Dynamics of SARS-CoV-2 VOC Neutralization and Novel mAb Reveal Protection against Omicron
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of SARS-CoV-2 Variants from Clinical Samples
2.2. Sequencing
2.2.1. Virus Growth Characterization
2.2.2. Monoclonal Antibody Expression and Purification
2.2.3. Antibody Neutralization Assay
2.2.4. Data Analysis
3. Results
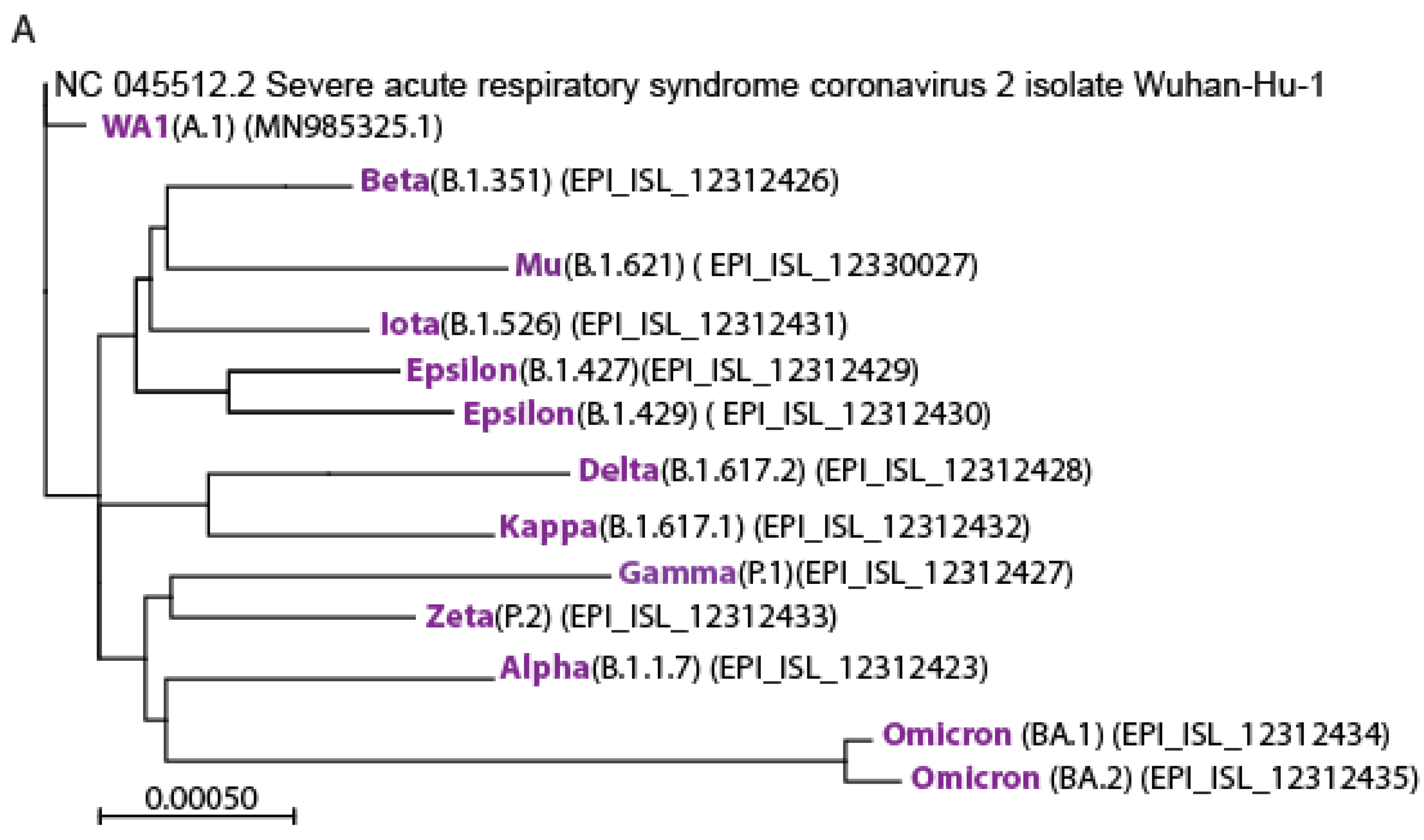
3.1. Isolation of the Variants of Concerns
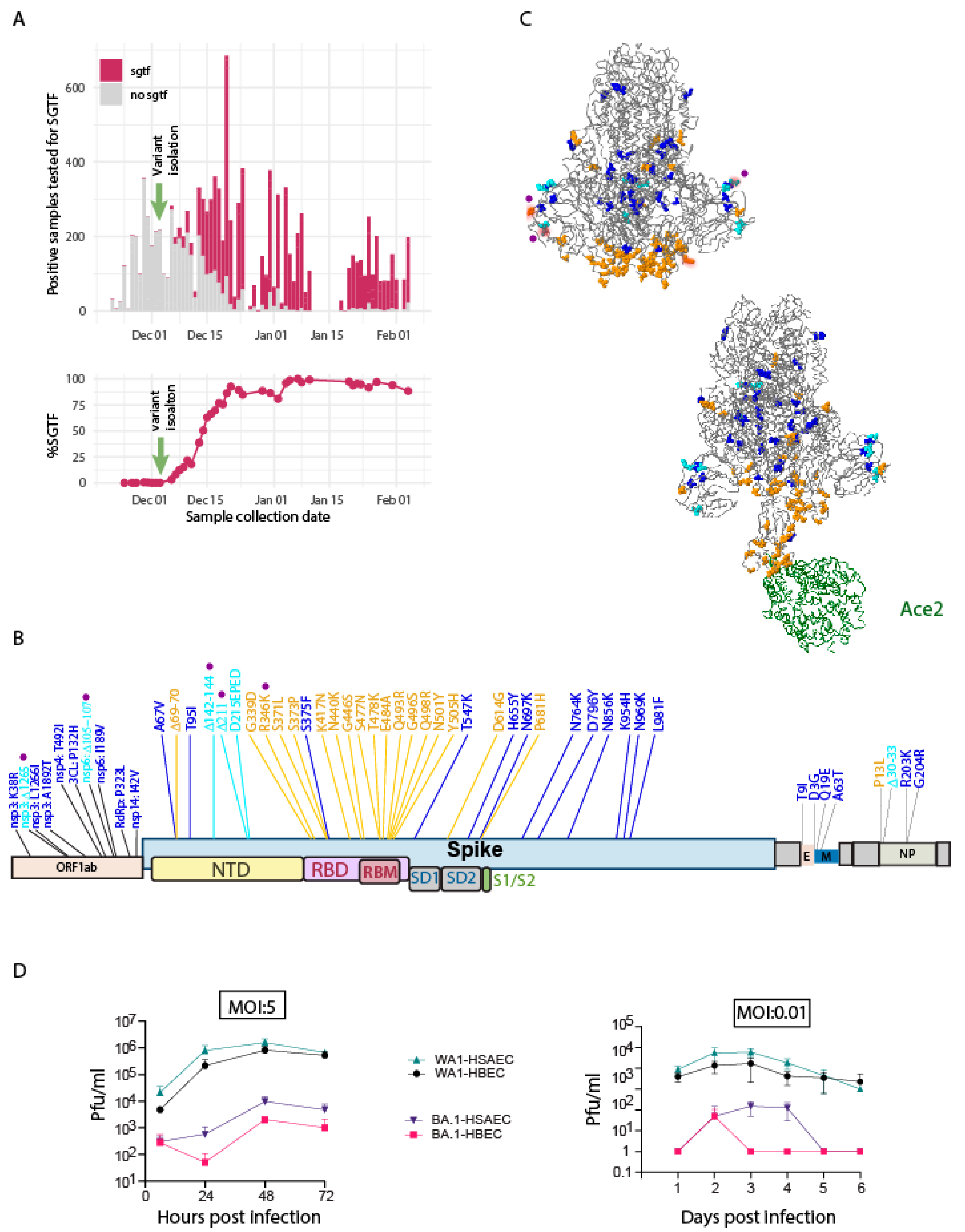
3.2. Identification and Characterization of Omicron BA.1
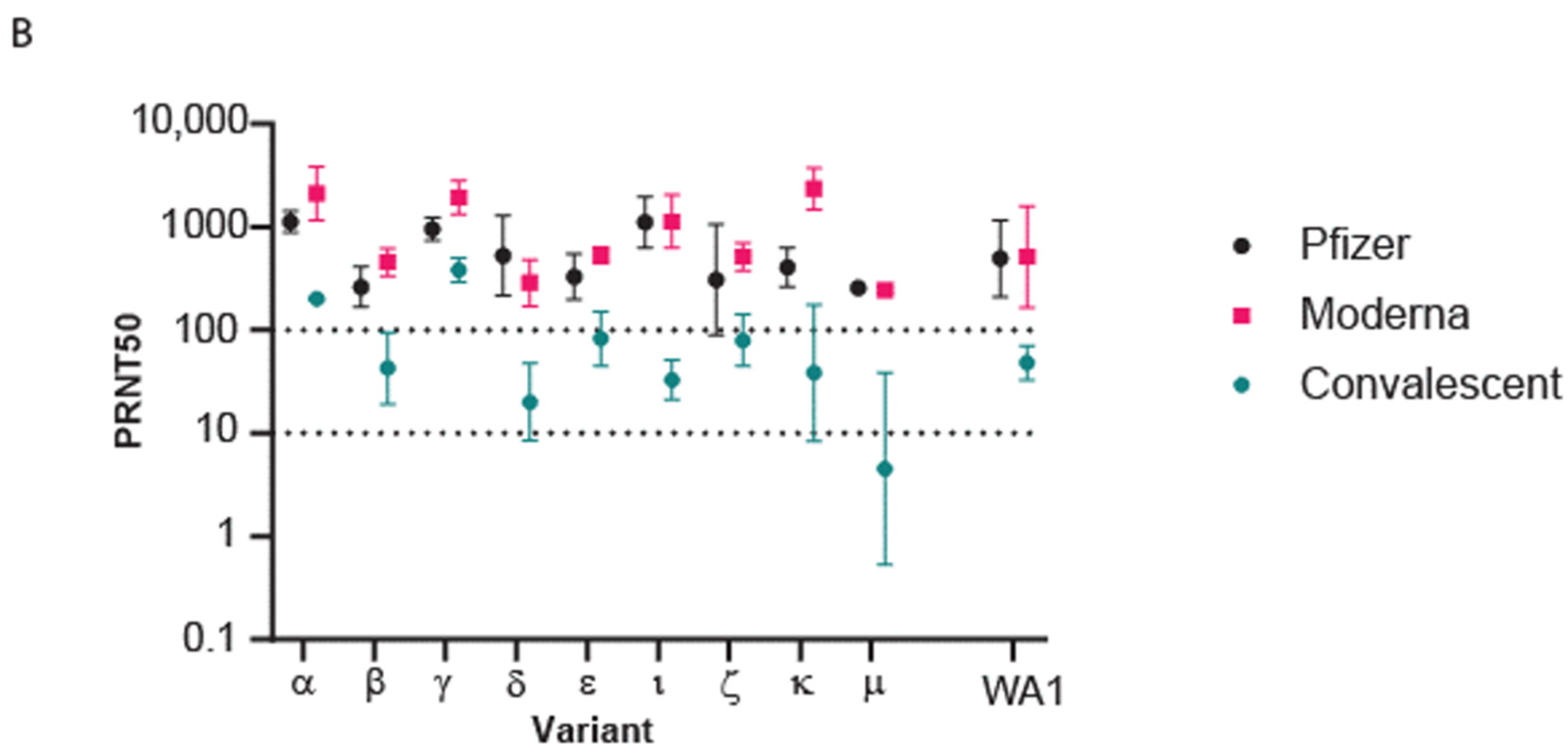
3.3. Population Immunity against Omicron BA.1
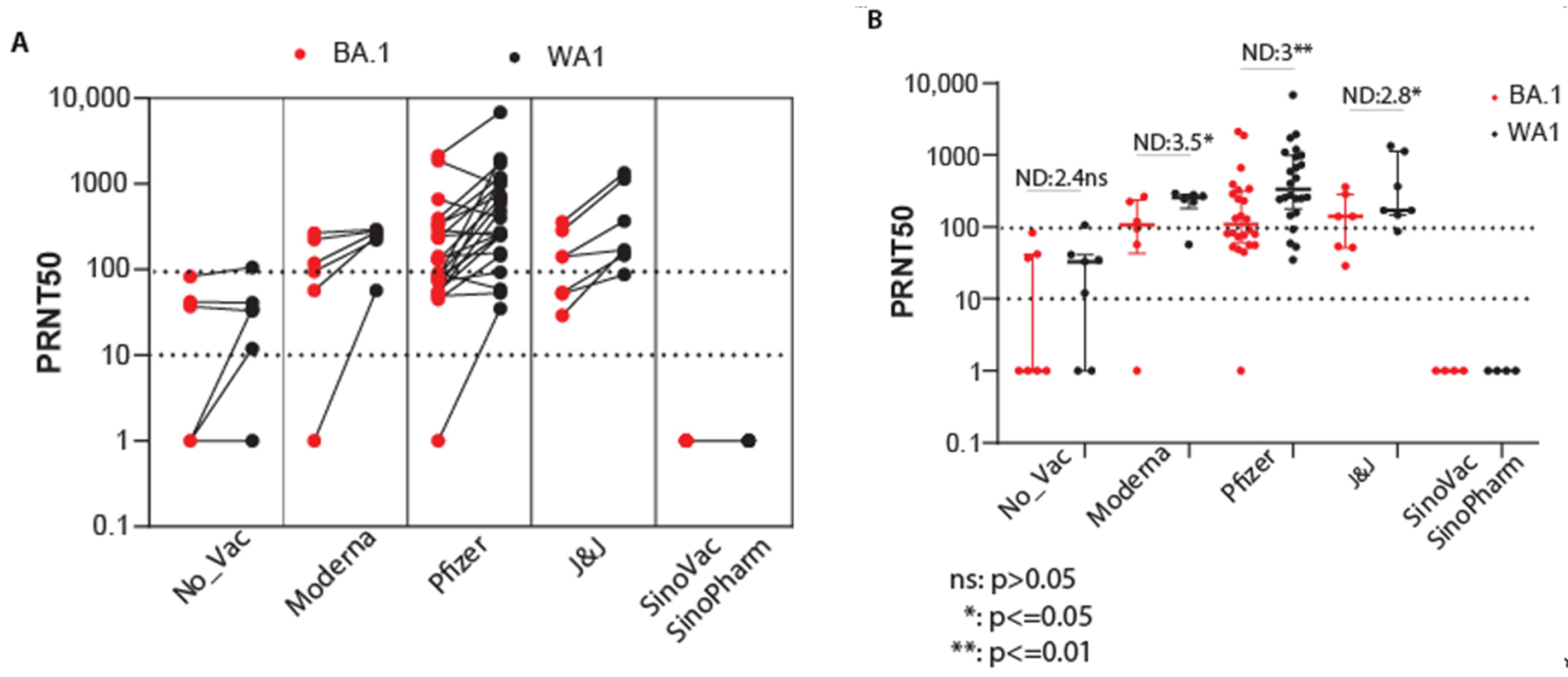
3.4. Efficacy of Vaccination against Omicron BA.1
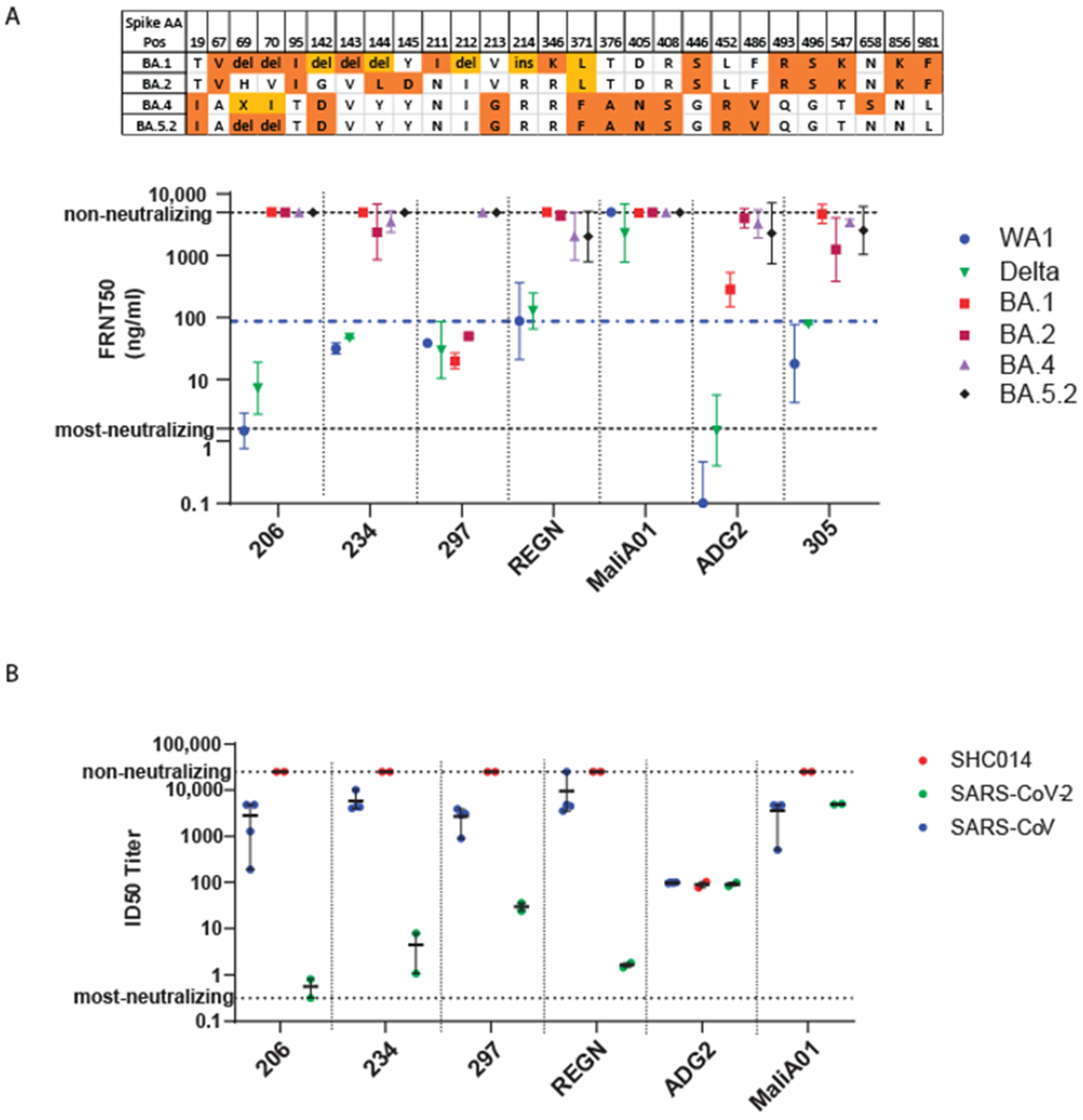
3.5. Efficacy of Monoclonal Antibody against Omicron
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; Plessis, L. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. medRxiv 2021. [Google Scholar] [CrossRef]
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 2022, 399, 924–944. [Google Scholar] [CrossRef]
- Bajema, K.L.; Dahl, R.M.; Evener, S.L.; Prill, M.M.; Rodriguez-Barradas, M.C.; Marconi, V.C.; Beenhouwer, D.O.; Holodniy, M.; Lucero-Obusan, C.; Brown, S.T.; et al. Comparative Effectiveness and Antibody Responses to Moderna and Pfizer-BioNTech COVID-19 Vaccines among Hospitalized Veterans-Five Veterans Affairs Medical Centers, United States, February 1-September 30, 2021, MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 1700–1705. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Goldberg, Y.; Mandel, M.; Bodenheimer, O.; Freedman, L.; Kalkstein, N.; Mizrahi, B.; Alroy-Preis, S.; Ash, N.; Milo, R.; et al. Protection of BNT162b2 Vaccine Booster against COVID-19 in Israel. N. Engl. J. Med. 2021, 385, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Cromer, D.; Steain, M.; Reynaldi, A.; Schlub, T.; Wheatley, A.; Juno, J.; Kent, S.; Triccas, J.; Khoury, D.; Davenport, M. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: A meta-analysis. Lancet Microbe 2022, 3, e52–e61. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. Reply. N. Engl. J. Med. 2021, 385, e92. [Google Scholar] [CrossRef] [PubMed]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; Denis, K.S.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383 e9. [Google Scholar] [CrossRef]
- Doria-Rose, N.A.; Shen, X.; Schmidt, S.D.; O’Dell, S.; McDanal, C.; Feng, W.; Tong, J.; Eaton, A.; Maglinao, M.; Tang, H.; et al. Booster of mRNA-1273 Strengthens SARS-CoV-2 Omicron Neutralization. medRxiv 2021. [Google Scholar] [CrossRef]
- Kissler, S.M.; Fauver, J.R.; Mack, C.; Tai, C.G.; Breban, M.I.; Watkins, A.E.; Samant, R.M.; Anderson, D.J.; Metti, J.; Khullar, G.; et al. Viral Dynamics of SARS-CoV-2 Variants in Vaccinated and Unvaccinated Persons. N. Engl. J. Med. 2021, 385, 2489–2491. [Google Scholar] [CrossRef] [PubMed]
- Altarawneh, H.N.; Chemaitelly, H.; Hasan, M.R.; Ayoub, H.H.; Qassim, S.; Coyle, P.; Yassine, H.M.; Al-Khatib, H.A.; Benslimane, F.M.; Al-Kanaani, Z.; et al. Protection against the Omicron Variant from Previous SARS-CoV-2 Infection. N. Engl. J. Med. 2022, 386, 1288–1290. [Google Scholar] [CrossRef] [PubMed]
- Lyngse, F.P.; Mortensen, L.H.; Denwood, M.J.; Christiansen, L.E.; Møller, C.H.; Skov, R.L.; Spiess, K.; Fomsgaard, A.; Lassaunière, M.M.; Rasmussen, M.; et al. SARS-CoV-2 Omicron VOC Transmission in Danish Households. medRxiv 2021. [Google Scholar] [CrossRef]
- Schmidt, F.; Schmidt, F.; Muecksch, F.; Weisblum, Y.; Da Silva, J.; Bednarski, E.; Cho, A.; Wang, Z.; Gaebler, C.; Caskey, M.; et al. Plasma Neutralization of the SARS-CoV-2 Omicron Variant. N. Engl. J. Med. 2022, 386, 599–601. [Google Scholar] [CrossRef] [PubMed]
- Hachmann, N.P.; Miller, J.; Collier, A.-r.Y.; Ventura, J.D.; Yu, J.; Rowe, M.; Bondzie, E.A.; Powers, O.; Surve, N.; Hall, K.; et al. Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022, 387, 86–88. [Google Scholar] [CrossRef]
- Khan, K.; Karim, F.; Ganga, Y.; Bernstein, M.; Jule, Z.; Reedoy, K.; Cele, S.; Lustig, G.; Amoako, D.; Wolter, N.; et al. Omicron sub-lineages BA.4/BA.5 escape BA.1 infection elicited neutralizing immunity. medRxiv 2022. [Google Scholar] [CrossRef]
- Khan, K.; Karim, F.; Cele, S.; Reedoy, K.; San, J.E.; Lustig, G.; Tegally, H.; Rosenberg, Y.; Bernstein, M.; Jule, Z.; et al. Omicron infection enhances Delta antibody immunity in vaccinated persons. Nature 2022, 607, 356–359. [Google Scholar] [CrossRef]
- Rossler, A.; Riepler, L.; Bante, D.; von Laer, D.; Kimpel, J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N. Engl. J. Med. 2022, 386, 698–700. [Google Scholar] [CrossRef]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.-H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef]
- Lu, L.; Mok, B.W.Y.; Chen, L.L.; Chan, J.M.C.; Tsang, O.T.Y.; Lam, B.H.S.; Chuang, V.W.M.; Chu, A.W.H.; Chan, W.M.; Ip, J.D.; et al. Neutralization of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant by Sera From BNT162b2 or CoronaVac Vaccine Recipients. Clin. Infect. Dis. 2021, 75, e822–e826. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.-W.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022, 602, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Denis, K.J.S.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef]
- Dejnirattisai, W. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet 2022, 399, 234–236. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Shaw, R.; Supasa, P.; Li, C.; Stuart, A.S.; Pollard, A.; Liu, X.; Lambe, T.; Crook, D.; Stuart, D.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.E.; Addetia, A.; Dang, H.V.; Stewart, C.; Brown, J.T.; Sharkey, W.K.; Sprouse, K.R.; Walls, A.C.; Mazzitelli, I.G.; Logue, J.K.; et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science 2022, 377, 890–894. [Google Scholar] [CrossRef]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.-M.; et al. COVID-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef]
- Cele, S.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, L.; Misasi, J.; Pegu, A.; Zhang, Y.; Harris, D.R.; Olia, A.S.; Yang, C.A.T.E.; Chen, M.; Choe, M.; et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science 2022, 376, eabn8897. [Google Scholar] [CrossRef] [PubMed]
- Hunt, A.C.; Case, J.B.; Park, Y.-J.; Cao, L.; Wu, K.; Walls, A.C.; Liu, Z.; Bowen, J.E.; Yeh, W.; Saini, S.; et al. Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Sci. Transl. Med. 2022, 14, eabn1252. [Google Scholar] [CrossRef]
- VanBlargan, L.A.; Errico, J.M.; Kafai, N.M.; Burgomaster, K.E.; Jethva, P.N.; Broeckel, R.M.; Meade-White, K.; Nelson, C.A.; Himansu, S.; Wang, D.; et al. Broadly neutralizing monoclonal antibodies protect against multiple tick-borne flaviviruses. J. Exp. Med. 2021, 218. [Google Scholar] [CrossRef]
- Rodda, L.B.; Netland, J.; Shehata, L.; Pruner, K.B.; Morawski, P.A.; Thouvenel, C.D.; Takehara, K.K.; Eggenberger, J.; Hemann, E.A.; Waterman, H.R.; et al. Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell 2021, 184, 169–183 e17. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Rathe, J.A.; Hemann, E.; Eggenberger, J.; Li, Z.; Knoll, M.; Stokes, C.; Hsiang, T.-Y.; Netland, J.; Takehara, K.; Pepper, M.; et al. SARS-CoV-2 Serologic Assays in Control and Unknown Populations Demonstrate the Necessity of Virus Neutralization Testing. J. Infect. Dis. 2021, 223, 1120–1131. [Google Scholar] [CrossRef]
- Addetia, A.; Lin, M.J.; Peddu, V.; Roychoudhury, P.; Jerome, K.R.; Greninger, A.L. Sensitive Recovery of Complete SARS-CoV-2 Genomes from Clinical Samples by Use of Swift Biosciences’ SARS-CoV-2 Multiplex Amplicon Sequencing Panel. J. Clin. Microbiol. 2020, 59. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Rodda, L.B.; Morawski, P.A.; Pruner, K.B.; Fahning, M.L.; Howard, C.A.; Franko, N.; Eggenberger, J.; Stokes, C.; Golez, I.; Hale, M.; et al. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. Cell 2022, 185, 1588–1601 e14. [Google Scholar] [CrossRef] [PubMed]
- SHAPIRO, S.S.; WILK, M. An analysis of variance test for normality (complete samples)†. Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Perchetti, G.A.; Zhu, H.; Mills, M.G.; Shrestha, L.; Wagner, C.; Bakhash, S.M.; Lin, M.J.; Xie, H.; Huang, M.-L.; Mathias, P.; et al. Specific allelic discrimination of N501Y and other SARS-CoV-2 mutations by ddPCR detects B.1.1.7 lineage in Washington State. J. Med. Virol. 2021, 93, 5931–5941. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef]
- Akova, M.; Unal, S. A randomized, double-blind, placebo-controlled phase III clinical trial to evaluate the efficacy and safety of SARS-CoV-2 vaccine (inactivated, Vero cell): A structured summary of a study protocol for a randomised controlled trial. Trials 2021, 22, 276. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Palacios, R.; Patiño, E.G.; de Oliveira Piorelli, R.; Conde, M.T.R.P.; Batista, A.P.; Zeng, G.; Xin, Q.; Kallas, E.G.; Flores, J.; Ockenhouse, C.F.; et al. Double-Blind, Randomized, Placebo-Controlled Phase III Clinical Trial to Evaluate the Efficacy and Safety of treating Healthcare Professionals with the Adsorbed COVID-19 (Inactivated) Vaccine Manufactured by Sinovac-PROFISCOV: A structured summary of a study protocol for a randomised controlled trial. Trials 2020, 21, 853. [Google Scholar] [PubMed]
- Thouvenel, C.D.; Fontana, M.F.; Netland, J.; Krishnamurty, A.T.; Takehara, K.K.; Chen, Y.; Singh, S.; Miura, K.; Keitany, G.J.; Lynch, E.M.; et al. Multimeric antibodies from antigen-specific human IgM+ memory B cells restrict Plasmodium parasites. J. Exp. Med. 2021, 218, e20200942. [Google Scholar] [CrossRef] [PubMed]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.P.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020, 369, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Yount, B.; Debbink, K.; Agnihothram, S.; Gralinski, L.; Plante, J.; Graham, R.; Scobey, T.; Ge, X.-Y.; Donaldson, E.; et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015, 21, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Rappazzo, C.G.; Tse, L.V.; Kaku, C.I.; Wrapp, D.; Sakharkar, M.; Huang, D.; Deveau, L.M.; Yockachonis, T.J.; Herbert, A.S.; Battles, M.B.; et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science 2021, 371, 823–829. [Google Scholar] [CrossRef]
- Yu, J.; Collier, A.-r.Y.; Rowe, M.; Mardas, F.; Ventura, J.D.; Wan, H.; Miller, J.; Powers, O.; Chung, B.; Siamatu, M.; et al. Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 Variants. N. Engl. J. Med. 2022, 386, 1579–1580. [Google Scholar] [CrossRef] [PubMed]
- Collie, S.; Nayager, J.; Bamford, L.; Bekker, L.-G.; Zylstra, M.; Gray, G. Effectiveness and Durability of the BNT162b2 Vaccine against Omicron Sublineages in South Africa. N. Engl. J. Med. 2022, 387, 1332–1333. [Google Scholar] [CrossRef] [PubMed]
- Hale, M.; Netland, J.; Chen, Y.; Thouvenel, C.D.; Smith, K.N.; Rich, L.M.; Vanderwall, E.R.; Miranda, M.C.; Eggenberger, J.; Hao, L.; et al. IgM antibodies derived from memory B cells are potent cross-variant neutralizers of SARS-CoV-2. J. Exp. Med. 2022, 219, e20220849. [Google Scholar] [CrossRef]
- Fan, C.; Cohen, A.A.; Park, M.; Hung, A.F.-H.; Keeffe, J.R.; Gnanapragasam, P.N.; Lee, Y.E.; Gao, H.; Kakutani, L.M.; Wu, Z.; et al. Neutralizing monoclonal antibodies elicited by mosaic RBD nanoparticles bind conserved sarbecovirus epitopes. Immunity 2022, 55, 2419–2435.e10. [Google Scholar] [CrossRef] [PubMed]
- Rappazzo, C.G.; Tse, L.V.; Kaku, C.I.; Wrapp, D.; Sakharkar, M.; Huang, D.; Deveau, L.M.; Yockachonis, T.J.; Herbert, A.S.; Battles, M.B.; et al. An Engineered Antibody with Broad Protective Efficacy in Murine Models of SARS and COVID-19. BioRxiv 2020. [Google Scholar] [CrossRef]
- Iketani, S.; Liu, L.; Guo, Y.; Liu, L.; Chan, J.F.-W.; Huang, Y.; Wang, M.; Luo, Y.; Yu, J.; Chu, H.; et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef] [PubMed]

| Vaccinated (Two-Dose) | Convalescent | |
|---|---|---|
| Characteristics | N = 15 | N = 13 |
| Sex | ||
| Male | 7 (46.7%) | 2 (15.4%) |
| Female | 8 (53.3%) | 11 (84.6%) |
| Age | ||
| Min–Max | 25–77 | 22–62 |
| Median (IQR) | 35.5 (30–47.8) | 45.5 (38–53) |
| Vaccine type | ||
| mRNA-1273 | 8 (53.3%) | |
| BNT162b2 | 7 (46.7%) | |
| None | 13 (100%) | |
| Time since | Recent vaccine dose | Symptom onset |
| Min-Max | 6–20 days | 157–366 days |
| Median (IQR) | 8 (8–12.5) days | 347 (289.5–360.5) days |
| Longitudinal Cohort | Inactivated Virus Vaccine Cohort | |
|---|---|---|
| Characteristics | N = 44 | N = 4 |
| Sex | ||
| Male | 25 (56.8%) | 3 (75%) |
| Female | 19 (43.2%) | 1 (25%) |
| Symptomatic | 41 (93.2%) | 1 (25%) |
| Age | ||
| Min–Max | 19–69 | 18–21 |
| Median (IQR) | 30 (25–46) | 19 (18–20.25) |
| Pangolin variant lineage | NA | |
| Alpha (B.1.1.7-like) | 10 (22.7%) | |
| Epsilon (B.1.427/429-like) | 1 (2.3%) | |
| B.1.2 | 9 (20.5%) | |
| Gamma (P.1-like) | 1 (2.3%) | |
| Not available | 23 (52.3) | |
| Vaccine type | ||
| mRNA-1273 | 6 (13.6%) | |
| BNT162b2 | 24 (54.5%) | |
| J&J-78436735 | 7 (15.9%) | |
| None-convalescent only | 7 (15.9%) | |
| SinoVac | 1 (25%) | |
| SinoPharm | 3 (75%) | |
| Time since symptom onset | NA | |
| Min–Max | 177–259 | |
| Median (IQR) | 185 (181–190) | |
| Time since recent vaccine dose | ||
| Min–Max | 93–177 days | 120–188 days |
| Median (IQR) | 130 (112–143) days | 165.5 (137.25–188) days |
| Two-Dose | Three-Dose | |
|---|---|---|
| Characteristics | N = 6 | N = 10 |
| Sex | ||
| Male | 2 (33.3%) | 2 (20%) |
| Female | 4 (66.7%) | 8 (80%) |
| Age | ||
| Min–Max | 31–61 | 22–63 |
| Median (IQR) | 50 (37–56.75) | 29 (24–36.75) |
| Vaccine type | ||
| BNT162b2, BNT162b2 | 6 (100%) | |
| mRNA-1273, mRNA-1273, mRNA-1273 | 8 (80%) | |
| BNT162b2, BNT162b2, mRNA-1273 | 2 (20%) | |
| Time since recent vaccine dose | ||
| Min–Max | 152–161 days | 19–50 days |
| Median (IQR) | 153.5 (153–157) | 32 (27–34.5) days |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, L.; Hsiang, T.-Y.; Dalmat, R.R.; Ireton, R.; Morton, J.F.; Stokes, C.; Netland, J.; Hale, M.; Thouvenel, C.; Wald, A.; et al. Dynamics of SARS-CoV-2 VOC Neutralization and Novel mAb Reveal Protection against Omicron. Viruses 2023, 15, 530. https://doi.org/10.3390/v15020530
Hao L, Hsiang T-Y, Dalmat RR, Ireton R, Morton JF, Stokes C, Netland J, Hale M, Thouvenel C, Wald A, et al. Dynamics of SARS-CoV-2 VOC Neutralization and Novel mAb Reveal Protection against Omicron. Viruses. 2023; 15(2):530. https://doi.org/10.3390/v15020530
Chicago/Turabian StyleHao, Linhui, Tien-Ying Hsiang, Ronit R. Dalmat, Renee Ireton, Jennifer F. Morton, Caleb Stokes, Jason Netland, Malika Hale, Chris Thouvenel, Anna Wald, and et al. 2023. "Dynamics of SARS-CoV-2 VOC Neutralization and Novel mAb Reveal Protection against Omicron" Viruses 15, no. 2: 530. https://doi.org/10.3390/v15020530
APA StyleHao, L., Hsiang, T.-Y., Dalmat, R. R., Ireton, R., Morton, J. F., Stokes, C., Netland, J., Hale, M., Thouvenel, C., Wald, A., Franko, N. M., Huden, K., Chu, H. Y., Sigal, A., Greninger, A. L., Tilles, S., Barrett, L. K., Van Voorhis, W. C., Munt, J., ... Gale, M., Jr. (2023). Dynamics of SARS-CoV-2 VOC Neutralization and Novel mAb Reveal Protection against Omicron. Viruses, 15(2), 530. https://doi.org/10.3390/v15020530






