SARS-CoV-2 Vaccine-Induced T-Cell Response after Three Doses in People Living with HIV on Antiretroviral Therapy Compared to Seronegative Controls (CTN 328 COVAXHIV Study)
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Vaccine Doses and Study Visits
2.3. Ex Vivo Immunophenotyping of T-Cells
2.4. Intracellular Cytokine Production upon In Vitro SARS-CoV-2 Peptide Stimulation
2.5. Ethical Statement
2.6. Statistical Analyses
3. Results
3.1. Study Population
3.2. Peak SARS-CoV-2-Specific CD4 T-Cell Cytokine Secretion Response Achieved after 2nd Vaccine Dose in PLWH
3.3. SARS-CoV-2 Vaccine Induces a Detectable CD8 T-Cell Response in PLWH
3.4. Changes in CD4 T-Cell Subsets following 3 Doses of SARS-CoV-2 Vaccination
3.5. Changes in CD8 T-Cell Subsets following 3 Doses of SARS-CoV-2 Vaccination
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spinelli, M.A.; Jones, B.L.H.; Gandhi, M. COVID-19 Outcomes and Risk Factors Among People Living with HIV. Curr. HIV/AIDS Rep. 2022, 19, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Alexandrova, Y.; Costiniuk, C.T.; Jenabian, M.A. Pulmonary Immune Dysregulation and Viral Persistence during HIV Infection. Front Immunol. 2021, 12, 808722. [Google Scholar] [CrossRef]
- Tebas, P.; Frank, I.; Lewis, M.; Quinn, J.; Zifchak, L.; Thomas, A.; Kenney, T.; Kappes, R.; Wagner, W.; Maffei, K.; et al. Poor immunogenicity of the H1N1 2009 vaccine in well controlled HIV-infected individuals. AIDS 2010, 24, 2187–2192. [Google Scholar] [CrossRef]
- Farooq, P.D.; Sherman, K.E. Hepatitis B Vaccination and Waning Hepatitis B Immunity in Persons Living with HIV. Curr. HIV/AIDS Rep. 2019, 16, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Alrubayyi, A.; Gea-Mallorquí, E.; Touizer, E.; Hameiri-Bowen, D.; Kopycinski, J.; Charlton, B.; Fisher-Pearson, N.; Muir, L.; Rosa, A.; Roustan, C.; et al. Characterization of humoral and SARS-CoV-2 specific T cell responses in people living with HIV. Nat. Commun. 2021, 12, 5839. [Google Scholar] [CrossRef] [PubMed]
- Antinori, A.; Cicalini, S.; Meschi, S.; Bordoni, V.; Lorenzini, P.; Vergori, A.; Lanini, S.; De Pascale, L.; Matusali, G.; Mariotti, D.; et al. Humoral and Cellular Immune Response Elicited by mRNA Vaccination against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in People Living with Human Immunodeficiency Virus Receiving Antiretroviral Therapy Based on Current CD4 T-Lymphocyte Count. Clin. Infect. Dis. 2022, 75, e552–e563. [Google Scholar]
- Coburn, S.B.; Humes, E.; Lang, R.; Stewart, C.; Hogan, B.C.; Gebo, K.A.; Napravnik, S.; Edwards, J.K.; Browne, L.E.; Park, L.S.; et al. Analysis of Postvaccination Breakthrough COVID-19 Infections among Adults with HIV in the United States. JAMA Netw. Open 2022, 5, e2215934. [Google Scholar] [CrossRef]
- Costiniuk, C.T.; Singer, J.; Langlois, M.A.; Kulic, I.; Needham, J.; Burchell, A.; Jenabian, M.A.; Walmsley, S.; Ostrowski, M.; Kovacs, C.; et al. CTN 328: Immunogenicity outcomes in people living with HIV in Canada following vaccination for COVID-19 (HIV-COV): Protocol for an observational cohort study. BMJ Open 2021, 11, e054208. [Google Scholar] [CrossRef]
- Costiniuk, C.T.; Singer, J.; Lee, T.; Langlois, M.-A.; Arnold, C.; Galipeau, Y.; Needham, J.; Kulic, I.; Jenabian, M.-A.; Burchell, A.N.; et al. COVID-19 vaccine immunogenicity in people living with HIV (CIHR Canadian HIV Trials Network 328). AIDS 2022, 37, F1–F10. [Google Scholar] [CrossRef]
- Cai, S.; Liao, G.; Yu, T.; Gao, Q.; Zou, L.; Zhang, H.; Xu, X.; Chen, J.; Lu, A.; Wu, Y.; et al. Immunogenicity and safety of an inactivated SARS-CoV-2 vaccine in people living with HIV: A cross-sectional study. J. Med. Virol. 2022, 94, 4224–4233. [Google Scholar] [CrossRef]
- Vergori, A.; Cozzi Lepri, A.; Cicalini, S.; Matusali, G.; Bordoni, V.; Lanini, S.; Meschi, S.; Iannazzo, R.; Mazzotta, V.; Colavita, F.; et al. Immunogenicity to COVID-19 mRNA vaccine third dose in people living with HIV. Nat. Commun. 2022, 13, 4922. [Google Scholar] [CrossRef]
- Collins, E.; Galipeau, Y.; Arnold, C.; Bosveld, C.; Heiskanen, A.; Keeshan, A.; Nakka, K.; Shir-Mohammadi, K.; St-Denis-Bissonnette, F.; Tamblyn, L.; et al. Cohort profile: Stop the Spread Ottawa (SSO)—A community-based prospective cohort study on antibody responses, antibody neutralisation efficiency and cellular immunity to SARS-CoV-2 infection and vaccination. BMJ Open 2022, 12, e062187. [Google Scholar] [CrossRef] [PubMed]
- Ao, L.; Lu, T.; Cao, Y.; Chen, Z.; Wang, Y.; Li, Z.; Ren, X.; Xu, P.; Peng, M.; Chen, M.; et al. Safety and immunogenicity of inactivated SARS-CoV-2 vaccines in people living with HIV. Emerg. Microbes Infect. 2022, 11, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Lamacchia, G.; Salvati, L.; Kiros, S.T.; Mazzoni, A.; Vanni, A.; Capone, M.; Carnasciali, A.; Farahvachi, P.; Lagi, F.; Di Lauria, N.; et al. Fourth Dose of mRNA COVID-19 Vaccine Transiently Reactivates Spike-Specific Immunological Memory in People Living with HIV (PLWH). Biomedicines 2022, 10, 3261. [Google Scholar] [CrossRef]
- Lang, R.; Humes, E.; Coburn, S.B.; Horberg, M.A.; Fathi, L.F.; Watson, E.; Jefferson, C.R.; Park, L.S.; Gordon, K.S.; Akgün, K.M.; et al. Analysis of Severe Illness after Postvaccination COVID-19 Breakthrough among Adults with and without HIV in the US. JAMA Netw. Open 2022, 5, e2236397. [Google Scholar] [CrossRef] [PubMed]
- De Pablo-Bernal, R.S.; Ruiz-Mateos, E.; Rosado, I.; Dominguez-Molina, B.; Alvarez-Ríos, A.I.; Carrillo-Vico, A.; De La Rosa, R.; Delgado, J.; Muñoz-Fernández, M.A.; Leal, M.; et al. TNF-α levels in HIV-infected patients after long-term suppressive cART persist as high as in elderly, HIV-uninfected subjects. J. Antimicrob. Chemother. 2014, 69, 3041–3046. [Google Scholar] [CrossRef]
- Collora, J.A.; Pinto-Santini, D.; Pasalar, S.; Ravindra, N.; Ganoza, C.; Lama, J.; Alfaro, R.; Chiarella, J.; Spudich, S.; van Dijk, D.; et al. Single-cell immune profiling reveals the impact of antiretroviral therapy on HIV-1-induced immune dysfunction, T cell clonal expansion, and HIV-1 persistence in vivo. bioRxiv 2021. [Google Scholar] [CrossRef]
- Osuji, F.N.; Onyenekwe, C.C.; Ahaneku, J.E.; Ukibe, N.R. The effects of highly active antiretroviral therapy on the serum levels of pro-inflammatory and anti-inflammatory cytokines in HIV infected subjects. J. Biomed. Sci. 2018, 25, 88. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M.L.; Shive, C.L.; Nguyen, T.P.; Younes, S.A.; Panigrahi, S.; Lederman, M.M. Cytokines and T-Cell Homeostasis in HIV Infection. J. Infect Dis. 2016, 214, S51–S57. [Google Scholar] [CrossRef]
- Day, C.L.; Kaufmann, D.E.; Kiepiela, P.; Brown, J.A.; Moodley, E.S.; Reddy, S.; Mackey, E.W.; Miller, J.D.; Leslie, A.J.; DePierres, C.; et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 2006, 443, 350–354. [Google Scholar] [CrossRef]
- Petrovas, C.; Casazza, J.P.; Brenchley, J.M.; Price, D.A.; Gostick, E.; Adams, W.C.; Precopio, M.L.; Schacker, T.; Roederer, M.; Douek, D.C.; et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 2006, 203, 2281–2292. [Google Scholar] [CrossRef]
- Thieme, C.J.; Anft, M.; Paniskaki, K.; Blazquez-Navarro, A.; Doevelaar, A.; Seibert, F.S.; Hoelzer, B.; Konik, M.J.; Brenner, T.; Tempfer, C.; et al. The SARS-CoV-2 T-cell immunity is directed against the spike, membrane, and nucleocapsid protein and associated with COVID 19 severity. medRxiv 2020. [Google Scholar] [CrossRef]
- Deming, M.E.; Lyke, K.E. A ‘mix and match’ approach to SARS-CoV-2 vaccination. Nat. Med. 2021, 27, 1510–1511. [Google Scholar] [CrossRef]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022, 185, 2434–2451.e17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, Y.; Kang, Y. Naïve T cells may be key to the low mortality of children with COVID-19. J. Evid.-Based Med. 2022, 15, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.D.; Emerson, S.G.; Punt, J.; Goff, W.D. Decreased Naïve T-cell Production Leading to Cytokine Storm as Cause of Increased COVID-19 Severity with Comorbidities. Aging Dis. 2020, 11, 742–745. [Google Scholar] [CrossRef]
- Vitalle, J.; Perez-Gomez, A.; Ostos, F.J.; Gasca-Capote, C.; Jimenez-Leon, M.R.; Bachiller, S.; Rivas-Jeremias, I.; Silva-Sanchez, M.D.M.; Ruiz-Mateos, A.M.; Martin-Sanchez, M.A.; et al. Immune defects associated with lower SARS-CoV-2 BNT162b2 mRNA vaccine response in aged people. JCI Insight 2022, 7, 17. [Google Scholar] [CrossRef]
- Amu, S.; Lantto Graham, R.; Bekele, Y.; Nasi, A.; Bengtsson, C.; Rethi, B.; Sorial, S.; Meini, G.; Zazzi, M.; Hejdeman, B.; et al. Dysfunctional phenotypes of CD4+ and CD8+ T cells are comparable in patients initiating ART during early or chronic HIV-1 infection. Medicine 2016, 95, 23. [Google Scholar] [CrossRef]
- Mold, J.E.; Réu, P.; Olin, A.; Bernard, S.; Michaëlsson, J.; Rane, S.; Yates, A.; Khosravi, A.; Salehpour, M.; Possnert, G.; et al. Cell generation dynamics underlying naive T-cell homeostasis in adult humans. PLoS Biol. 2019, 17, e3000383. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Schulte, S.; Wildner, N.H.; Wittner, M.; Brehm, T.T.; Ramharter, M.; Woost, R.; Lohse, A.W.; Jacobs, T.; Schulze zur Wiesch, J. Analysis of Co-inhibitory Receptor Expression in COVID-19 Infection Compared to Acute Plasmodium falciparum Malaria: LAG-3 and TIM-3 Correlate with T Cell Activation and Course of Disease. Front. Immunol. 2020, 11, 1870. [Google Scholar] [CrossRef] [PubMed]
- Yoshie, O.; Matsushima, K. CCR4 and its ligands: From bench to bedside. Int. Immunol. 2015, 27, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Spoerl, S.; Kremer, A.N.; Aigner, M.; Eisenhauer, N.; Koch, P.; Meretuk, L.; Löffler, P.; Tenbusch, M.; Maier, C.; Überla, K.; et al. Upregulation of CCR4 in activated CD8+ T cells indicates enhanced lung homing in patients with severe acute SARS-CoV-2 infection. Eur. J. Immunol. 2021, 51, 1436–1448. [Google Scholar] [CrossRef] [PubMed]
- Guerrera, G.; Picozza, M.; D’Orso, S.; Placido, R.; Pirronello, M.; Verdiani, A.; Termine, A.; Fabrizio, C.; Giannessi, F.; Sambucci, M.; et al. BNT162b2 vaccination induces durable SARS-CoV-2–specific T cells with a stem cell memory phenotype. Sci. Immunol. 2021, 6, eabl5344. [Google Scholar] [CrossRef] [PubMed]
- Ewer, K.J.; Barrett, J.R.; Belij-Rammerstorfer, S.; Sharpe, H.; Makinson, R.; Morter, R.; Flaxman, A.; Wright, D.; Bellamy, D.; Bittaye, M.; et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021, 27, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.G.; Mendoza, A.; Hemmers, S.; Moltedo, B.; Niec, R.E.; Schizas, M.; Hoyos, B.E.; Putintseva, E.V.; Chaudhry, A.; Dikiy, S.; et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 2017, 546, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Fu, J.; Xing, S.; Fu, B.; Zhang, Z.; Shi, M.; Wang, X.; Zhang, J.; Jin, L.; Kang, F.; et al. The decrease of regulatory T cells correlates with excessive activation and apoptosis of CD8+ T cells in HIV-1-infected typical progressors, but not in long-term non-progressors. Immunology 2009, 128, e366–e375. [Google Scholar] [CrossRef]
- Machicote, A.; Belén, S.; Baz, P.; Billordo, L.A.; Fainboim, L. Human CD8. Front. Immunol. 2018, 9, 2788. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.K.; Kagari, T.; Clingan, J.M.; Matloubian, M. Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc. Natl. Acad. Sci. USA 2011, 108, E118–E127. [Google Scholar] [CrossRef] [PubMed]
- Valente, M.; Dölen, Y.; van Dinther, E.; Vimeux, L.; Fallet, M.; Feuillet, V.; Figdor, C.G. Cross-talk between iNKT cells and CD8 T cells in the spleen requires the IL-4/CCL17 axis for the generation of short-lived effector cells. Proc. Natl. Acad. Sci. USA 2019, 116, 25816–25827. [Google Scholar] [CrossRef]
- Bayry, J. Regulatory T cells as adjuvant target for enhancing the viral disease vaccine efficacy. VirusDisease 2014, 25, 18–25. [Google Scholar] [CrossRef]
- Xu, Z.; Jiang, X.; Dai, X.; Li, B. The Dynamic Role of FOXP3(+) Tregs and Their Potential Therapeutic Applications During SARS-CoV-2 Infection. Front. Immunol. 2022, 13, 916411. [Google Scholar] [CrossRef] [PubMed]
- Kidani, Y.; Nogami, W.; Yasumizu, Y.; Kawashima, A.; Tanaka, A.; Sonoda, Y.; Tona, Y.; Nashiki, K.; Matsumoto, R.; Hagiwara, M.; et al. CCR8-targeted specific depletion of clonally expanded Treg cells in tumor tissues evokes potent tumor immunity with long-lasting memory. Proc. Natl. Acad. Sci. USA 2022, 119, e2114282119. [Google Scholar] [CrossRef]
- Moore, A.C.; Gallimore, A.; Draper, S.J.; Watkins, K.R.; Gilbert, S.C.; Hill, A.V.S. Anti-CD25 Antibody Enhancement of Vaccine-Induced Immunogenicity: Increased Durable Cellular Immunity with Reduced Immunodominance1. J. Immunol. 2005, 175, 7264–7273. [Google Scholar] [CrossRef] [PubMed]
- Antonioli, L.; Pacher, P.; Vizi, E.S.; Haskó, G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013, 19, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Yero, A.; Farnos, O.; Rabezanahary, H.; Racine, G.; Estaquier, J.; Jenabian, M.A. Differential Dynamics of Regulatory T-Cell and Th17 Cell Balance in Mesenteric Lymph Nodes and Blood following Early Antiretroviral Initiation during Acute Simian Immunodeficiency Virus Infection. J. Virol. 2019, 93, e00371-19. [Google Scholar] [CrossRef] [PubMed]
- Carrière, M.; Lacabaratz, C.; Kök, A.; Benne, C.; Jenabian, M.A.; Casartelli, N.; Hüe, S.; Hocqueloux, L.; Lelièvre, J.D.; Lévy, Y. HIV “elite controllers” are characterized by a high frequency of memory CD8+ CD73+ T cells involved in the antigen-specific CD8+ T-cell response. J. Infect Dis. 2014, 209, 1321–1330. [Google Scholar] [CrossRef]
- Gupta, P.K.; Godec, J.; Wolski, D.; Adland, E.; Yates, K.; Pauken, K.E.; Cosgrove, C.; Ledderose, C.; Junger, W.G.; Robson, S.C.; et al. CD39 Expression Identifies Terminally Exhausted CD8+ T Cells. PLoS Pathog. 2015, 11, e1005177. [Google Scholar] [CrossRef]
- Bai, A.; Moss, A.; Rothweiler, S.; Serena Longhi, M.; Wu, Y.; Junger, W.G.; Robson, S.C. NADH oxidase-dependent CD39 expression by CD8+ T cells modulates interferon gamma responses via generation of adenosine. Nat. Commun. 2015, 6, 8819. [Google Scholar] [CrossRef]
- Steiner, K.; Waase, I.; Rau, T.; Dietrich, M.; Fleischer, B.; Bröker, B.M. Enhanced expression of CTLA-4 (CD152) on CD4+ T cells in HIV infection. Clin. Exp. Immunol. 1999, 115, 451–457. [Google Scholar] [CrossRef]
- Jubel, J.M.; Barbati, Z.R.; Burger, C.; Wirtz, D.C.; Schildberg, F.A. The Role of PD-1 in Acute and Chronic Infection. Front. Immunol. 2020, 11, 487. [Google Scholar] [CrossRef] [PubMed]
- Kusnadi, A.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Meckiff, B.J.; Simon, H.; Pelosi, E.; Seumois, G.; Ay, F.; Vijayanand, P.; et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021, 6, eabe4782. [Google Scholar] [CrossRef]
- Lin, H.-S.; Lin, X.-H.; Wang, J.-W.; Wen, D.-N.; Xiang, J.; Fan, Y.-Q.; Li, H.-D.; Wu, J.; Lin, Y.; Lin, Y.-L.; et al. Exhausting T Cells During HIV Infection May Improve the Prognosis of Patients with COVID-19. Front. Cell. Infect. Microbiol. 2021, 11, 564938. [Google Scholar] [CrossRef]
- Rha, M.-S.; Jeong, H.W.; Ko, J.-H.; Choi, S.J.; Seo, I.-H.; Lee, J.S.; Sa, M.; Kim, A.R.; Joo, E.-J.; Ahn, J.Y.; et al. PD-1-Expressing SARS-CoV-2-Specific CD8+ T Cells Are Not Exhausted, but Functional in Patients with COVID-19. Immunity 2021, 54, 44–52.e3. [Google Scholar] [CrossRef]
- Garcia-Bates, T.M.; Palma, M.L.; Shen, C.; Gambotto, A.; Macatangay, B.J.C.; Ferris, R.L.; Rinaldo, C.R.; Mailliard, R.B. Contrasting Roles of the PD-1 Signaling Pathway in Dendritic Cell-Mediated Induction and Regulation of HIV-1-Specific Effector T Cell Functions. J. Virol. 2019, 93, e02035-18. [Google Scholar] [CrossRef] [PubMed]
- Day, C.L.; Abrahams, D.A.; Bunjun, R.; Stone, L.; de Kock, M.; Walzl, G.; Wilkinson, R.J.; Burgers, W.A.; Hanekom, W.A. PD-1 Expression on Mycobacterium tuberculosis-Specific CD4 T Cells Is Associated with Bacterial Load in Human Tuberculosis. Front. Immunol. 2018, 9, 1995. [Google Scholar] [CrossRef] [PubMed]
- Breton, G.; Chomont, N.; Takata, H.; Fromentin, R.; Ahlers, J.; Filali-Mouhim, A.; Riou, C.; Boulassel, M.R.; Routy, J.P.; Yassine-Diab, B.; et al. Programmed death-1 is a marker for abnormal distribution of naive/memory T cell subsets in HIV-1 infection. J. Immunol. 2013, 191, 2194–2204. [Google Scholar] [CrossRef]
- Jin, J.-M.; Bai, P.; He, W.; Wu, F.; Liu, X.-F.; Han, D.-M.; Liu, S.; Yang, J.-K. Gender Differences in Patients with COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.J.; Swadling, L.; Gibbons, J.M.; Pade, C.; Jensen, M.P.; Diniz, M.O.; Schmidt, N.M.; Butler, D.K.; Amin, O.E.; Bailey, S.N.L.; et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 2020, 5, eabf3698. [Google Scholar] [CrossRef]

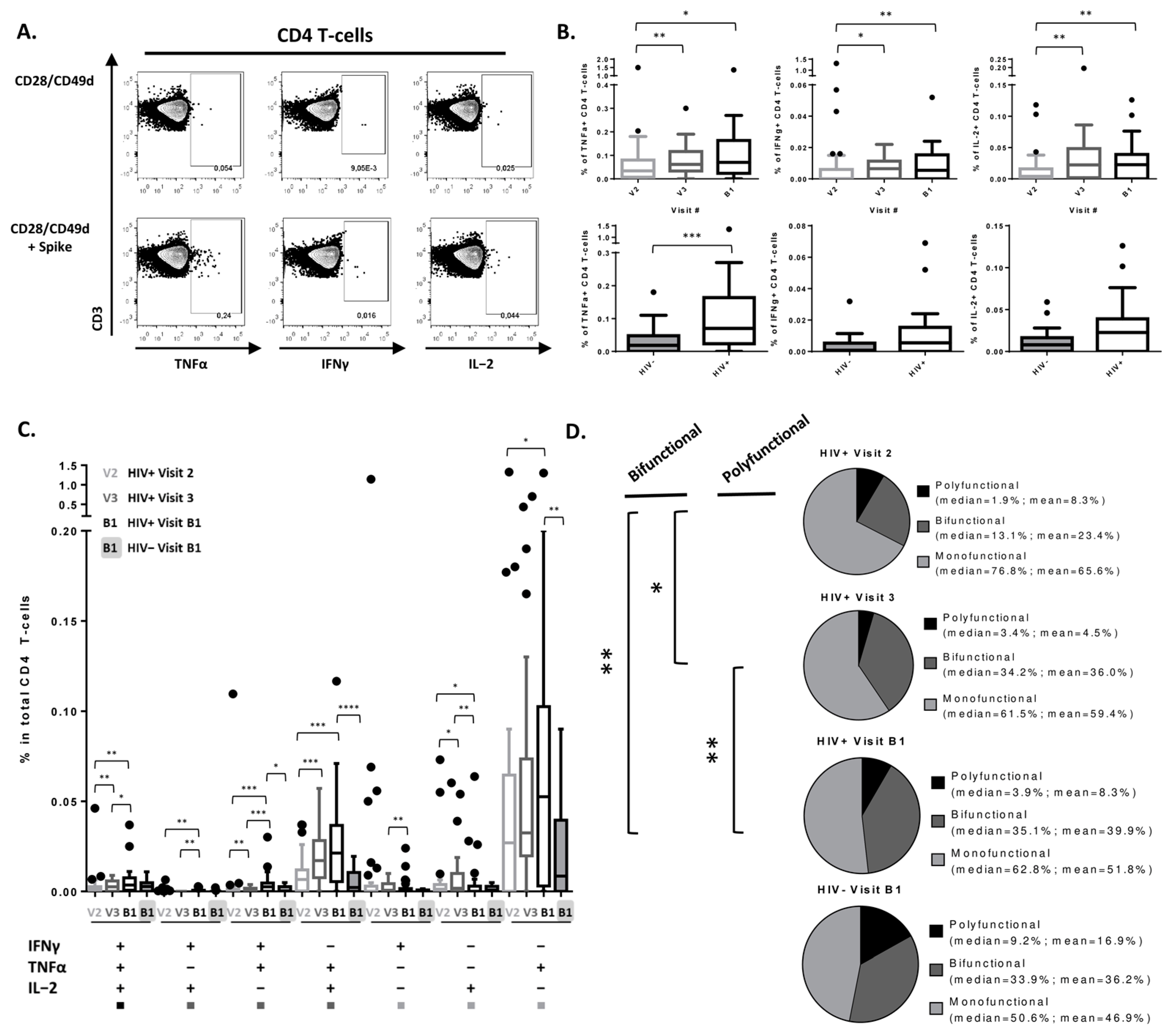
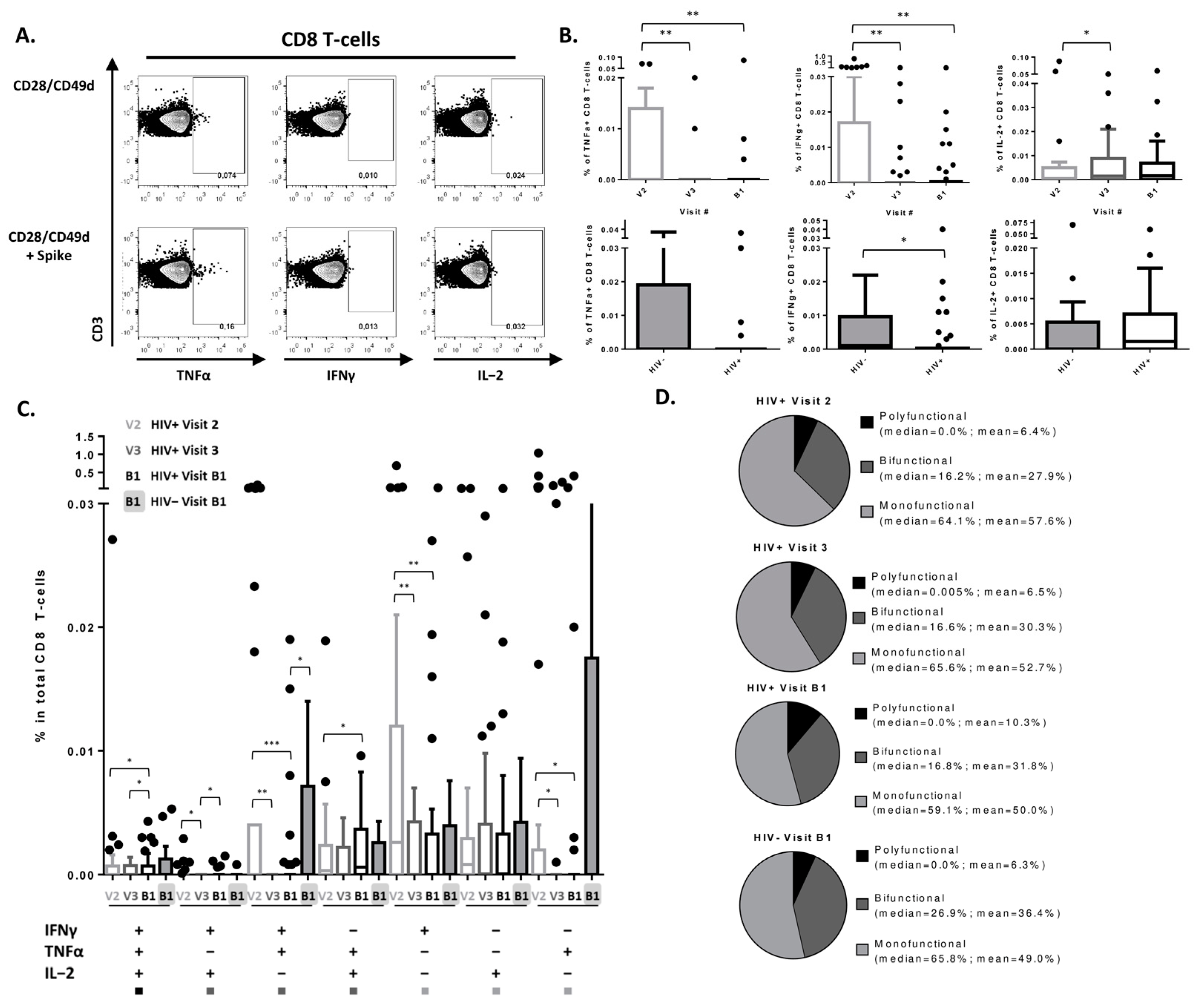
| Characteristics | PLWH, N = 38 1 | Uninfected, N = 24 1 | p-Value |
|---|---|---|---|
| Age, years, median (IQR) | 43 (36, 57) | 44 (38, 56) | 0.8 2 |
| Male sex, n (%) | 33 (87%) | 12 (50%) | 0.002 3 |
| Ethnicity, n (%) | 0.008 4 | ||
| Caucasian | 21 (55%) | 24 (100%) | |
| Black | 2 (5.3%) | 0 (0%) | |
| Chinese | 4 (11%) | 0 (0%) | |
| Filipino | 2 (5.3%) | 0 (0%) | |
| Japanese | 1 (2.6%) | 0 (0%) | |
| Latin American | 5 (13%) | 0 (0%) | |
| Other | 3 (7.9%) | 0 (0%) | |
| SARS-CoV2 vaccine type received, n (%) | 0.019 4 | ||
| 3 doses of mRNA (Pfizer or Moderna) | 38 (100%) | 20 (83%) | |
| 1st dose Adenovirus vector (Astrazeneca), 2nd/3d dose mRNA (Pfizer or Moderna) | 0 (0%) | 4 (17%) | |
| CD4 count (n = 35), cells/mm3, median (IQR) | 700 (480, 839) | N/A | N/A |
| CD4/CD8 ratio (n = 35) | 0.81 (0.59, 1.01) | ||
| CD4 nadir, cells/mm3, median (IQR) (n = 37) | 290 (167, 420) | ||
| Duration of HIV infection, years (IRQ) (n = 37) | 6.5 (5.0, 21.0) | ||
| Undetectable viral load for at least 6 months, n (%) | 30 (79%) | ||
| If detectable, highest viral load over past 6 months (n = 8), median (IQR) (copies/mL) | 182 (56, 2, 040) | ||
| ART regimen, n (%) | |||
| INSTI-based regimen | 28 (74%) | N/A | |
| NNRTI-based regimen | 5 (13%) | ||
| NRTI-based regimen | 2 (5.3%) | ||
| Other | 3 (7.9%) | ||
| Coinfections, n (%) | N/A | ||
| Hepatitis B virus | 0 (0%) | 0 (0%) | |
| Hepatitis C virus | 0 (0%) | 0 (0%) | |
| Characteristics | PLWH Longitudinal | Uninfected Longitudinal | PLWH vs. Uninfected Cross-Sectional | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 N = 38 1 | V2 N = 38 1 | V3 N = 38 1 | B1 N = 38 1 | V1 vs. V2 2 | V1 vs. V3 2 | V1 vs. B1 2 | V1 N = 24 1 | B1 N = 24 1 | V1 vs. B1 2 | V1 3 | B1 3 | |
| Memory T-Cell Subsets | ||||||||||||
| Naive (CD45RA+CD28+CCR7+) CD4 T-cells | 19.55 (14.62, 27.73) | 26.35 (19.67, 34.05) | 29.05 (18.27, 35.15) | 28.45 (21.33, 33.70) | <0.001 | <0.001 | <0.001 | 11.45 (6.65, 17.12) | 21.60 (8.47, 29.63) | 0.001 | 0.002 | 0.017 |
| Central Memory (CD45RA-CD28+CCR7+) CD4 T-cells | 42.35 (35.00, 48.13) | 42.60 (37.00, 49.88) | 42.90 (38.30, 50.08) | 44.10 (37.55, 48.95) | 0.14 | 0.3 | 0.5 | 48.05 (40.00, 51.85) | 48.20 (39.70, 55.08) | 0.2 | 0.048 | 0.043 |
| Transitional Memory (CD45RA-CD28+CCR7-) CD4 T-cells | 19.65 (15.50, 22.30) | 17.20 (14.83, 20.15) | 16.55 (14.33, 19.70) | 16.70 (13.55, 21.20) | 0.027 | 0.001 | <0.001 | 20.45 (16.25, 22.65) | 16.50 (13.78, 21.20) | 0.075 | 0.8 | 0.9 |
| Effector Memory (CD45RA-CD28-CCR7-) CD4 T-cells | 3.23 (1.62, 6.33) | 2.00 (1.27, 4.22) | 1.68 (1.01, 3.31) | 1.65 (0.98, 3.40) | <0.001 | <0.001 | <0.001 | 3.98 (2.61, 5.62) | 1.37 (0.76, 3.43) | 0.008 | 0.5 | 0.8 |
| Terminally Differentiated (CD45RA+CD28-CCR7-) CD4 T-cells | 0.35 (0.12, 0.70) | 0.23 (0.11, 0.45) | 0.23 (0.07, 0.49) | 0.16 (0.08, 0.43) | 0.004 | <0.001 | <0.001 | 0.28 (0.19, 0.47) | 0.12 (0.05, 0.32) | 0.025 | 0.7 | 0.4 |
| T-cell function | ||||||||||||
| % HLA-DR+CD38+ in CD4 T-cells | 2.32 (1.93, 3.67) | 2.18 (1.65, 2.86) | 2.04 (1.55, 2.69) | 2.23 (1.74, 2.96) | 0.010 | 0.016 | 0.013 | 2.57 (1.49, 4.92) | 2.36 (1.34, 3.83) | 0.2 | >0.9 | 0.6 |
| % Ki-67+ in CD4 T-cells | 2.70 (2.19, 3.98) | 2.30 (2.00, 3.11) | 2.32 (1.86, 2.72) | 2.39 (1.87, 2.85) | 0.074 | <0.001 | <0.001 | 2.36 (1.97, 3.03) | 2.12 (1.59, 2.80) | 0.3 | 0.070 | 0.4 |
| % PD-1+ in CD4 T-cells | 18.60 (14.67, 25.33) | 19.55 (15.12, 23.98) | 22.20 (16.35, 25.65) | 20.70 (16.12, 25.97) | 0.8 | 0.006 | 0.032 | 9.32 (6.68, 13.03) | 12.15 (11.38, 16.00) | 0.005 | <0.001 | <0.001 |
| % CTLA-4+ in CD4 T-cells | 2.96 (2.30, 3.94) | 3.16 (2.16, 4.29) | 2.73 (2.15, 3.79) | 2.70 (2.15, 3.56) | 0.2 | >0.9 | 0.7 | 2.44 (1.90, 3.15) | 2.84 (1.87, 3.89) | 0.023 | 0.056 | 0.8 |
| % Senescent (CD28-CD57+) in CD4 T-cells | 4.16 (0.98, 9.00) | 2.50 (0.70, 6.60) | 2.15 (0.71, 4.10) | 1.61 (0.85, 4.82) | <0.001 | <0.001 | <0.001 | 1.18 (0.37, 3.22) | 0.32 (0.13, 3.24) | 0.009 | 0.030 | 0.025 |
| % CD39+ in CD4 T-cells | 4.92 (3.66, 7.37) | 4.92 (3.73, 6.81) | 5.34 (4.00, 7.69) | 5.07 (3.65, 6.98) | 0.8 | 0.6 | 0.3 | 2.39 (1.90, 3.60) | 2.89 (2.29, 4.36) | 0.037 | <0.001 | 0.006 |
| % CD73+ in CD4 T-cells | 8.59 (5.22, 12.35) | 8.45 (5.77, 12.40) | 9.46 (6.10, 13.03) | 9.12 (5.98, 13.55) | 0.9 | 0.048 | 0.005 | 11.05 (9.23, 14.12) | 12.45 (8.56, 15.95) | 0.079 | 0.062 | 0.036 |
| Chemokine receptors | ||||||||||||
| % CCR4+ in CD4 T-cells | 17.05 (12.20, 22.48) | 16.95 (14.90, 23.58) | 19.15 (15.85, 22.48) | 18.75 (16.72, 22.87) | 0.5 | 0.006 | 0.073 | 16.35 (6.43, 23.18) | 16.25 (10.28, 24.33) | 0.2 | 0.4 | 0.5 |
| % CCR6+ in CD4 T-cells | 38.90 (36.12, 41.20) | 38.60 (35.75, 40.25) | 39.50 (36.97, 41.25) | 39.45 (37.45, 41.25) | 0.4 | 0.7 | 0.7 | 29.35 (19.60, 32.05) | 31.30 (22.10, 34.67) | 0.064 | <0.001 | <0.001 |
| % CXCR3+ in CD4 T-cells | 19.45 (13.15, 26.20) | 17.60 (13.48, 21.32) | 15.70 (13.62, 18.17) | 16.90 (12.60, 18.77) | 0.13 | 0.004 | 0.018 | 35.40 (25.35, 44.88) | 19.40 (12.20, 26.30) | <0.001 | <0.001 | 0.3 |
| Th subsets | ||||||||||||
| % Th17 in CD4 T-cells (CD45RA-CCR4+CCR6+ CXCR3-) | 4.46 (3.26, 6.66) | 4.99 (3.09, 7.38) | 6.10 (5.26, 8.09) | 6.11 (4.54, 7.16) | 0.3 | <0.001 | 0.010 | 1.45 (1.07, 2.12) | 3.19 (2.19, 4.48) | <0.001 | <0.001 | <0.001 |
| % Th1-Th17 in CD4 T-cells (CD45RA-CCR4-CCR6+CXCR3+) | 5.06 (2.71, 6.87) | 3.85 (2.49, 5.55) | 3.97 (2.91, 5.03) | 4.12 (2.78, 5.41) | 0.030 | 0.007 | 0.059 | 4.28 (2.42, 6.52) | 2.68 (2.04, 4.70) | 0.010 | 0.6 | 0.040 |
| % Th2 in CD4 T-cells (CD45RA-CCR4+CCR6-CXCR3-) | 6.36 (4.86, 8.41) | 7.88 (5.95, 10.06) | 8.66 (6.64, 10.31) | 9.11 (6.32, 10.40) | 0.053 | <0.001 | 0.014 | 4.97 (2.86, 9.53) | 8.57 (4.80, 10.48) | 0.004 | 0.2 | 0.8 |
| % Th1 in CD4 T-cells (CD45RA-CCR4-CCR6-CXCR3+) | 9.97 (5.84, 12.55) | 9.00 (5.82, 10.52) | 7.26 (5.65, 9.19) | 8.14 (5.84, 8.92) | 0.2 | 0.002 | 0.013 | 23.90 (15.40, 27.60) | 11.15 (6.63, 16.00) | <0.001 | <0.001 | 0.037 |
| Regulatory T-cells | ||||||||||||
| % Treg (CD25hi CD127lo FoxP3+) in CD4 T-cells | 0.82 (0.52, 1.23) | 1.03 (0.76, 1.95) | 1.44 (1.15, 2.17) | 1.56 (1.11, 1.97) | 0.003 | <0.001 | <0.001 | 0.64 (0.38, 1.05) | 1.21 (0.74, 2.24) | <0.001 | 0.14 | 0.5 |
| % CD39+ Treg in CD4 T-cells | 0.47 (0.28, 0.65) | 0.49 (0.30, 0.72) | 0.58 (0.43, 0.85) | 0.64 (0.34, 0.86) | 0.4 | 0.002 | 0.003 | 0.29 (0.13, 0.49) | 0.35 (0.22, 0.88) | 0.004 | 0.032 | 0.3 |
| % CD73+ Treg in CD4 T-cells | 0.04 (0.02, 0.07) | 0.05 (0.03, 0.11) | 0.09 (0.05, 0.15) | 0.08 (0.05, 0.14) | 0.022 | <0.001 | <0.001 | 0.05 (0.01, 0.10) | 0.13 (0.02, 0.18) | <0.001 | 0.8 | 0.7 |
| % CD39+CD73+ Treg in CD4 T-cells | 0.01 (0.01, 0.03) | 0.01 (0.01, 0.04) | 0.02 (0.02, 0.05) | 0.03 (0.01, 0.04) | 0.4 | <0.001 | 0.002 | 0.02 (0.00, 0.04) | 0.03 (0.01, 0.06) | 0.071 | 0.8 | 0.6 |
| Characteristics | PLWH Longitudinal | Uninfected Longitudinal | PLWH vs. Uninfected Cross-Sectional | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 N = 38 1 | V2 N = 38 1 | V3 N = 38 1 | B1 N = 38 1 | V1 vs. V2 2 | V1 vs. V3 2 | V1 vs. B1 2 | V1 N = 24 1 | B1 N = 24 1 | V1 vs. B1 2 | V1 3 | B1 3 | |
| Memory Subsets | ||||||||||||
| Naive (CD45RA+CD28+CCR7+) CD8 T-cells | 8.33 (5.07, 15.63) | 12.95 (8.91, 20.60) | 15.40 (8.79, 23.02) | 15.75 (7.96, 26.08) | <0.001 | <0.001 | <0.001 | 8.25 (4.69, 15.15) | 20.90 (8.97, 39.67) | <0.001 | 0.8 | 0.2 |
| Central Memory (CD45RA-CD28+CCR7+) CD8 T-cells | 7.70 (4.30, 13.47) | 8.49 (5.41, 16.05) | 10.90 (6.95, 16.50) | 10.90 (6.40, 16.65) | 0.027 | <0.001 | 0.003 | 8.01 (5.77, 10.50) | 12.45 (8.38, 14.82) | <0.001 | 0.9 | 0.6 |
| Transitional Memory (CD45RA-CD28+CCR7-) CD8 T-cells | 15.35 (10.60, 17.50) | 17.05 (11.28, 21.02) | 17.15 (13.12, 20.73) | 16.40 (12.22, 21.25) | 0.040 | 0.002 | 0.009 | 12.40 (9.21, 14.72) | 13.95 (11.88, 20.95) | 0.007 | 0.2 | 0.5 |
| Effector Memory (CD45RA-CD28-CCR7-) CD8 T-cells | 24.40 (17.35, 34.98) | 20.95 (14.72, 27.80) | 15.25 (11.77, 26.12) | 17.80 (12.00, 24.70) | 0.009 | <0.001 | <0.001 | 30.25 (21.68, 35.25) | 14.10 (9.51, 24.65) | <0.001 | 0.4 | 0.5 |
| Terminally Differentiated (CD45RA+CD28-CCR7-) CD8 T-cells | 13.90 (8.24, 19.17) | 11.90 (7.04, 15.90) | 9.93 (6.80, 16.45) | 9.82 (7.18, 12.60) | 0.001 | <0.001 | 0.021 | 8.25 (5.56, 11.25) | 5.74 (4.45, 8.03) | 0.010 | 0.009 | <0.001 |
| T-cell function | ||||||||||||
| % HLA-DR+CD38+ in CD8 T-cells | 3.55 (2.18, 7.49) | 3.91 (2.42, 6.18) | 3.72 (2.47, 5.25) | 3.97 (2.65, 7.11) | 0.5 | 0.7 | 0.4 | 3.02 (2.67, 4.22) | 3.15 (2.77, 3.88) | 0.9 | 0.4 | 0.12 |
| % Ki-67+ in CD8 T-cells | 1.90 (1.63, 2.55) | 2.13 (1.57, 2.66) | 1.83 (1.48, 2.24) | 1.84 (1.47, 2.46) | 0.3 | 0.045 | 0.023 | 2.20 (1.95, 2.51) | 1.95 (1.47, 2.23) | 0.042 | 0.2 | 0.9 |
| % PD-1+ in CD8 T-cells | 22.20 (17.20, 27.95) | 22.90 (15.57, 28.25) | 25.90 (18.68, 31.77) | 24.75 (17.45, 30.62) | 0.7 | 0.011 | 0.045 | 16.95 (11.62, 23.92) | 17.85 (15.23, 26.67) | 0.037 | 0.078 | 0.12 |
| % CTLA-4+ in CD8 T-cells | 1.16 (0.87, 1.52) | 1.13 (0.91, 1.41) | 1.15 (0.87, 1.40) | 1.11 (0.88, 1.57) | 0.9 | 0.4 | 0.6 | 1.23 (1.06, 1.59) | 1.11 (0.93, 1.39) | 0.4 | 0.4 | 0.7 |
| % Senescent (CD28-CD57+) in CD8 T-cells | 36.90 (25.90, 51.78) | 31.20 (23.35, 44.67) | 27.60 (17.48, 43.40) | 28.50 (18.40, 44.52) | <0.001 | <0.001 | <0.001 | 30.00 (15.23, 36.53) | 13.45 (9.50, 31.17) | 0.001 | 0.022 | 0.007 |
| % CD73+ in CD8 T-cells | 17.75 (11.53, 31.97) | 23.40 (14.95, 32.12) | 24.45 (18.42, 39.15) | 27.05 (19.02, 38.48) | 0.005 | <0.001 | <0.001 | 28.80 (22.80, 45.13) | 45.10 (35.62, 57.58) | <0.001 | 0.007 | <0.001 |
| % CD39+ in CD8 T-cells | 0.93 (0.70, 1.33) | 1.00 (0.66, 1.44) | 0.95 (0.61, 1.27) | 0.88 (0.47, 1.63) | 0.5 | 0.13 | 0.5 | 1.41 (0.96, 1.94) | 1.97 (1.21, 2.71) | 0.002 | 0.023 | <0.001 |
| Chemokine receptors | ||||||||||||
| % CCR4+ in CD8 T-cells | 35.45 (28.53, 46.92) | 33.00 (24.93, 43.13) | 22.55 (16.70, 28.25) | 23.40 (17.42, 35.88) | 0.4 | <0.001 | <0.001 | 39.65 (28.15, 52.00) | 35.25 (21.50, 46.45) | 0.005 | 0.4 | 0.059 |
| % CCR6+ in CD8 T-cells | 28.25 (26.80, 29.35) | 28.05 (25.83, 29.73) | 27.80 (25.15, 29.58) | 28.00 (25.85, 30.05) | >0.9 | 0.3 | >0.9 | 12.98 (5.14, 23.40) | 14.91 (4.63, 25.52) | 0.2 | <0.001 | <0.001 |
| % CXCR3+ in CD8 T-cells | 21.20 (12.85, 36.65) | 18.60 (12.85, 26.40) | 15.05 (12.05, 19.82) | 17.80 (11.47, 22.77) | 0.2 | <0.001 | 0.004 | 36.55 (30.50, 46.33) | 19.55 (13.40, 24.70) | <0.001 | <0.001 | 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandrova, Y.; Yero, A.; Mboumba Bouassa, R.-S.; Comeau, E.; Samarani, S.; Brumme, Z.L.; Hull, M.; Crawley, A.M.; Langlois, M.-A.; Angel, J.B.; et al. SARS-CoV-2 Vaccine-Induced T-Cell Response after Three Doses in People Living with HIV on Antiretroviral Therapy Compared to Seronegative Controls (CTN 328 COVAXHIV Study). Viruses 2023, 15, 575. https://doi.org/10.3390/v15020575
Alexandrova Y, Yero A, Mboumba Bouassa R-S, Comeau E, Samarani S, Brumme ZL, Hull M, Crawley AM, Langlois M-A, Angel JB, et al. SARS-CoV-2 Vaccine-Induced T-Cell Response after Three Doses in People Living with HIV on Antiretroviral Therapy Compared to Seronegative Controls (CTN 328 COVAXHIV Study). Viruses. 2023; 15(2):575. https://doi.org/10.3390/v15020575
Chicago/Turabian StyleAlexandrova, Yulia, Alexis Yero, Ralph-Sydney Mboumba Bouassa, Eve Comeau, Suzanne Samarani, Zabrina L. Brumme, Mark Hull, Angela M. Crawley, Marc-André Langlois, Jonathan B. Angel, and et al. 2023. "SARS-CoV-2 Vaccine-Induced T-Cell Response after Three Doses in People Living with HIV on Antiretroviral Therapy Compared to Seronegative Controls (CTN 328 COVAXHIV Study)" Viruses 15, no. 2: 575. https://doi.org/10.3390/v15020575
APA StyleAlexandrova, Y., Yero, A., Mboumba Bouassa, R.-S., Comeau, E., Samarani, S., Brumme, Z. L., Hull, M., Crawley, A. M., Langlois, M.-A., Angel, J. B., Cooper, C. L., Needham, J., Lee, T., Singer, J., Anis, A. H., Costiniuk, C. T., & Jenabian, M.-A. (2023). SARS-CoV-2 Vaccine-Induced T-Cell Response after Three Doses in People Living with HIV on Antiretroviral Therapy Compared to Seronegative Controls (CTN 328 COVAXHIV Study). Viruses, 15(2), 575. https://doi.org/10.3390/v15020575








