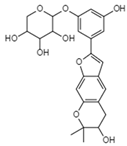 | Unidentified fungus | Inhibited HIV-1 protease in a dose-dependent manner | <7.8 µg/mL | [56,122] |
Structures presented in Table 1 were adapted from The Natural Product databases: Coconut [122] or Natural Product Atlas [123].
10. Conclusions
The recent progress in identifying antiviral compounds have potential to yield novel HIV therapeutics that can escape current treatment barriers. The recent genomic evolution has shown that fungal species holds a much greater biosynthetic potential with the revelation of several biosynthetic genes which are cryptic or transiently expressed under laboratory conditions. Here, we have shown that there is a concerted effort in developing unbiased antiviral screening assays. The challenge that limit early discovery efforts in identifying hit-to-lead compounds is the access to standardized antiviral screening systems that can map the inhibition impact of these antiviral compounds against various HIV target sites, including the HIV-1 proteins, cellular pathways, host factors and protein–protein interactions [59]. This suggests a need to standardize and validate currently existing antiviral screening assays and develop more comprehensive and high-throughput whole replication antiviral screening platforms to fully exploit the diverse antiviral mechanisms of small metabolites such as those produced by fungal species. Furthermore, the success of fungal derived antiviral compounds will only be realized if the hit compounds proceed from early drug development phases to advanced phases such as clinical trials. This will require a collaborative investment from research initiatives and Government sectors and pharmaceutical companies.
Author Contributions
Conceptualization, S.I.N. and N.P.M.; initial draft writing, B.N.; writing, review and editing, B.N., J.P.M., N.P.M. and S.I.N. All authors have read and agreed to the published version of the manuscript.
Funding
SIN was funded by the National Research Foundation (NRF) (Grant No. 129778), and NPM was funded by the Poliomyelitis Research Foundation (PRF) (Grant No. 21/76).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data availability statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
HIV: Human Immunodeficiency Virus; CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/ with associated Cas9 endonuclease); LRA, latency reversing agent; FDA, food and drug administration; RNA, ribonucleic acid; ds/DNA, double-stranded /Deoxyribonucleic acid; AZT, azidothymidine; HAART, highly active antiretroviral therapy; AIDS, acquired immunodeficiency syndrome; NRTI, nucleoside/nucleotide reverse transcriptase inhibitor; ARV, antiretroviral; INSTIs, integrase strand transfer inhibitors; BIC, bictegravir; WHO, World Health Organisation; RT, reverse transcriptase; ATP, adenosine triphosphate; MIP, macrophage inflammatory proteins; RAL, raltegravir; EVG, elvitergravir; DTG, dolutegravir; HSP90, heat shock protein 90; cART, combination antiretroviral therapy; HDAC, histone deacetylase; DNMT, DNA methyl transferases, HMT, histone methyl transferases; Aza-CdR, 5-aza-2 deoxycytidine; CpG, cytoside phosphate guanine; P-TEFb, positive transcription elongation factor b, HMBA, hexamethylene bisacetamide; CPE, cytopathic effect; HEK, human embryonic kidney; GFP, green fluorescence protein; PBMC, peripheral blood mononuclear cells; ELISA, enzyme immunosorbent assay.
References
- Global HIV & AIDS statistics. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 13 April 2023).
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar] [CrossRef]
- Andersen, R.J.; Ntie-Kang, F.; Tietjen, I. Natural product-derived compounds in HIV suppression, remission, and eradication strategies. Antivir. Res. 2018, 158, 63–77. [Google Scholar] [CrossRef]
- Reed, J.C.; Solas, D.; Kitaygorodskyy, A.; Freeman, B.; Ressler, D.T.B.; Phuong, D.J.; Swain, J.V.; Matlack, K.; Hurt, C.R.; Lingappa, V.R.; et al. Identification of an Antiretroviral Small Molecule That Appears to Be a Host-Targeting Inhibitor of HIV-1 Assembly. J. Virol. 2021, 95, e00883-20. [Google Scholar] [CrossRef] [PubMed]
- Andrae-Marobela, K.; Ghislain, F.W.; Okatch, H.; Majinda, R.R. Polyphenols: A diverse class of multi-target anti-HIV-1 agents. Curr. Drug Metab. 2013, 14, 392–413. [Google Scholar] [CrossRef] [PubMed]
- Farooq, T.; Hameed, A.; Rehman, K.; Ibrahim, M.; Qadir, M.I.; Akash, M.S. Antiretroviral Agents: Looking for the Best Possible Chemotherapeutic Options to Conquer HIV. Crit. Rev. Eukaryot. Gene Expr. 2016, 26, 363–381. [Google Scholar] [CrossRef]
- Joly, V.; Flandre, P.; Meiffredy, V.; Leturque, N.; Harel, M.; Aboulker, J.P.; Yeni, P. Increased risk of lipoatrophy under stavudine in HIV-1-infected patients: Results of a substudy from a comparative trial. Aids 2002, 16, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Povar-Echeverría, M.; Comet-Bernad, M.; Gasso-Sánchez, A.; Ger-Buil, A.; Navarro-Aznarez, H.; Martínez-Álvarez, R.; Arazo-Garcés, P. Neuropsychiatric adverse effects of dolutegravir in real-life clinical practice. Enferm. Infecc. Microbiol. Clin. 2021, 39, 78–82. [Google Scholar] [CrossRef]
- Desai, M.; Iyer, G.; Dikshit, R.K. Antiretroviral drugs: Critical issues and recent advances. Indian J. Pharmacol. 2012, 44, 288–298. [Google Scholar] [CrossRef]
- Alexaki, A.; Liu, Y.; Wigdahl, B. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr. HIV Res. 2008, 6, 388–400. [Google Scholar] [CrossRef]
- Siliciano, J.D.; Siliciano, R.F. A long-term latent reservoir for HIV-1: Discovery and clinical implications. J. Antimicrob. Chemother. 2004, 54, 6–9. [Google Scholar] [CrossRef]
- Sharapov, A.D.; Fatykhov, R.F.; Khalymbadzha, I.A.; Zyryanov, G.V.; Chupakhin, O.N.; Tsurkan, M.V. Plant Coumarins with Anti-HIV Activity: Isolation and Mechanisms of Action. Int. J. Mol. Sci. 2023, 24, 2839. [Google Scholar] [CrossRef]
- Gunatilaka, A.A. Natural products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 2006, 69, 509–526. [Google Scholar] [CrossRef]
- Hashemi, P.; Sadowski, I. Diversity of small molecule HIV-1 latency reversing agents identified in low- and high-throughput small molecule screens. Med. Res. Rev. 2020, 40, 881–908. [Google Scholar] [CrossRef]
- Linnakoski, R.; Reshamwala, D.; Veteli, P.; Cortina-Escribano, M.; Vanhanen, H.; Marjomäki, V. Antiviral Agents from Fungi: Diversity, Mechanisms and Potential Applications. Front. Microbiol. 2018, 9, 2325. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.G. Potential of small-molecule fungal metabolites in antiviral chemotherapy. Antivir. Chem. Chemother. 2017, 25, 20–52. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.A.; Dalgleish, A.G.; Loveday, C.; Pillay, D. Immunodeficiency Viruses. In Principles and Practice of Clinical Virology; John Wiley and Sons: Hoboken, NJ, USA, 2004; pp. 721–757. [Google Scholar]
- Bhatti, A.B.; Usman, M.; Kandi, V. Current Scenario of HIV/AIDS, Treatment Options, and Major Challenges with Compliance to Antiretroviral Therapy. Cureus 2016, 8, e515. [Google Scholar] [CrossRef]
- Kapila, A.; Chaudhary, S.; Sharma, R.B.; Vashist, H.; Sisodia, S.S.; Gupta, A. A REVIEW on: HIV AIDS. Indian J. Pharm. Biol. Res. 2016, 4, 69–73. [Google Scholar] [CrossRef]
- Singh, I.P.; Bharate, S.; Bhutani, K. Anti-HIV natural products. Curr. Sci. 2005, 89, 269–290. [Google Scholar]
- Mesters, J.R.; Tan, J.; Hilgenfeld, R. Viral enzymes. Curr. Opin. Struct. Biol. 2006, 16, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Kräusslich, H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect Med. 2012, 2, a006924. [Google Scholar] [CrossRef]
- Li, L.; Li, H.S.; Pauza, C.D.; Bukrinsky, M.; Zhao, R.Y. Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions. Cell Res. 2005, 15, 923–934. [Google Scholar] [CrossRef]
- Rheinemann, L.; Sundquist, W.I. Virus. Budding. In Encyclopedia of Virology; Academic Press: Cambridge, MA, USA, 2021; pp. 519–528. [Google Scholar] [CrossRef]
- Kessl, J.J.; Kutluay, S.B.; Townsend, D.; Rebensburg, S.; Slaughter, A.; Larue, R.C.; Shkriabai, N.; Bakouche, N.; Fuchs, J.R.; Bieniasz, P.D.; et al. HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis. Cell 2016, 166, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Piot, P.; Abdool Karim, S.S.; Hecht, R.; Legido-Quigley, H.; Buse, K.; Stover, J.; Resch, S.; Ryckman, T.; Møgedal, S.; Dybul, M.; et al. Defeating AIDS—Advancing global health. Lancet 2015, 386, 171–218. [Google Scholar] [CrossRef]
- Broder, S. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antivir. Res. 2010, 85, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Serna-Arbeláez, M.S.; Florez-Sampedro, L.; Orozco, L.P.; Ramírez, K.; Galeano, E.; Zapata, W. Natural Products with Inhibitory Activity against Human Immunodeficiency Virus Type 1. Adv. Virol. 2021, 2021, 5552088. [Google Scholar] [CrossRef]
- Painter, G.R.; Almond, M.R.; Mao, S.; Liotta, D.C. Biochemical and mechanistic basis for the activity of nucleoside analogue inhibitors of HIV reverse transcriptase. Curr. Top Med. Chem. 2004, 4, 1035–1044. [Google Scholar] [CrossRef]
- Blanco, J.L.; Whitlock, G.; Milinkovic, A.; Moyle, G. HIV integrase inhibitors: A new era in the treatment of HIV. Expert Opin. Pharmacother. 2015, 16, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Konvalinka, J.; Kräusslich, H.G.; Müller, B. Retroviral proteases and their roles in virion maturation. Virology 2015, 479–480, 403–417. [Google Scholar] [CrossRef]
- Peters, H.; Francis, K.; Sconza, R.; Horn, A.; Peckham, C.S.; Tookey, P.A.; Thorne, C. UK Mother-to-Child HIV Transmission Rates Continue to Decline: 2012–2014. Clin. Infect. Dis. 2017, 64, 527–528. [Google Scholar] [CrossRef]
- Fantauzzi, A.; Mezzaroma, I. Dolutegravir: Clinical efficacy and role in HIV therapy. Ther. Adv. Chronic Dis. 2014, 5, 164–177. [Google Scholar] [CrossRef]
- Cutrell, J.; Bedimo, R. Single-Tablet Regimens in the Treatment of HIV-1 Infection. Fed. Pract. 2016, 33, 24s–30s. [Google Scholar]
- Günthard, H.F.; Saag, M.S.; Benson, C.A.; del Rio, C.; Eron, J.J.; Gallant, J.E.; Hoy, J.F.; Mugavero, M.J.; Sax, P.E.; Thompson, M.A.; et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2016 Recommendations of the International Antiviral Society-USA Panel. JAMA 2016, 316, 191–210. [Google Scholar] [CrossRef] [PubMed]
- Eggleton, J.S.; Nagalli, S. Highly Active Antiretroviral Therapy (HAART). In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- Montessori, V.; Press, N.; Harris, M.; Akagi, L.; Montaner, J.S. Adverse effects of antiretroviral therapy for HIV infection. Cmaj 2004, 170, 229–238. [Google Scholar] [PubMed]
- Margolis, A.M.; Heverling, H.; Pham, P.A.; Stolbach, A. A review of the toxicity of HIV medications. J. Med. Toxicol. 2014, 10, 26–39. [Google Scholar] [CrossRef]
- Das, K.; Arnold, E. HIV-1 reverse transcriptase and antiviral drug resistance. Part 2. Curr. Opin. Virol. 2013, 3, 119–128. [Google Scholar] [CrossRef]
- Fobofou, S.A.T.; Franke, K.; Brandt, W.; Manzin, A.; Madeddu, S.; Serreli, G.; Sanna, G.; Wessjohann, L.A. Bichromonol, a dimeric coumarin with anti-HIV activity from the stem bark of Hypericum roeperianum. Nat. Prod. Res. 2022, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Richman, D.D.; Nathanson, N. Chapter 20—Antiviral Therapy. In Viral Pathogenesis, 3rd ed.; Katze, M.G., Korth, M.J., Law, G.L., Nathanson, N., Eds.; Academic Press: Boston, MA, USA, 2016; pp. 271–287. [Google Scholar]
- Payne, S. Methods to Study Viruses. In Viruses; Elsevier Inc.: Houston, TX, USA, 2017; pp. 37–52. [Google Scholar] [CrossRef]
- Hudson, J.B.; Graham, E.A.; Towers, G.H. Antiviral assays on phytochemicals: The influence of reaction parameters. Planta Med. 1994, 60, 329–332. [Google Scholar] [CrossRef]
- Cao, J.; Isaacson, J.; Patick, A.K.; Blair, W.S. High-throughput human immunodeficiency virus type 1 (HIV-1) full replication assay that includes HIV-1 Vif as an antiviral target. Antimicrob. Agents Chemother. 2005, 49, 3833–3841. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.A.; Lyddon, T.D.; Gil, H.M.; Evans, D.T.; Kuzmichev, Y.V.; Richard, J.; Finzi, A.; Welbourn, S.; Rasmussen, L.; Nebane, N.M.; et al. Novel Compound Inhibitors of HIV-1NL4-3 Vpu. Viruses 2022, 14, 817. [Google Scholar] [CrossRef]
- Daelemans, D.; Pauwels, R.; De Clercq, E.; Pannecouque, C. A time-of-drug addition approach to target identification of antiviral compounds. Nat. Protoc. 2011, 6, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Adamson, C.S.; Chibale, K.; Goss, R.J.M.; Jaspars, M.; Newman, D.J.; Dorrington, R.A. Antiviral drug discovery: Preparing for the next pandemic. Chem. Soc. Rev. 2021, 50, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Nzimande, B.; Kumalo, H.M.; Ndlovu, S.I.; Mkhwanazi, N.P. Secondary metabolites produced by endophytic fungi, Alternaria alternata, as potential inhibitors of the human immunodeficiency virus. Front. Genet. 2022, 13, 13.1–14. [Google Scholar] [CrossRef]
- McMahon, M.A.; Shen, L.; Siliciano, R.F. New approaches for quantitating the inhibition of HIV-1 replication by antiviral drugs in vitro and in vivo. Curr. Opin. Infect. Dis. 2009, 22, 574–582. [Google Scholar] [CrossRef]
- Shen, L.; Peterson, S.; Sedaghat, A.R.; McMahon, M.A.; Callender, M.; Zhang, H.; Zhou, Y.; Pitt, E.; Anderson, K.S.; Acosta, E.P.; et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat. Med. 2008, 14, 762–766. [Google Scholar] [CrossRef]
- Banning, C.; Votteler, J.; Hoffmann, D.; Koppensteiner, H.; Warmer, M.; Reimer, R.; Kirchhoff, F.; Schubert, U.; Hauber, J.; Schindler, M. A Flow Cytometry-Based FRET Assay to Identify and Analyse Protein-Protein Interactions in Living Cells. PLoS ONE 2010, 5, e9344. [Google Scholar] [CrossRef]
- Arhel, N.; Kirchhoff, F. Host proteins involved in HIV infection: New therapeutic targets. Biochim. Biophys. Acta 2010, 1802, 313–321. [Google Scholar] [CrossRef]
- Passaes, C.; Delagreverie, H.M.; Avettand-Fenoel, V.; David, A.; Monceaux, V.; Essat, A.; Müller-Trutwin, M.; Duffy, D.; De Castro, N.; Wittkop, L.; et al. Ultrasensitive Detection of p24 in Plasma Samples from People with Primary and Chronic HIV-1 Infection. J. Virol. 2021, 95, e0001621. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.R.; Bain, R.; Varsaneux, O.; Peeling, R.W.; Stevens, M.M.; McKendry, R.A. A landscape review of antigen detection for early HIV diagnosis. Aids 2018, 32, 2089–2102. [Google Scholar] [CrossRef]
- Vora, J.; Velhal, S.; Sinha, S.; Patel, V.; Shrivastava, N. Bioactive phytocompound mulberroside C and endophytes of Morus alba as potential inhibitors of HIV-1 replication: A mechanistic evaluation. HIV Med. 2021, 22, 690–704. [Google Scholar] [CrossRef]
- Cao, S.; Woodrow, K.A. Nanotechnology approaches to eradicating HIV reservoirs. Eur. J. Pharm. Biopharm. 2019, 138, 48–63. [Google Scholar] [CrossRef]
- Sinha, C.; Nischal, A.; Bandaru, S.; Kasera, P.; Rajput, A.; Nayarisseri, A.; Khattri, S. An in silico approach for identification of novel inhibitors as a potential therapeutics targeting HIV-1 viral infectivity factor. Curr. Top Med. Chem. 2015, 15, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; De Clercq, E. Overview of Antiviral Drug Discovery and Development: ViralVersusHost Targets. In Antiviral Discovery for Highly Pathogenic Emerging Viruses; Royal Society of Chemistry: Piccadilly, Australia, 2021. [Google Scholar]
- Zhang, D.W.; Xie, L.; Xu, X.S.; Li, Y.; Xu, X. A Broad-Spectrum Antiviral Molecule, Protoporphyrin IX, Acts as a Moderator of HIV-1 Capsid Assembly by Targeting the Capsid Hexamer. Microbiol. Spectr. 2023, 11, e0266322. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, Z.; Liu, Z.; Pan, Q.; Wang, J.; Li, X.; Guo, F.; Liang, C.; Hu, L.; Zhou, J.; et al. Identification of small molecule compounds targeting the interaction of HIV-1 Vif and human APOBEC3G by virtual screening and biological evaluation. Sci. Rep. 2018, 8, 8067. [Google Scholar] [CrossRef]
- Meng, J.; Lai, M.-T.; Munshi, V.; Grobler, J.; McCauley, J.; Zuck, P.; Johnson, E.N.; Uebele, V.N.; Hermes, J.D.; Adam, G.C. Screening of HIV-1 protease using a combination of an ultra-high-throughput fluorescent-based assay and RapidFire mass spectrometry. J. Biomol. Screen. 2015, 20, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Doi, N.; Koma, T.; Adachi, A.; Nomaguchi, M. Novel In Vitro Screening System Based on Differential Scanning Fluorimetry to Search for Small Molecules against the Disassembly or Assembly of HIV-1 Capsid Protein. Front. Microbiol. 2017, 8, 1413. [Google Scholar] [CrossRef] [PubMed]
- Eglen, M.R.; Singh, R. Galactosidase Enzyme Fragment Complementation as A Novel Technology for High Throughput Screening. Comb. Chem. High Throughput Screen. 2003, 6, 381–387. [Google Scholar] [CrossRef]
- Ding, J.; Zhao, J.; Yang, Z.; Ma, L.; Mi, Z.; Wu, Y.; Guo, J.; Zhou, J.; Li, X.; Guo, Y.; et al. Microbial Natural Product Alternariol 5-O-Methyl Ether Inhibits HIV-1 Integration by Blocking Nuclear Import of the Pre-Integration Complex. Viruses 2017, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Sillapachaiyaporn, C.; Chuchawankul, S. HIV-1 protease and reverse transcriptase inhibition by tiger milk mushroom (Lignosus rhinocerus) sclerotium extracts: In vitro and in silico studies. J. Tradit. Complement Med. 2020, 10, 396–404. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Zhang, W.; Li, B.; Zhu, Y.; Hu, Q.; Yang, Y.; Zhang, X.; Yan, H.; Zeng, Y. Inhibition of Human Immunodeficiency Virus Type 1 Entry by a Keggin Polyoxometalate. Viruses 2018, 10, 265. [Google Scholar] [CrossRef]
- Emert-Sedlak, L.; Kodama, T.; Lerner, E.C.; Dai, W.; Foster, C.; Day, B.W.; Lazo, J.S.; Smithgall, T.E. Chemical library screens targeting an HIV-1 accessory factor/host cell kinase complex identify novel antiretroviral compounds. ACS Chem. Biol. 2009, 4, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jiang, S. High throughput screening and characterization of HIV-1 entry inhibitors targeting gp41: Theories and techniques. Curr. Pharm. Des. 2004, 10, 1827–1843. [Google Scholar] [CrossRef] [PubMed]
- Shui, X.; Lu, X.; Gao, Y.; Liu, C.; Ren, F.; Jiang, Q.; Zhang, H.; Zhao, B.; Zheng, Z. A mammalian two-hybrid system-based assay for small-molecular HIV fusion inhibitors targeting gp41. Antivir. Res. 2011, 90, 54–63. [Google Scholar] [CrossRef]
- Lin, P.F.; Blair, W.; Wang, T.; Spicer, T.; Guo, Q.; Zhou, N.; Gong, Y.F.; Wang, H.G.; Rose, R.; Yamanaka, G.; et al. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc. Natl. Acad. Sci. USA 2003, 100, 11013–11018. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.-J.; Brik, A.; Wong, C.-H.; Kan, C.-C. Model System for High-Throughput Screening of Novel Human Immunodeficiency Virus Protease Inhibitors in Escherichia coli. Antimicrob. Agents Chemother. 2004, 48, 2437–2447. [Google Scholar] [CrossRef]
- Omoruyi, B.E.; Ighodaro, D.I.; Afolayan, A.J.; Bradley, G. Inhibition of HIV-1 Protease by Carpobrotus edulis (L.). Evid. Based Complement Altern. Med. 2020, 2020, 9648056. [Google Scholar] [CrossRef]
- Painter, M.M.; Zimmerman, G.E.; Merlino, M.S.; Robertson, A.W.; Terry, V.H.; Ren, X.; McLeod, M.R.; Gomez-Rodriguez, L.; Garcia, K.A.; Leonard, J.A.; et al. Concanamycin A counteracts HIV-1 Nef to enhance immune clearance of infected primary cells by cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 2020, 117, 23835–23846. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Nair, D.N.; Padmavathy, S. Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef]
- Gakuubi, M.M.; Munusamy, M.; Liang, Z.X.; Ng, S.B. Fungal Endophytes: A Promising Frontier for Discovery of Novel Bioactive Compounds. J. Fungi 2021, 7, 786. [Google Scholar] [CrossRef]
- Stierle, A.A.; Stierle, D.B. Bioactive Secondary Metabolites Produced by the Fungal Endophytes of Conifers. Nat. Prod. Commun. 2015, 10, 1671–1682. [Google Scholar] [CrossRef]
- Wellensiek, B.P.; Ramakrishnan, R.; Bashyal, B.P.; Eason, Y.; Gunatilaka, A.A.; Ahmad, N. Inhibition of HIV-1 Replication by Secondary Metabolites from Endophytic Fungi of Desert Plants. Open Virol. J. 2013, 7, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Raut, J.K. Mushroom: A potent source of natural antiviral drugs. Appl. Sci. Technol. Ann. 2020, 1, 81–91. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Rossman, A.Y. Where are all the undescribed fungi? Phytopathology 1997, 87, 888–891. [Google Scholar] [CrossRef]
- Schmit, J.; Mueller, G. An estimate of the lower limit of global fungal diversity. Biodivers. Conserv. 2007, 16, 99–111. [Google Scholar] [CrossRef]
- Yang, S.W.; Mierzwa, R.; Terracciano, J.; Patel, M.; Gullo, V.; Wagner, N.; Baroudy, B.; Puar, M.; Chan, T.M.; Chu, M. Sch 213766, a novel chemokine receptor CCR-5 inhibitor from Chaetomium globosum. J. Antibiot. 2007, 60, 524–528. [Google Scholar] [CrossRef]
- Henrich, T.J.; Kuritzkes, D.R. HIV-1 entry inhibitors: Recent development and clinical use. Curr. Opin. Virol. 2013, 3, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Tanaka, H.; Matsuzaki, K.; Ikeda, H.; Masuma, R. Isochromophilones I and II, novel inhibitors against gp120-CD4 binding from Penicillium sp. J. Antibiot. 1993, 46, 1908–1911. [Google Scholar] [CrossRef]
- Luo, X.; Lin, X.; Tao, H.; Wang, J.; Li, J.; Yang, B.; Zhou, X.; Liu, Y. Isochromophilones A–F, Cytotoxic Chloroazaphilones from the Marine Mangrove Endophytic Fungus Diaporthe sp. SCSIO 41011. J. Nat. Prod. 2018, 81, 934–941. [Google Scholar] [CrossRef]
- Yoganathan, K.; Rossant, C.; Glover, R.P.; Cao, S.; Vittal, J.J.; Ng, S.; Huang, Y.; Buss, A.D.; Butler, M.S. Inhibition of the Human Chemokine Receptor CCR5 by Variecolin and Variecolol and Isolation of Four New Variecolin Analogues, Emericolins A−D, from Emericella aurantiobrunnea. J. Nat. Prod. 2004, 67, 1681–1684. [Google Scholar] [CrossRef] [PubMed]
- Min, B.S.; Miyashiro, H.; Hattori, M. Inhibitory effects of quinones on RNase H activity associated with HIV-1 reverse transcriptase. Phytother. Res. 2002, 16, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Anstett, K.; Brenner, B.; Mesplède, T.; Wainberg, M.A. HIV-1 Resistance to Dolutegravir Is Affected by Cellular Histone Acetyltransferase Activity. J. Virol. 2017, 91, e00912-17. [Google Scholar] [CrossRef] [PubMed]
- Bashyal, B.P.; Wellensiek, B.P.; Ramakrishnan, R.; Faeth, S.H.; Ahmad, N.; Gunatilaka, A.A. Altertoxins with potent anti-HIV activity from Alternaria tenuissima QUE1Se, a fungal endophyte of Quercus emoryi. Bioorg. Med. Chem. 2014, 22, 6112–6116. [Google Scholar] [CrossRef]
- Melappa, G.; Hemmanur, K.; Nithin, S.; Poojari, C.; Kakunje, G.; Ryavalad, C. In Vitro Anti-HIV Activity of Partially Purified Coumarin(s) Isolated from Fungal Endophyte, Alternaria Species of Calophyllum inophyllum. Pharmacol. Pharm. 2015, 6, 321–328. [Google Scholar] [CrossRef]
- Richetta, C.; Tu, N.Q.; Delelis, O. Different Pathways Conferring Integrase Strand-Transfer Inhibitors Resistance. Viruses 2022, 14, 2591. [Google Scholar] [CrossRef]
- Singh, S.B.; Zink, D.L.; Guan, Z.; Collado, J.; Pelaez, F.; Felock, P.J.; Hazuda, D.J. Isolation, structure, and HIV-1 integrase inhibitory activity of Xanthoviridicatin E and F, two novel fungal metabolites produced by Penicillium chrysogenum. Helv. Chim. Acta 2003, 86, 3380–3385. [Google Scholar] [CrossRef]
- Jayasuriya, H.; Guan, Z.; Polishook, J.D.; Dombrowski, A.W.; Felock, P.J.; Hazuda, D.J.; Singh, S.B. Isolation, structure, and HIV-1 integrase inhibitory activity of Cytosporic acid, a fungal metabolite produced by a Cytospora sp. J. Nat. Prod. 2003, 66, 551–553. [Google Scholar] [CrossRef] [PubMed]
- Mandala, S.M.; Thornton, R.A.; Frommer, B.R.; Curotto, J.E.; Rozdilsky, W.; Kurtz, M.B.; Giacobbe, R.A.; Bills, G.F.; Cabello, M.A.; Martín, I.; et al. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. 1995, 48, 349–356. [Google Scholar] [CrossRef]
- Guo, Z. The modification of natural products for medical use. Acta Pharm Sin B. 2017, 7, 119–136. [Google Scholar] [CrossRef]
- Shiomi, K.; Matsui, R.; Isozaki, M.; Chiba, H.; Sugai, T.; Yamaguchi, Y.; Masuma, R.; Tomoda, H.; Chiba, T.; Yan, H.; et al. Fungal phenalenones inhibit HIV-1 integrase. J. Antibiot. 2005, 58, 65–68. [Google Scholar] [CrossRef]
- Venugopala, K.N.; Rashmi, V.; Odhav, B. Review on Natural Coumarin Lead Compounds for Their Pharmacological Activity. BioMed. Res. Int. 2013, 2013, 963248. [Google Scholar] [CrossRef] [PubMed]
- Ryang, J.; Yan, Y.; Song, Y.; Liu, F.; Ng, T.B. Anti-HIV, antitumor and immunomodulatory activities of paclitaxel from fermentation broth using molecular imprinting technique. AMB Express 2019, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.B.; Ondeyka, J.G.; Tsipouras, N.; Ruby, C.; Sardana, V.; Schulman, M.; Sanchez, M.; Pelaez, F.; Stahlhut, M.W.; Munshi, S.; et al. Hinnuliquinone, a C2-symmetric dimeric non-peptide fungal metabolite inhibitor of HIV-1 protease. Biochem. Biophys. Res. Commun. 2004, 324, 108–113. [Google Scholar] [CrossRef]
- Sato, N.; Ma, C.M.; Komatsu, K.; Hattori, M. Triterpene-farnesyl hydroquinone conjugates from Ganoderma sinense. J. Nat. Prod. 2009, 72, 958–961. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, N.T.; Collins, K.L. Targeting HIV latency: Resting memory T cells, hematopoietic progenitor cells and future directions. Expert Rev. Anti-Infect. Ther. 2014, 12, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Fromentin, R.; Chomont, N. HIV persistence in subsets of CD4+ T cells: 50 shades of reservoirs. Semin. Immunol. 2021, 51, 101438. [Google Scholar] [CrossRef]
- Haigentz, M.; Moore, P.; Bimali, M.; Cooley, T.; Sparano, J.; Rudek, M.; Ratner, L.; Henry, D.; Ramos, J.; Deeken, J.; et al. Safety and Tolerability of Carboplatin and Paclitaxel in Cancer Patients with HIV (AMC-078), an AIDS Malignancy Consortium (AMC) Study. Oncologist 2022, 27, e623–e624. [Google Scholar] [CrossRef] [PubMed]
- Abbas, U.L.; Glaubius, R.; Mubayi, A.; Hood, G.; Mellors, J.W. Antiretroviral therapy and pre-exposure prophylaxis: Combined impact on HIV transmission and drug resistance in South Africa. J. Infect. Dis. 2013, 208, 224–234. [Google Scholar] [CrossRef]
- Churchill, M.J.; Deeks, S.G.; Margolis, D.M.; Siliciano, R.F.; Swanstrom, R. HIV reservoirs: What, where and how to target them. Nat. Rev. Microbiol. 2016, 14, 55–60. [Google Scholar] [CrossRef]
- Niu, S.; Liu, D.; Shao, Z.; Proksch, P.; Lin, W. Eutypellazines N−S, new thiodiketopiperazines from a deep sea sediment derived fungus Eutypella sp. with anti-VRE activities. Tetrahedron Lett. 2017, 58, 3695–3699. [Google Scholar] [CrossRef]
- Stoszko, M.; Al-Hatmi, A.M.S.; Skriba, A.; Roling, M.; Ne, E.; Crespo, R.; Mueller, Y.M.; Najafzadeh, M.J.; Kang, J.; Ptackova, R.; et al. Gliotoxin, identified from a screen of fungal metabolites, disrupts 7SK snRNP, releases P-TEFb, and reverses HIV-1 latency. Sci. Adv. 2020, 6, eaba6617. [Google Scholar] [CrossRef]
- Xing, S.; Siliciano, R.F. Targeting HIV latency: Pharmacologic strategies toward eradication. Drug Discov. Today 2013, 18, 541–551. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; del-Val, E.; Larsen, J. Ecological functions of Trichoderma spp. and their secondary metabolites in the rhizosphere: Interactions with plants. FEMS Microbiol. Ecol. 2012, 18, 541–551. [Google Scholar] [CrossRef]
- Spivak, A.M.; Planelles, V. Novel Latency Reversal Agents for HIV-1 Cure. Annu. Rev. Med. 2018, 69, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.N. Strategies for inhibiting function of HIV-1 accessory proteins: A necessary route to AIDS therapy. Curr. Med. Chem. 2009, 16, 267–286. [Google Scholar] [CrossRef]
- Sugui, J.A.; Pardo, J.; Chang, Y.C.; Zarember, K.A.; Nardone, G.; Galvez, E.M.; Müllbacher, A.; Gallin, J.I.; Simon, M.M.; Kwon-Chung, K.J. Gliotoxin is a virulence factor of Aspergillus fumigatus: GliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 2007, 6, 1562–1569. [Google Scholar] [CrossRef]
- Scharf, D.H.; Heinekamp, T.; Remme, N.; Hortschansky, P.; Brakhage, A.A.; Hertweck, C. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 2012, 93, 467–472. [Google Scholar] [CrossRef]
- Watanabe, N.; Nishihara, Y.; Yamaguchi, T.; Koito, A.; Miyoshi, H.; Kakeya, H.; Osada, H. Fumagillin suppresses HIV-1 infection of macrophages through the inhibition of Vpr activity. FEBS Lett. 2006, 580, 2598–2602. [Google Scholar] [CrossRef] [PubMed]
- Kamata, M.; Wu, R.P.; An, D.S.; Saxe, J.P.; Damoiseaux, R.; Phelps, M.E.; Huang, J.; Chen, I.S. Cell-based chemical genetic screen identifies damnacanthal as an inhibitor of HIV-1 Vpr induced cell death. Biochem. Biophys. Res. Commun. 2006, 348, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- van den Heever, J.P.; Thompson, T.S.; Curtis, J.M.; Ibrahim, A.; Pernal, S.F. Fumagillin: An overview of recent scientific advances and their significance for apiculture. J. Agric. Food Chem. 2014, 62, 2728–2737. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Kaul, S.; Gupta, S.; Ahmed, M.; Dhar, M.K. Endophytic fungi from medicinal plants: A treasure hunt for bioactive metabolites. Phytochem. Rev. 2012, 11, 487–505. [Google Scholar] [CrossRef]
- Truax, N.J.; Romo, D. Bridging the gap between natural product synthesis and drug discovery. Nat. Prod. Rep. 2020, 37, 1436–1453. [Google Scholar] [CrossRef]
- Frediansyah, A.; Sofyantoro, F.; Alhumaid, S.; Al Mutair, A.; Albayat, H.; Altaweil, H.I.; Al-Afghani, H.M.; AlRamadhan, A.A.; AlGhazal, M.R.; Turkistani, S.A.; et al. Microbial Natural Products with Antiviral Activities, Including Anti-SARS-CoV-2: A Review. Molecules 2022, 27, 4305. [Google Scholar] [CrossRef]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of Open Natural Products database. J. Cheminf. 2021, 13, 2. [Google Scholar] [CrossRef]
- van Santen, J.; Poynton, E.; Iskakova, D.; McMann, E.; Alsup, T.; Clark, T.; Fergusson, C.; Fewer, D.; Hughes, A.; McCadden, C.; et al. The Natural Products Atlas 2.0: A Database of microbially-derived natural products. Nucleic Acids Res. 2022, 50, D1317–D1323. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Yang, B.; Liu, J.; Xun, T.; Liu, Y.; Zhou, X. Penicillixanthone A, a marine-derived dual-coreceptor antagonist as anti-HIV-1 agent. Nat. Prod. Res. 2019, 33, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, L.; Zhu, T.; Ba, M.; Li, G.; Gu, Q.; Guo, Y.; Li, D. Phenylspirodrimanes with Anti-HIV Activity from the Sponge-Derived Fungus Stachybotrys chartarum MXH-X73. J. Nat. Prod. 2013, 76, 2298–2306. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.-W.; Sun, B.-D.; Chen, X.-L.; Liu, X.-Z.; Che, Y.-S. Helotialins A—C, Anti-HIV Metabolites from a Helotialean Ascomycete. Chin. J. Nat. Med. 2009, 7, 140–144. [Google Scholar] [CrossRef]
- Hamano, K.; Kinoshita-Okami, M.; Minagawa, K.; Haruyama, H.; Kinoshita, T.; Hosoya, T.; Furuya, K.; Kinoshita, K.; Tabata, K.; Hemmi, A.; et al. Aquastatin A, an inhibitor of mammalian adenosine triphosphatases from Fusarium aquaeductuum. Taxonomy, fermentation, isolation, structure determination and biological properties. J. Antibiot. 1993, 46, 1648–1657. [Google Scholar] [CrossRef]
- Singh, S.B.; Zink, D.L.; Bills, G.F.; Pelaez, F.; Teran, A.; Collado, J.; Silverman, K.C.; Lingham, R.B.; Felock, P.; Hazuda, D.J. Discovery, structure and HIV-1 integrase-inhibitory activities of integracins, novel dimeric alkyl aromatics from Cytonaema. Tetrahedron Lett. 2002, 43, 1617–1620. [Google Scholar] [CrossRef]
- Srisapoomi, T.; Ichiyanagi, T.; Nakajima, H.; Aimi, T.; Boonlue, S. Biological Activities of Integric Acid Isolated from the Wood-Decay Fungus Xylaria feejeensis 2FB-PPM08M. Chiang Mai J. Sci. 2015, 42, 71–79. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).