Nipah Virus Impairs Autocrine IFN Signaling by Sequestering STAT1 and STAT2 into Inclusion Bodies
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Infection
2.2. Plasmids and Transfection
2.3. Confocal Immunofluorescence Analysis
2.4. Image Analysis to Quantify Nuclear Translocation of STATs
2.5. Image Analysis to Quantify Nuclear pSTAT Levels
2.6. Quantitative Real-Time Polymerase Chain Reaction
3. Results
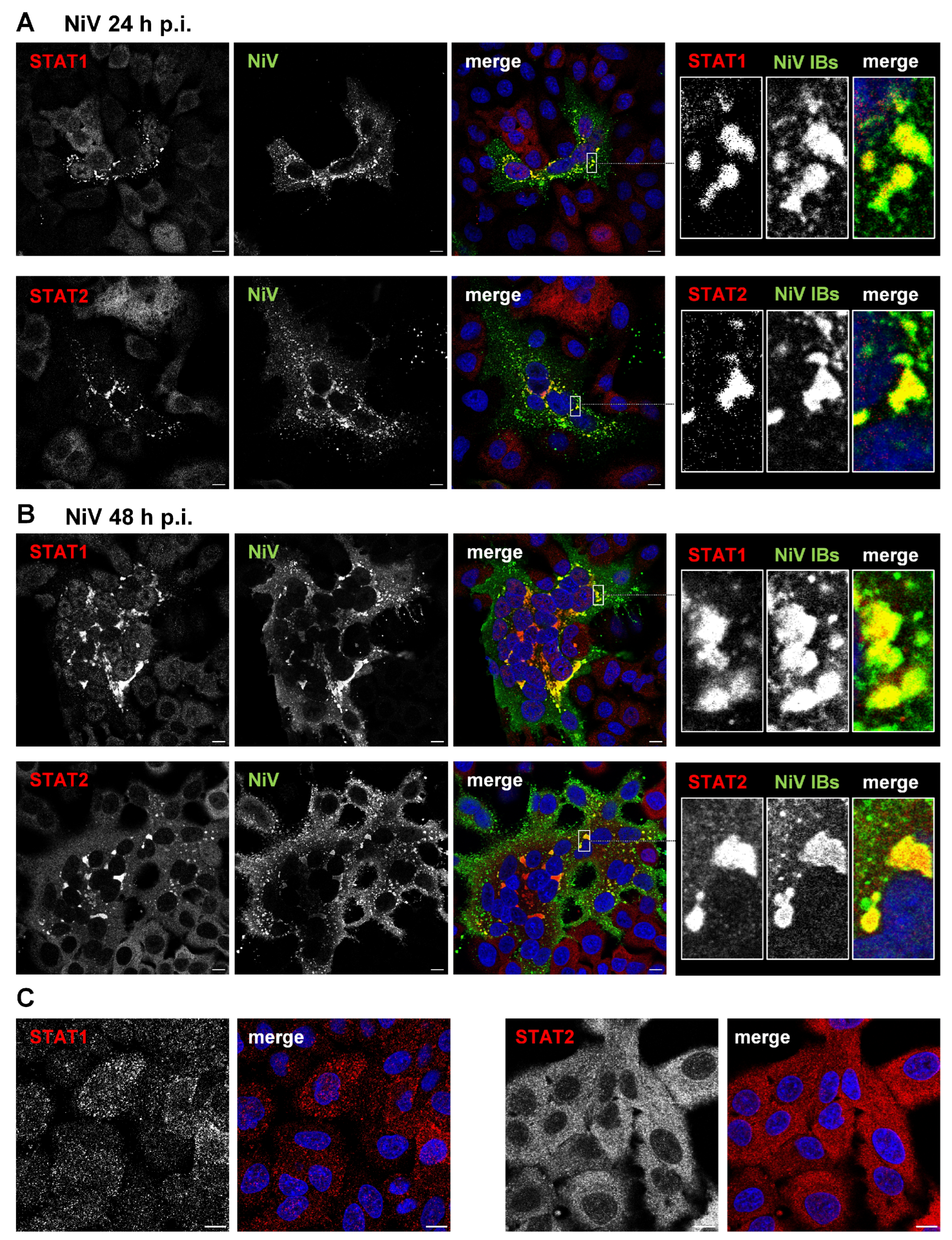
3.1. STAT1 and STAT2 Are Sequestered to Cytosolic IBs in NiV-Infected Cells
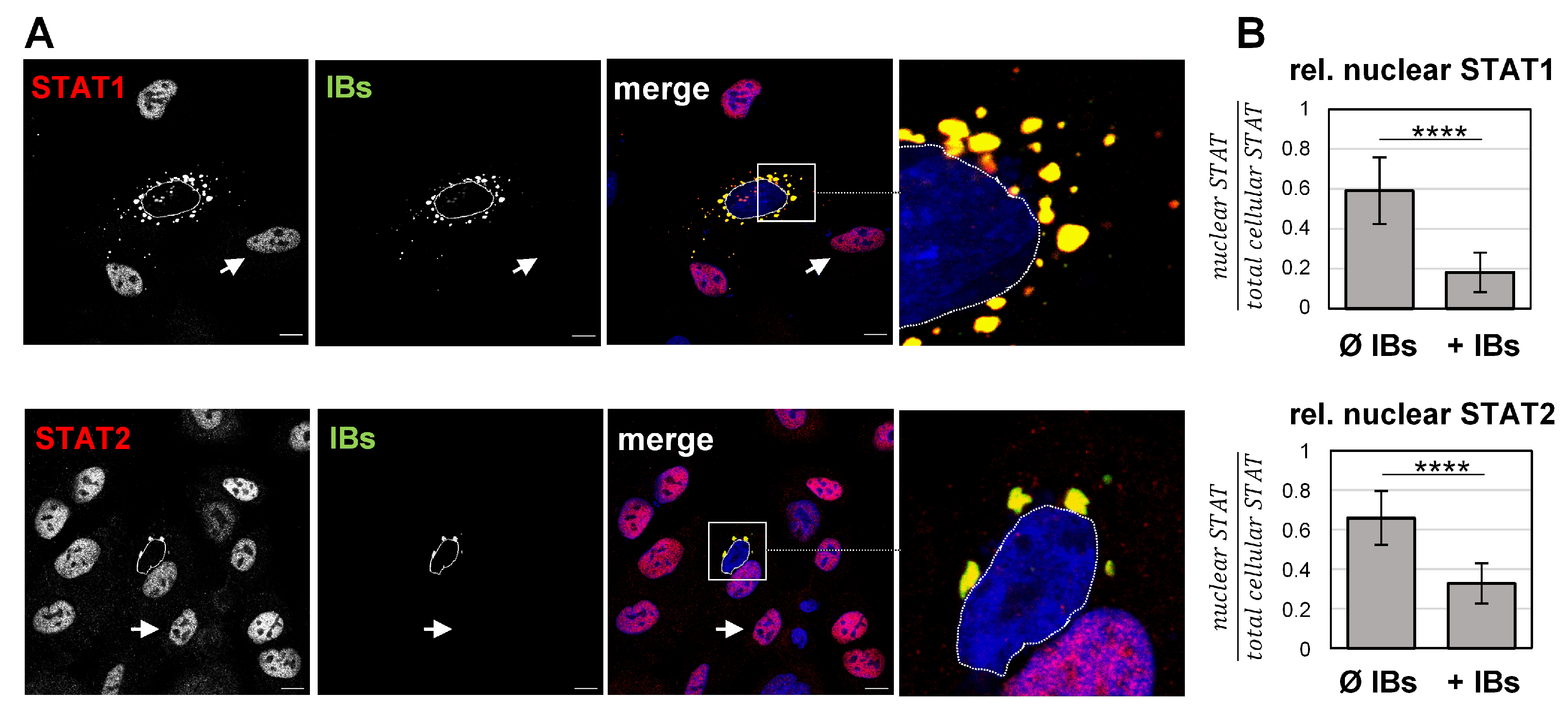
3.2. Recruitment to IBs Blocks IFN-Triggered Nuclear Accumulation of STAT1 and STAT2
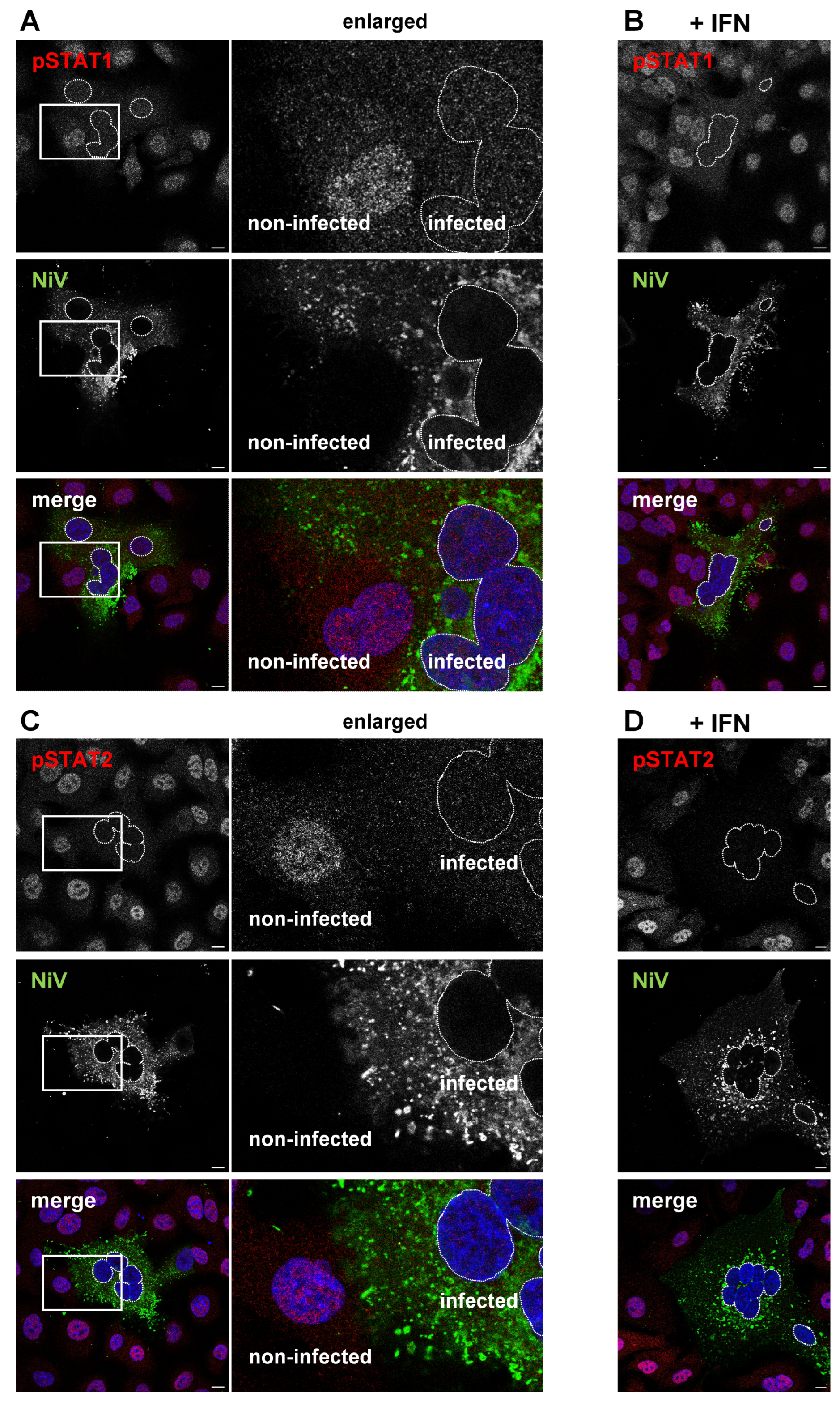
3.3. IFN-Induced Formation of pSTAT Is Prevented in the Presence of IBs
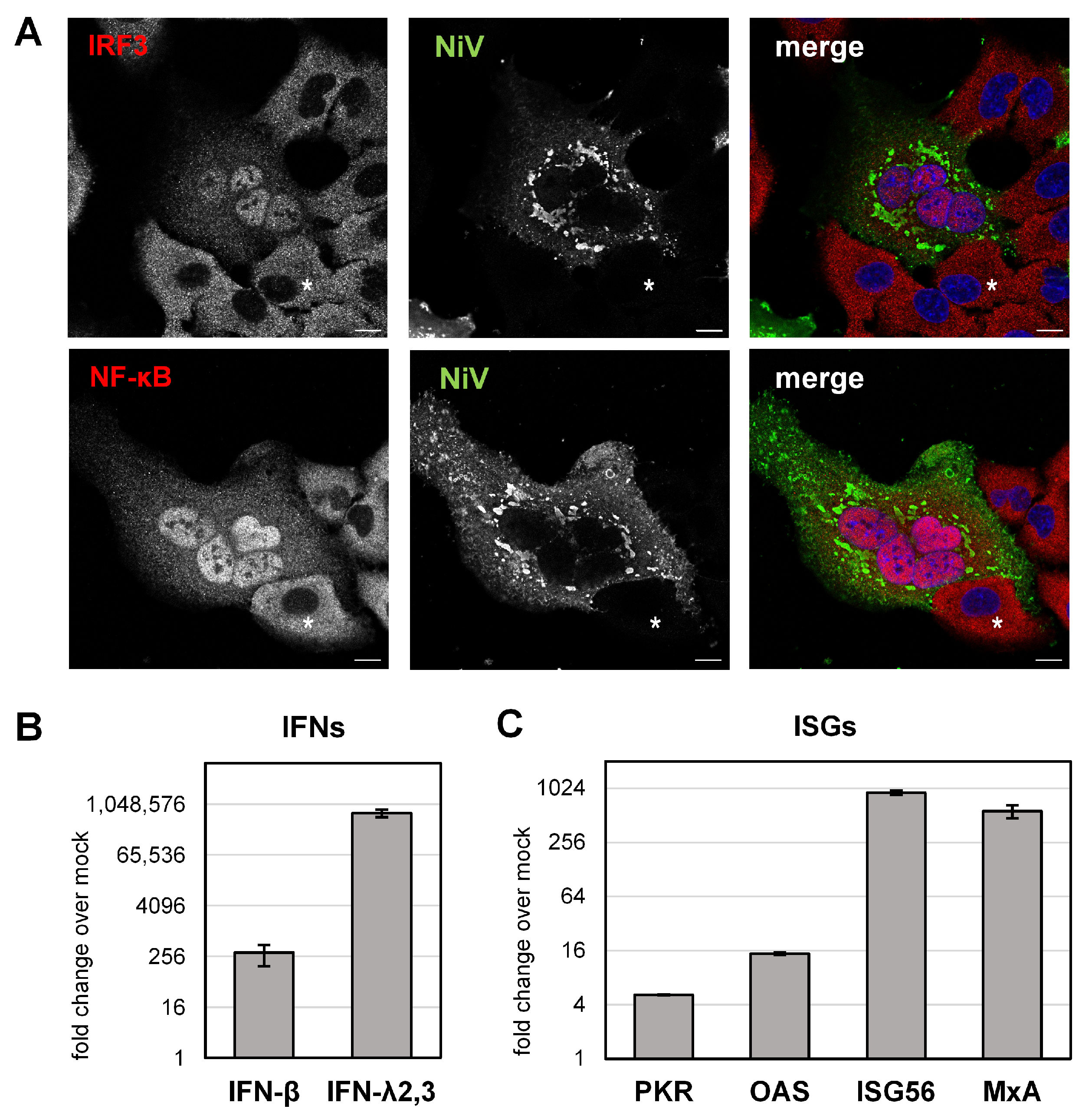
3.4. NiV Induces IFN and ISG Upregulation in Infected Cell Cultures
3.5. Autocrine IFN Signaling Is Blocked in NiV-Infected Cells
3.6. Model for NiV IB-Mediated Blockade of Autocrine IFN Signaling
4. Discussion
4.1. Targeting of STAT Proteins by NiV P Gene Products
4.2. Blockade of STAT Complex Formation by NiV N
4.3. STAT Sequestration to IBs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chua, K.B.; Goh, K.J.; Wong, K.T.; Kamarulzaman, A.; Tan, P.S.; Ksiazek, T.G.; Zaki, S.R.; Paul, G.; Lam, S.K.; Tan, C.T. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 1999, 354, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.H.; Anthony, S.J.; Islam, A.; Kilpatrick, A.M.; Ali Khan, S.; Balkey, M.D.; Ross, N.; Smith, I.; Zambrana-Torrelio, C.; Tao, Y.; et al. Nipah virus dynamics in bats and implications for spillover to humans. Proc. Natl. Acad. Sci. USA 2020, 117, 29190–29201. [Google Scholar] [CrossRef] [PubMed]
- Yob, J.M.; Field, H.; Rashdi, A.M.; Morrissy, C.; van der Heide, B.; Rota, P.; bin Adzhar, A.; White, J.; Daniels, P.; Jamaluddin, A.; et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001, 7, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.P.G.; Sugunan, A.P.; Yadav, P.; Kurup, K.K.; Aarathee, R.; Manickam, P.; Bhatnagar, T.; Radhakrishnan, C.; Thomas, B.; Kumar, A.; et al. Infections among Contacts of Patients with Nipah Virus, India. Emerg. Infect. Dis. 2019, 25, 1007–1010. [Google Scholar] [CrossRef] [PubMed]
- Luby, S.P.; Gurley, E.S.; Hossain, M.J. Transmission of Human Infection with Nipah Virus. Clin. Infect. Dis. 2009, 49, 1743–1748. [Google Scholar] [CrossRef]
- Soman Pillai, V.; Krishna, G.; Valiya Veettil, M. Nipah Virus: Past Outbreaks and Future Containment. Viruses 2020, 12, 465. [Google Scholar] [CrossRef]
- Johnson, K.; Vu, M.; Freiberg, A.N. Recent advances in combating Nipah virus. Fac. Rev. 2021, 10, 74. [Google Scholar] [CrossRef]
- Dietzel, E.; Kolesnikova, L.; Sawatsky, B.; Heiner, A.; Weis, M.; Kobinger, G.P.; Becker, S.; von Messling, V.; Maisner, A. Nipah Virus Matrix Protein Influences Fusogenicity and Is Essential for Particle Infectivity and Stability. J. Virol. 2015, 90, 2514–2522. [Google Scholar] [CrossRef]
- Harcourt, B.H.; Tamin, A.; Ksiazek, T.G.; Rollin, P.E.; Anderson, L.J.; Bellini, W.J.; Rota, P.A. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 2000, 271, 334–349. [Google Scholar] [CrossRef]
- Bonaparte, M.I.; Dimitrov, A.S.; Bossart, K.N.; Crameri, G.; Mungall, B.A.; Bishop, K.A.; Choudhry, V.; Dimitrov, D.S.; Wang, L.F.; Eaton, B.T.; et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 2005, 102, 10652–10657. [Google Scholar] [CrossRef]
- Negrete, O.A.; Levroney, E.L.; Aguilar, H.C.; Bertolotti-Ciarlet, A.; Nazarian, R.; Tajyar, S.; Lee, B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 2005, 436, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Negrete, O.A.; Wolf, M.C.; Aguilar, H.C.; Enterlein, S.; Wang, W.; Muhlberger, E.; Su, S.V.; Bertolotti-Ciarlet, A.; Flick, R.; Lee, B. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006, 2, e7. [Google Scholar] [CrossRef] [PubMed]
- Diederich, S.; Maisner, A. Molecular characteristics of the Nipah virus glycoproteins. Ann. N. Y. Acad. Sci. 2007, 1102, 39–50. [Google Scholar] [CrossRef]
- Lee, B.; Ataman, Z.A. Modes of paramyxovirus fusion: A Henipavirus perspective. Trends. Microbiol. 2011, 19, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Gamble, A.; Yeo, Y.Y.; Butler, A.A.; Tang, H.; Snedden, C.E.; Mason, C.T.; Buchholz, D.W.; Bingham, J.; Aguilar, H.C.; Lloyd-Smith, J.O. Drivers and Distribution of Henipavirus-Induced Syncytia: What Do We Know? Viruses 2021, 13, 1755. [Google Scholar] [CrossRef] [PubMed]
- Middleton, D.J.; Westbury, H.A.; Morrissy, C.J.; van der Heide, B.M.; Russell, G.M.; Braun, M.A.; Hyatt, A.D. Experimental Nipah virus infection in pigs and cats. J. Comp. Pathol. 2002, 126, 124–136. [Google Scholar] [CrossRef]
- Wong, K.T.; Shieh, W.J.; Kumar, S.; Norain, K.; Abdullah, W.; Guarner, J.; Goldsmith, C.S.; Chua, K.B.; Lam, S.K.; Tan, C.T.; et al. Nipah virus infection: Pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am. J. Pathol. 2002, 161, 2153–2167. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Whistler, T.; Rollin, P.E.; Ksiazek, T.G.; Rota, P.A.; Bellini, W.J.; Daszak, P.; Wong, K.T.; Shieh, W.J.; Zaki, S.R. Elucidation of Nipah virus morphogenesis and replication using ultrastructural and molecular approaches. Virus Res. 2003, 92, 89–98. [Google Scholar] [CrossRef]
- Geisbert, T.W.; Daddario-DiCaprio, K.M.; Hickey, A.C.; Smith, M.A.; Chan, Y.P.; Wang, L.F.; Mattapallil, J.J.; Geisbert, J.B.; Bossart, K.N.; Broder, C.C. Development of an acute and highly pathogenic nonhuman primate model of Nipah virus infection. PLoS ONE 2010, 5, e10690. [Google Scholar] [CrossRef]
- Ringel, M.; Heiner, A.; Behner, L.; Halwe, S.; Sauerhering, L.; Becker, N.; Dietzel, E.; Sawatsky, B.; Kolesnikova, L.; Maisner, A. Nipah virus induces two inclusion body populations: Identification of novel inclusions at the plasma membrane. PLoS Pathog. 2019, 15, e1007733. [Google Scholar] [CrossRef]
- Heinrich, B.S.; Cureton, D.K.; Rahmeh, A.A.; Whelan, S.P. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog. 2010, 6, e1000958. [Google Scholar] [CrossRef] [PubMed]
- Hoenen, T.; Shabman, R.S.; Groseth, A.; Herwig, A.; Weber, M.; Schudt, G.; Dolnik, O.; Basler, C.F.; Becker, S.; Feldmann, H. Inclusion bodies are a site of ebolavirus replication. J. Virol. 2012, 86, 11779–11788. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, J.; Civas, A.; Lama, Z.; Lagaudrière-Gesbert, C.; Blondel, D. Rabies Virus Infection Induces the Formation of Stress Granules Closely Connected to the Viral Factories. PLoS Pathog. 2016, 12, e1005942. [Google Scholar] [CrossRef] [PubMed]
- Nevers, Q.; Albertini, A.A.; Lagaudrière-Gesbert, C.; Gaudin, Y. Negri bodies and other virus membrane-less replication compartments. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118831. [Google Scholar] [CrossRef]
- Zhou, Y.; Su, J.M.; Samuel, C.E.; Ma, D. Measles Virus Forms Inclusion Bodies with Properties of Liquid Organelles. J. Virol. 2019, 93, e00948-19. [Google Scholar] [CrossRef]
- Dolnik, O.; Gerresheim, G.K.; Biedenkopf, N. New Perspectives on the Biogenesis of Viral Inclusion Bodies in Negative-Sense RNA Virus Infections. Cells 2021, 10, 1460. [Google Scholar] [CrossRef]
- Boggs, K.B.; Edmonds, K.; Cifuentes-Munoz, N.; El Najjar, F.; Ossandón, C.; Roe, M.; Wu, C.; Moncman, C.L.; Creamer, T.P.; Amarasinghe, G.K.; et al. Human Metapneumovirus Phosphoprotein Independently Drives Phase Separation and Recruits Nucleoprotein to Liquid-Like Bodies. mBio 2022, 13, e0109922. [Google Scholar] [CrossRef]
- Becker, N.; Heiner, A.; Maisner, A. Cytosolic Nipah Virus Inclusion Bodies Recruit Proteins Without Using Canonical Aggresome Pathways. Front. Virol. 2022, 1, 821004. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Pervolaraki, K.; Boulant, S. Differential Regulation of Type I and Type III Interferon Signaling. Int. J. Mol. Sci. 2019, 20, 1445. [Google Scholar] [CrossRef]
- Levy, D.E.; Marié, I.J.; Durbin, J.E. Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 2011, 1, 476–486. [Google Scholar] [CrossRef]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Young, D.F.; Didcock, L.; Goodbourn, S.; Randall, R.E. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 2000, 269, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Andrejeva, J.; Poole, E.; Young, D.F.; Goodbourn, S.; Randall, R.E. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 2002, 76, 11379–11386. [Google Scholar] [CrossRef] [PubMed]
- Young, D.F.; Andrejeva, J.; Li, X.; Inesta-Vaquera, F.; Dong, C.; Cowling, V.H.; Goodbourn, S.; Randall, R.E. Human IFIT1 Inhibits mRNA Translation of Rubulaviruses but Not Other Members of the Paramyxoviridae Family. J. Virol. 2016, 90, 9446–9456. [Google Scholar] [CrossRef] [PubMed]
- Pisanelli, G.; Pagnini, U.; Iovane, G.; García-Sastre, A. Type I and Type II Interferon Antagonism Strategies Used by Paramyxoviridae: Previous and New Discoveries, in Comparison. Viruses 2022, 14, 1107. [Google Scholar] [CrossRef]
- Pelissier, R.; Iampietro, M.; Horvat, B. Recent advances in the understanding of Nipah virus immunopathogenesis and anti-viral approaches. F1000Research 2019, 8, 1763. [Google Scholar] [CrossRef]
- Shaw, M.L.; Garcia-Sastre, A.; Palese, P.; Basler, C.F. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 2004, 78, 5633–5641. [Google Scholar] [CrossRef]
- Keiffer, T.R.; Ciancanelli, M.J.; Edwards, M.R.; Basler, C.F. Interactions of the Nipah Virus P, V, and W Proteins across the STAT Family of Transcription Factors. mSphere 2020, 5, e00449-20. [Google Scholar] [CrossRef]
- Shaw, M.L.; Cardenas, W.B.; Zamarin, D.; Palese, P.; Basler, C.F. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 2005, 79, 6078–6088. [Google Scholar] [CrossRef]
- Ciancanelli, M.J.; Volchkova, V.A.; Shaw, M.L.; Volchkov, V.E.; Basler, C.F. Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. J. Virol. 2009, 83, 7828–7841. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.J.; Parisien, J.P.; Horvath, C.M. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 2002, 76, 11476–11483. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, M.; Guillaume, V.; Sato, H.; Fujita, K.; Georges-Courbot, M.C.; Ikeda, F.; Omi, M.; Muto-Terao, Y.; Wild, T.F.; Kai, C. The nonstructural proteins of Nipah virus play a key role in pathogenicity in experimentally infected animals. PLoS ONE 2010, 5, e12709. [Google Scholar] [CrossRef] [PubMed]
- Satterfield, B.A.; Cross, R.W.; Fenton, K.A.; Agans, K.N.; Basler, C.F.; Geisbert, T.W.; Mire, C.E. The immunomodulating V and W proteins of Nipah virus determine disease course. Nat. Commun 2015, 6, 7483. [Google Scholar] [CrossRef]
- Satterfield, B.A.; Borisevich, V.; Foster, S.L.; Rodriguez, S.E.; Cross, R.W.; Fenton, K.A.; Agans, K.N.; Basler, C.F.; Geisbert, T.W.; Mire, C.E. Antagonism of STAT1 by Nipah virus P gene products modulates disease course but not lethal outcome in the ferret model. Sci. Rep. 2019, 9, 16710. [Google Scholar] [CrossRef]
- Jensen, M.R.; Yabukarski, F.; Communie, G.; Condamine, E.; Mas, C.; Volchkova, V.; Tarbouriech, N.; Bourhis, J.M.; Volchkov, V.; Blackledge, M.; et al. Structural Description of the Nipah Virus Phosphoprotein and Its Interaction with STAT1. Biophys. J. 2020, 118, 2470–2488. [Google Scholar] [CrossRef]
- Omi-Furutani, M.; Yoneda, M.; Fujita, K.; Ikeda, F.; Kai, C. Novel phosphoprotein-interacting region in Nipah virus nucleocapsid protein and its involvement in viral replication. J. Virol. 2010, 84, 9793–9799. [Google Scholar] [CrossRef]
- Moll, M.; Diederich, S.; Klenk, H.D.; Czub, M.; Maisner, A. Ubiquitous activation of the Nipah virus fusion protein does not require a basic amino acid at the cleavage site. J. Virol. 2004, 78, 9705–9712. [Google Scholar] [CrossRef]
- Diederich, S.; Thiel, L.; Maisner, A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology 2008, 375, 391–400. [Google Scholar] [CrossRef]
- Sauerhering, L.; Zickler, M.; Elvert, M.; Behner, L.; Matrosovich, T.; Erbar, S.; Matrosovich, M.; Maisner, A. Species-specific and individual differences in Nipah virus replication in porcine and human airway epithelial cells. J. Gen. Virol. 2016, 97, 1511–1519. [Google Scholar] [CrossRef]
- Elvert, M.; Sauerhering, L.; Maisner, A. Cytokine Induction in Nipah Virus-Infected Primary Human and Porcine Bronchial Epithelial Cells. J. Infect. Dis. 2020, 221, S395–S400. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Harcourt, B.H.; Mungall, B.A.; Tamin, A.; Peeples, M.E.; Bellini, W.J.; Rota, P.A. Determination of the henipavirus phosphoprotein gene mRNA editing frequencies and detection of the C, V and W proteins of Nipah virus in virus-infected cells. J. Gen. Virol. 2009, 90, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, W.; Duggan, E.S.; Booth, J.L.; Zou, M.H.; Metcalf, J.P. RIG-I and TLR3 are both required for maximum interferon induction by influenza virus in human lung alveolar epithelial cells. Virology 2015, 482, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Sauerhering, L.; Müller, H.; Behner, L.; Elvert, M.; Fehling, S.K.; Strecker, T.; Maisner, A. Variability of interferon-λ induction and antiviral activity in Nipah virus infected differentiated human bronchial epithelial cells of two human donors. J. Gen. Virol. 2017, 98, 2447–2453. [Google Scholar] [CrossRef]
- Rodriguez, J.J.; Cruz, C.D.; Horvath, C.M. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 2004, 78, 5358–5367. [Google Scholar] [CrossRef]
- Yabukarski, F.; Lawrence, P.; Tarbouriech, N.; Bourhis, J.M.; Delaforge, E.; Jensen, M.R.; Ruigrok, R.W.; Blackledge, M.; Volchkov, V.; Jamin, M. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat. Struct. Mol. Biol. 2014, 21, 754–759. [Google Scholar] [CrossRef]
- Lo, M.K.; Peeples, M.E.; Bellini, W.J.; Nichol, S.T.; Rota, P.A.; Spiropoulou, C.F. Distinct and overlapping roles of Nipah virus P gene products in modulating the human endothelial cell antiviral response. PLoS ONE 2012, 7, e47790. [Google Scholar] [CrossRef]
- Eaton, B.T.; Broder, C.C.; Middleton, D.; Wang, L.F. Hendra and Nipah viruses: Different and dangerous. Nat. Rev. Microbiol. 2006, 4, 23–35. [Google Scholar] [CrossRef]
- Seto, J.; Qiao, L.; Guenzel, C.A.; Xiao, S.; Shaw, M.L.; Hayot, F.; Sealfon, S.C. Novel Nipah virus immune-antagonism strategy revealed by experimental and computational study. J. Virol. 2010, 84, 10965–10973. [Google Scholar] [CrossRef]
- Sugai, A.; Sato, H.; Takayama, I.; Yoneda, M.; Kai, C. Nipah and Hendra Virus Nucleoproteins Inhibit Nuclear Accumulation of Signal Transducer and Activator of Transcription 1 (STAT1) and STAT2 by Interfering with Their Complex Formation. J. Virol. 2017, 91, e01136-17. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Sakai, M.; Shimojima, M.; Saijo, M.; Itoh, M.; Gotoh, B. Nonstructural protein of severe fever with thrombocytopenia syndrome phlebovirus targets STAT2 and not STAT1 to inhibit type I interferon-stimulated JAK-STAT signaling. Microbes Infect. 2018, 20, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.Q.; Shi, C.; Yao, T.; Feng, K.; Mo, Q.; Deng, F.; Wang, H.; Ning, Y.J. The Nonstructural Protein of Guertu Virus Disrupts Host Defenses by Blocking Antiviral Interferon Induction and Action. ACS Infect. Dis. 2020, 6, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Dolnik, O.; Kolesnikova, L.; Welsch, S.; Strecker, T.; Schudt, G.; Becker, S. Interaction with Tsg101 is necessary for the efficient transport and release of nucleocapsids in marburg virus-infected cells. PLoS Pathog. 2014, 10, e1004463. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.V.; Schmidt, K.M.; Deflubé, L.R.; Doğanay, S.; Banadyga, L.; Olejnik, J.; Hume, A.J.; Ryabchikova, E.; Ebihara, H.; Kedersha, N.; et al. Ebola Virus Does Not Induce Stress Granule Formation during Infection and Sequesters Stress Granule Proteins within Viral Inclusions. J. Virol. 2016, 90, 7268–7284. [Google Scholar] [CrossRef]
- Brandt, J.; Wendt, L.; Bodmer, B.S.; Mettenleiter, T.C.; Hoenen, T. The Cellular Protein CAD is Recruited into Ebola Virus Inclusion Bodies by the Nucleoprotein NP to Facilitate Genome Replication and Transcription. Cells 2020, 9, 1126. [Google Scholar] [CrossRef]
- Wendt, L.; Brandt, J.; Bodmer, B.S.; Reiche, S.; Schmidt, M.L.; Traeger, S.; Hoenen, T. The Ebola Virus Nucleoprotein Recruits the Nuclear RNA Export Factor NXF1 into Inclusion Bodies to Facilitate Viral Protein Expression. Cells 2020, 9, 187. [Google Scholar] [CrossRef]
- Ménager, P.; Roux, P.; Mégret, F.; Bourgeois, J.P.; Le Sourd, A.M.; Danckaert, A.; Lafage, M.; Préhaud, C.; Lafon, M. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri Bodies. PLoS Pathog. 2009, 5, e1000315. [Google Scholar] [CrossRef]
- Lifland, A.W.; Jung, J.; Alonas, E.; Zurla, C.; Crowe, J.E.; Santangelo, P.J. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 2012, 86, 8245–8258. [Google Scholar] [CrossRef]
- Jobe, F.; Simpson, J.; Hawes, P.; Guzman, E.; Bailey, D. Respiratory Syncytial Virus Sequesters NF-κB Subunit p65 to Cytoplasmic Inclusion Bodies To Inhibit Innate Immune Signaling. J. Virol. 2020, 94, e01380-20. [Google Scholar] [CrossRef]
- He, B.; Paterson, R.G.; Stock, N.; Durbin, J.E.; Durbin, R.K.; Goodbourn, S.; Randall, R.E.; Lamb, R.A. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: The multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 2002, 303, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Palosaari, H.; Parisien, J.P.; Rodriguez, J.J.; Ulane, C.M.; Horvath, C.M. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 2003, 77, 7635–7644. [Google Scholar] [CrossRef] [PubMed]
- Devaux, P.; von Messling, V.; Songsungthong, W.; Springfeld, C.; Cattaneo, R. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 2007, 360, 72–83. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becker, N.; Maisner, A. Nipah Virus Impairs Autocrine IFN Signaling by Sequestering STAT1 and STAT2 into Inclusion Bodies. Viruses 2023, 15, 554. https://doi.org/10.3390/v15020554
Becker N, Maisner A. Nipah Virus Impairs Autocrine IFN Signaling by Sequestering STAT1 and STAT2 into Inclusion Bodies. Viruses. 2023; 15(2):554. https://doi.org/10.3390/v15020554
Chicago/Turabian StyleBecker, Nico, and Andrea Maisner. 2023. "Nipah Virus Impairs Autocrine IFN Signaling by Sequestering STAT1 and STAT2 into Inclusion Bodies" Viruses 15, no. 2: 554. https://doi.org/10.3390/v15020554
APA StyleBecker, N., & Maisner, A. (2023). Nipah Virus Impairs Autocrine IFN Signaling by Sequestering STAT1 and STAT2 into Inclusion Bodies. Viruses, 15(2), 554. https://doi.org/10.3390/v15020554




