Abstract
Cyanobacterial expansion is harmful to the environment, the ecology of Lake Baikal and the economy of nearby regions and can be dangerous to people and animals. Since 2011, the process of colonisation of the lake with potentially toxic cyanobacteria belonging to the genus Tychonema has continued. An understanding of the mechanism of successful expansion of Tychonema requires scrutiny of biological and genomic features. Tychonema sp. BBK16 was isolated from the coastal zone of Lake Baikal. The morphology of BBK16 biofilm was studied with light, scanning electron and confocal microscopy. The biofilm is based on filaments of cyanobacteria, which are intertwined like felt; there are also dense fascicles of rope-like twisted filaments that impart heterogeneity to the surface of the biofilm. Genome sequencing, intergenomic comparisons and phylogenetic analyses indicated that Tychonema sp. BBK16 represent a new species related to planktic cyanobacterium Tychonema bourrellyi, isolated from Alpine lentic freshwater. Genome investigation revealed the genes possibly responsible for the mixotrophic lifestyle. The presence of CRISPR-Cas and restriction modification defence mechanisms allowed to suggest the existence of phages infecting Tychonema sp. BBK16. Analysis of CRISPR spacers and prophage-derived regions allowed to suggest related cyanophages. Genomic analysis supported the assumption that mobile elements and horizontal transfer participate in shaping the Tychonema sp. BBK16 genome. The findings of the current research suggest that the aptitude of Tychonema sp. BBK16 for biofilm formation and, possibly, its mixotrophic lifestyle provide adaptation advantages that lead to the successful expansion of this cyanobacterium in the Baikal’s conditions of freshwater lake environments.
1. Introduction
The global expansion of harmful cyanobacterial blooms in eutrophic waters constitutes a serious threat to freshwater ecology and public health [1,2]. The process of colonisation of Lake Baikal by cyanobacteria, initially named Phormidium spp., was first noticed in 2011 [3]. Further studies showed that this cyanobacterium belonged to the genus Tychonema (family Microcoleaceae, order Oscillatoriales), which was new to Baikal. The potentially toxic genus Tychonema proliferates in the benthos of Lake Baikal and now prevails in the biofilm ulcers of the Baikal sponge and other biofouling. The occupation of new ecological niches by these cyanobacteria raises questions about the biological and genetic mechanisms of such an evolutionary success.
Biofilm formation is an important evolutionary step in the colonisation of new ecological niches [4,5]. Cyanobacterial mats, which are laminated biofilms, are the oldest communities on Earth, dating back 3.5 billion years [6]. Although cyanobacterial mats formed by members of the genus Tychonema are rarely mentioned, it is known that they can cause problems, leading to animal poisoning with anatoxin, as in Lake Tegel and Reservoir Mandichosee, Germany, or the mass death of endemic sponges, as in Lake Baikal [3,7,8].
Bolshiye Koty is a small settlement on the coast of Lake Baikal, located within the territory of the Pribaikalsky National Park. It is fairly isolated, which facilitates nature preservation. There is no road, and the village can only be reached by water or on foot. The settlement is located on a popular tourist route—the Great Baikal Trail. Since 2009, the coastal zone of Bolshiye Koty has been included in interdisciplinary ecological studies of the splash zone of Lake Baikal in order to study the natural course of hydrobiological processes in this area, including the composition of cyanobacteria [9]. The appearance of biofilms uncharacteristic of Lake Baikal on the surface of stones and wooden substrates in this area has been noted since 2010 [3].
Cyanobacteria are characterised by the ability to carry out oxygenic photosynthesis [10], but the capability of mixotrophy has also been reported [11,12]. Mixotrophy has traditionally been defined as an alternative form of carbon uptake but may also involve the acquisition of molecules containing nitrogen, phosphorus, trace minerals, vitamins and high-energy compounds such as ATP. Sensu lato mixotrophy can be considered a combination of different nutritional pathways in one organism [12,13]. The capacity for mixotrophy provides a significant competitive advantage, enabling mixotrophic bacteria to dominate broader aquatic environments since they can utilise more resources than photoautotrophic bacteria [14].
It is important that Lake Baikal is characterised by its oligotrophy [15]. However, the concentration of total organic carbon (Corg) in bottom water in the coastal zone of Lake Baikal is 1.5–2.0 mg C/L, and the concentration of total organic nitrogen is 0.33–0.47 mg/L [16], exceeding such values in the oligotrophic pelagial zone of the lake by two times. In biofilms, the content of Corg is much higher due to the activity of photoautotrophic organisms (cyanobacteria, algae and macrophytes) and chemoautotrophic bacteria capable of synthesising organic carbon. Thus, in biofilms taken near the Bolshiye Koty settlement, the Corg was found to be 35 mg/L, total phosphorus to be 0.022 mg/L and total nitrogen 0.154 mg/L. [16]. In the bottom water at the same station, the concentration of total carbon was 5.6 mg/L, total phosphorus 0.007 mg/L and total nitrogen 0.142 mg/L. These conditions may contribute to the evolutionary success of mixotrophic cyanobacteria.
It is possible to suggest that Baikal cyanobacteria might also use the advantages of mixotrophy to expand their ecological niche. The ability to uptake organic compounds is related to the presence of transport proteins, and genes genomic analysis of various cyanobacteria, including marine cyanobacteria Prochlorococcus and Synechococcus, has demonstrated the presence of genes encoding amino acid, phosphate and sugar transporters [12,17,18,19]. It would be interesting to check for the presence of mixotrophy-related genes in the genomes of Baikal cyanobacteria and related ones.
The environmental adaptation of bacteria, in particular, is affected by bacteriophages (aka phages) [20], viruses that infect bacteria. Cyanobacterial phages (cyanophages) contribute to the evolution of cyanobacteria and can be considered a reservoir of genes important for the ecological adaptation of cyanobacteria [21]. These genes can alter the physiology of an infected cyanobacterium, increasing the production of progeny phage [22]. At the beginning of 2022, the NCBI Genome database contained about 500 sequences of complete genomes of cyanophages, most of which were isolated from marine Synechococcus and Prochlorococcus (order Synechococcales), but there are only a few examples of isolated and published cyanophages infecting Oscillatoriales cyanobacteria [23,24,25,26,27,28,29], and no Tychonema phages are described to date. However, some information about phage–host interactions can be recovered using the analysis of genomic CRISPR arrays [30,31,32]. Moreover, temperate phages that insert their genome into the host chromosome can leave traces of phage infection [33,34,35,36].
The purpose of this study was to analyse the biofilm formed by the benthic Tychonema sp. strain BBK16 isolated in the coastal zone of Lake Baikal and to characterise the general genomics of the cyanobacterium, including the analysis of CRISPR spacers and prophage-derived regions. Another intention of this research was to investigate the possibility of the mixotrophy of Tychonema sp. BBK16, using bioinformatic analysis, to reveal the mechanisms that can assist the expansion of Tychonema in the lake.
2. Materials and Methods
2.1. Sampling Location
Biofilm samples were obtained in 2015–2016 from the underwater part of the wooden pier located in the area of the Scientific Research Station “Bolshiye Koty” of the Limnological Institute (Bolshiye Koty Settlement, 51.883333° N, 105.05° E) (Figure 1). Native biofilm macro photography was carried out using a Pentax WG-3 GPS camera (Ricoh Imaging, Tokyo, Japan).
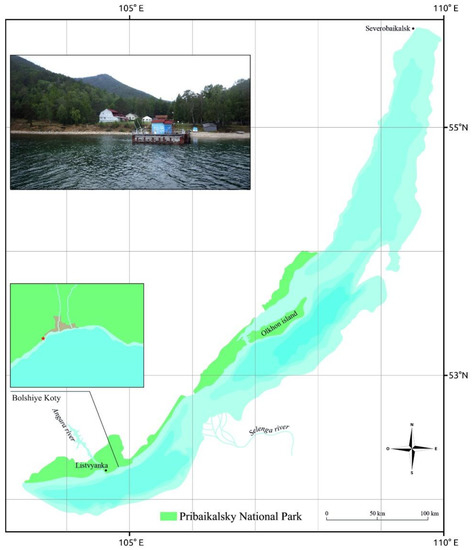
Figure 1.
Map of Lake Baikal, Pribaikalsky National Park, and Bolshiye Koty, from where samples were obtained.
2.2. Cultivation and Biofilm Visual Analysis
During cultivation, biofilm fragments were washed with sterile Z-8 mineral medium [37], crushed into smaller pieces and placed on agar plates with the same medium in an incubator. The cultivation conditions were as follows: illumination 1200 lux, the light mode consisted of alternating night and day periods 16:8, temperature 11–12 °C. Next, individual trichomes were sterilely excised from agar, suspended in a liquid medium and cultured again to obtain a unialgal culture.
Microphotographs of Tychonema sp. BBK16 were obtained using an Axio Imager light microscope (Carl Zeiss, Jena, Germany). The structure of the biofilm surface was visualised using scanning electron microscopy (SEM). First, thin sterile glasses were introduced into the liquid medium with cyanobacteria, which, after fouling, were fixed with 2% formaldehyde and dehydrated in an ethanol concentration gradient. Subsequently, the glasses with Tychonema biofouling were dried at 40 °C, coated in gold using a Balzers SCD 004 sputter-coater (Bal-Tec AG, Balzers, Liechtenstein) and examined using SEM Quanta 200 (FEI Co., Hillsboro, OR, USA). Scanning laser confocal microscopy studies were carried out on an LSM 710 microscope (Carl Zeiss) equipped with a helium–neon laser (561 nm), which causes the autofluorescence of cyanobacterial pigments. Biofilm images were obtained using ZEN 2010 software (Carl Zeiss), and 3D reconstruction was achieved with Imaris software (Bitplane Scientific Software, Zürich, Switzerland).
2.3. DNA Extraction and Sequencing
The total DNA was extracted by enzymatic lysis using lysozyme (Roche, Basel, Switzerland), proteinase K (Thermo Scientific, Waltham, MA, USA) and sodium dodecyl sulphate (VWR Life Science, Radnor, PA, USA), followed by phenol and chloroform (Medigen, Novosibirsk, Russia) extraction [38]. The NEBNextUltra DNA library prep kit for Illumina (New England BioLabs, Ipswich, MA, USA) was used for DNA library construction. DNA samples were sequenced to generate 300 bp paired-end reads using the Illumina MiSeq platform. De novo genome assembly was performed using SPAdes 3.12 [39], binning was conducted using MaxBin 2.0 [40], and the assembly was manually curated using a BLASTN [41] search with default settings against the NCBI Bacterial GenBank Database [42]. The resulting genome was deposited in GenBank (Accession # JAKJHX000000000).
2.4. Genome Annotation and Prediction of Gene Functions
The assembled genome was annotated using the Prokaryotic Genome Annotation Pipeline (PGAP) [43] with default settings. Additionally, annotations of all genomic regions shown in this study were manually curated. Manual curation included checking the positions of open reading frames (ORFs) and functional assignment. Checking the ORF positions was conducted using Geneious Prime 2022.0.1 tools (Biomatters, Inc., Auckland, New Zealand) [44] and Gimmer 3.0.2 [45]. Gene functional assignment was performed using the BLAST search with nr/nt database and HMM-HMM motif comparison. The parameters of BLAST search were: BLASTP algorithm, E-value < 1 × 10−5 and other parameters were default. The HMM-HMM motif comparison was performed using the HHpred server [46] and PDB, SCOPe, CATH and UniProt-SwissProt-viral databases. Pfam domains were identified using pfam_scan 1.6 tool [47] applying default settings. Clusters of orthologous groups of proteins (COGs) were identified using the eggNOG-mapper 2 server [48] applying the “Genomic” settings.
2.5. Average Nucleotide Identity Calculations and Phylogenetic Analysis
Cyanobacterial genome and gene sequences were downloaded from the NCBI GenBank. The relevance of species names was checked in Algaebase (https://www.algaebase.org, accessed on 10 January 2023). The average nucleotide identity (ANI) matrix was calculated using ANI/AAI-Matrix Genome-based distance matrix calculator (Kostas lab, Atlanta, GA, USA) and Bio-NJ clustering [49]. Alignments of 16S rDNA sequences and protein sequences were performed with MAFFT 7.48 [50] with default settings and using the L-INS-i algorithm. 16S rDNA and single protein phylogenetic trees were constructed using RAxML-NG 1.1.0 [51] and the raxmlGUI 2.0.10 graphic interface [52] with (--tree rand{10} --bs-trees 1000) settings and applying the best protein model found with ModelTest-NG 0.1.7 [53]. The robustness of the RAxML-NG 1.1.0 trees was assessed using bootstrapping and calculations of transfer bootstrap estimation (TBE) support [54].
Alignment of concatenated sequences of orthologous proteins was obtained with PhyloPhlAn 3.0 applying (-d phylophlan-diversity medium -f supermatrix_aa.cfg) settings. The tree was constructed using RAxML-NG 1.1.0 with (-tree rand{1}-bs-trees 100) settings. Other details of the phylogenetic analysis using concatenated alignments were the same as described above.
2.6. Identification of CRISPR Loci and Prophage-Derived Regions
CRISPR loci were identified with MinCED [55] using “-gffFull” settings. Spacer sequences were extracted with MinCED using the “-spacers” settings. Prophage-derived regions were found using the PHASTER server [56] applying default settings. Similarity of genes of supposedly prophage origin was estimated using built-in PHASTER utilities.
3. Results
3.1. Biofilm Characterisation
The fouling that was the source of strain isolation was a dense, leathery, olive-green biofilm covering the underwater surface of the wooden pier (Figure 2a). SEM visualisation of the structure of a biofilm formed by Tychonema sp. BBK16 in a liquid medium showed that it was based on cyanobacterial filaments intertwined like felt and the presence of dense fascicles of rope-like twisted filaments that imparted heterogeneity to the biofilm surface, as observed in nature (Figure 2b). Light microscopy showed that the trichomes of the strain were straight and that they were enclosed in thin, transparent polysaccharide sheaths, which strengthened the structure of the biofilm (Figure 2c). Since the sheaths were thin, the cells were clearly visible, even in SEM (Figure 2b). The autofluorescence of chlorophyll and the phycobiliproteins of cyanobacteria enabled visualisation of the 3D structure of the biofilm using laser scanning microscopy without additional staining (Figure 2d). Confocal microscopy showed that the biofilm consisted of randomly intertwined threads and had a multilayer structure.
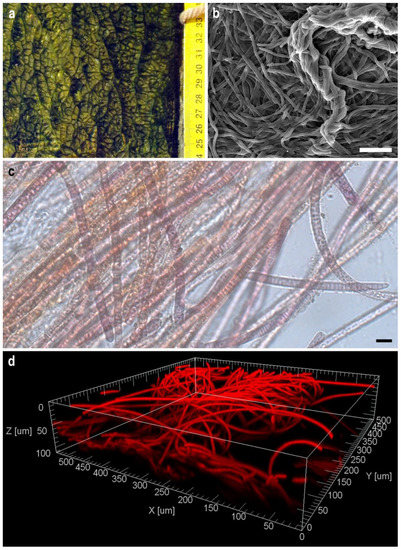
Figure 2.
Macro photography of a natural biofilm from the fouling of the pier, the source of isolation of the strain Tychonema sp. BBK16 (a). Morphology of the biofilm of Tychonema sp. BBK16 formed in culture: (b) scanning electron microscopy. Scale bar = 50 µm; (c) light microscopy. Scale bar =10 µm; (d) confocal microscopy.
3.2. General Genomic Features and Intergenomic Comparisons
The total size of the Tychonema sp. BBK16 draft genome is 5,267,730 nucleotides (nt), which is close to the size of the genome of Tychonema bourrellyi FEM_GT703 (5,081,867 nt) isolated from a freshwater sample taken from Lake Garda [57] and less than the genome size of other Tychonema genomes contained in the NCBI GenBank Database (Table 1). The GC-content of 44.3% is close to the GC-content of other Tychonema genomes and is slightly closer to the GC-content of T. bourrellyi FEM_GT703 (44.7%) (Table 1). All Tychonema genomes were deposited as draft sequences.

Table 1.
General features of Tychonema genomes.
The average nucleotide identity (ANI) calculations were performed using all available genomes of Tychonema and other related cyanobacterial strains found with BLAST search using BBK16 rDNA sequences. The maximum ANI value of about 91% was found for the Tychonema sp. BBK16–T. bourrellyi FEM_GT703 pair (Supplementary Figure S1). This value is lower than the 95–96% ANI cutoff, the standard most frequently used for prokaryotic species demarcation using complete or nearly complete genomes [58,59,60]. The next highest ANI values of about 83% corresponded to other Tychonema strains and Microcoleus sp. LEGE 07076. This value is higher than the estimated prokaryotic mean demarcation boundaries of the genus, which is about 74% [61].
3.3. Phylogenetic Analyses
Phylogenetic analysis was performed using 16S rDNA and concatenated alignments of orthologous proteins. The 16S rDNA representative sequences were selected using the most similar sequences found with the BLAST search and NCBI nt databases using the sequences of Oscillatoriales species mentioned in [62] and sequences of more distantly related organisms used in [63]. The 16S rDNA phylogenetic tree (Figure 3) places all the Tychonema strains and several Microcoleus, Phormidium and Oscillatoria strains into a clade with a TBE support of 0.81. Phylogenetic analysis using the PhyloPhlAn pipeline, which employs 400 most conserved proteins, can show better resolution and higher bootstrap support. The PhyloPhlAn phylogenetic tree (Figure 4) using the sequences representing most of the available Tychonema/Microcoleus/Phormidium full genomic sequences and other cyanobacterial groups placed Tychonema sp. BBK16 and T. bourrellyi FEM_GT703 into a distinct clade and depicted a group of Tychonema strains as a paraphyletic group.
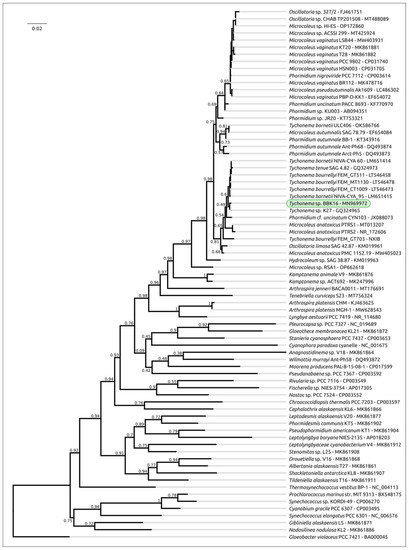
Figure 3.
Best-scoring ML phylogenetic tree constructed with 75 nucleotide sequences of 16S rDNA. The NCBI accession number is shown to the right of the organism’s name. Gloeobacter violaceus PCC 7421 was used as an outgroup. The numbers near the tree branches indicate the TBE values. The total number of bootstrap trees was 1000. The scale bar shows 0.02 estimated substitutions per site.
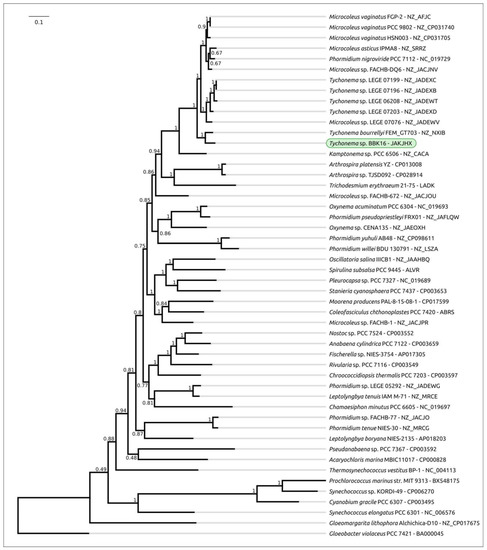
Figure 4.
Best-scoring ML phylogenetic tree constructed with 50 concatenated amino acid sequences of conserved proteins found with PhyloPhlAn. The NCBI accession is shown to the right of the organism’s name. Gloeobacter violaceus PCC 7421 was used as an outgroup. The numbers near the tree branches indicate the TBE values. The total number of bootstrap trees was 100. The scale bar shows 0.1 estimated substitutions per site.
3.4. General Proteome Features
The Tychonema strains deposited in GenBank clearly exhibit diversity in genome size and the number of predicted proteins (Table 1). There are 4708 protein-coding sequences predicted in the BBK16 genome, which is close to the number of proteins encoded in the FEM_GT703 genome (4629) and less than in other Tychonema genomes. Comparison of the distribution of clusters of orthologous groups of proteins (COGs) of Tychonema and several other cyanobacterial strains indicated that the number of orthologous groups assigned to categories V (defence mechanisms), N (cell motility), L (replication, recombination and repair), K (transcription) and to several other categories in Tychonema sp. BBK16 was similar to that in T. bourrellyi FEM_GT703, seemingly indicating the relatedness of these species (Figure 5).
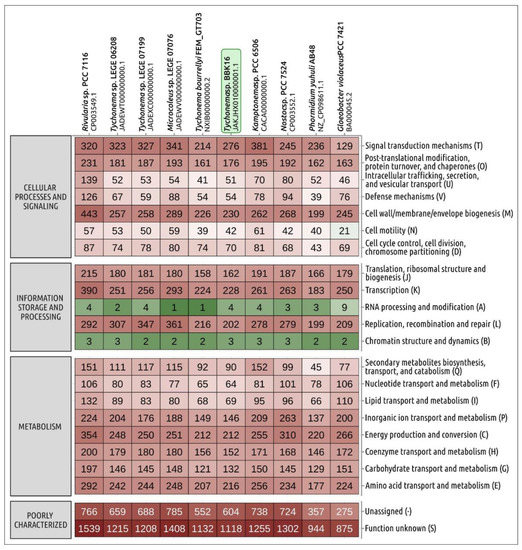
Figure 5.
Heatmap showing the distribution of clusters of orthologous groups of proteins of Tychonema sp. BBK16 and other cyanobacteria obtained using eggNOG-mapper. The numbers displayed in cells indicate the number of proteins belonging to the orthologous groups.
Transposases are of particular importance for bacteria. In freshwater, cyanobacteria provide the basis for rapid adaptation and survival in harsh freshwater environments [64]. The Tychonema sp. BBK16 genome content analysis also indicated the presence of 29 transposase-encoding genes, which is somewhat lower than in genomes of other analysed Tychonema strains (32–42 genes) and noticeably lower than in Microcoleus sp. LEGE 07076 (52 genes) (Table 2). Interestingly, a similar situation is observed for restriction modification enzymes, which are important for protecting the cell from phages [65,66]. The Tychonema sp. BBK16 genome contains 62 genes of restriction modification enzymes, while other analysed Tychonema strains have 81–110 genes, and Microcoleus sp. LEGE 07076 has 110 such genes (Table 2).

Table 2.
Number of genes of transposes and restriction modification enzymes in cyanobacterial genomes predicted using eggNOG-mapper 2.
3.5. Mixotrophy-Associated Proteins
The ability to assimilate organic nutrients detected in both marine and freshwater cyanobacteria is related to transporters needed for the uptake of organic compounds [12,67]. Pfam domain search predicted that the genome of Tychonema sp. BBK16 contains four genes to encode the proteins containing sugar-transporter-like domains. Experimentally, the uptake of sugars in cyanobacteria has been shown to be associated with the presence of GlcH permease importer, a high-affinity glucose transporter (called “Pro1404” in Prochlorococcus marinus SS120, where it was first studied) [68,69,70] and GlcP permease [71,72].
The BLAST search conducted using the Pro1404 amino acid sequence, NCBI nr database and 50 cyanobacterial genomes used in phylogenetic analysis (Figure 4) indicated the presence of genes encoding GlcH in all 50 genomes mentioned above and in numerous other cyanobacteria (E-value < 1 × 10−5). Some of the 50 genomes contained two or three copies of the glcH-like gene. These results are consistent with a result published earlier using another dataset [12]. The genomes of all the analysed strains of Tychonema, Microcoleus and Kamptonema carry a single copy of the glcH-like gene.
Phylogenetic analysis using GlcH-like amino cyanobacterial acid and sequences of the closest homologues from other bacterial taxa indicate the complex evolutionary history of this protein (Supplementary Figure S2). The large clade containing Pro1404 also includes sequences from noncyanobacterial taxa, and some strains appear to carry genes resulting from duplication events.
Glucose:H+ symporters GlcP were searched in a similar way but using a sequence of experimentally studied permease from Nostoc punctiforme [72,73]. In contrast, genes encoding GlcP-like proteins have only been found in 14 of the 50 genomes mentioned above. Interestingly, no genomes of Tychonema, with the exception of BBK16, carry the glcP gene. Phylogenetic analysis indicated, with high bootstrap support (TBE 0.93), a close relatedness of Tychonema sp. BBK16, Microcoleus sp. FACHB-1, Nostoc punctiforme PCC 73102 and Pleurocapsa sp. CCALA 161 proteins (Figure 6). The phylogeny of GlcP may indicate multiple events of horizontal transfer accompanying the evolution of GlcP, which is consistent with a result published earlier [72].
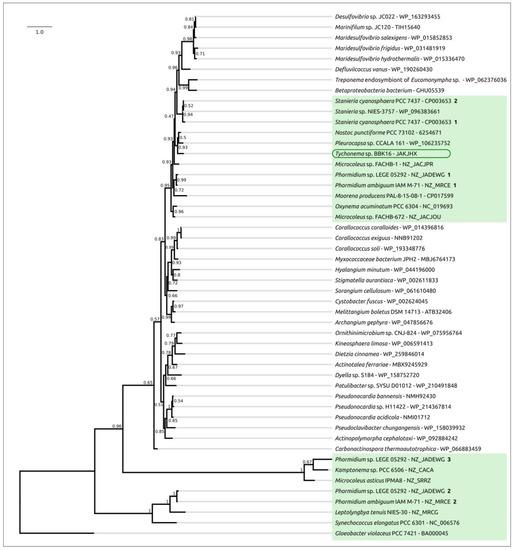
Figure 6.
Best-scoring ML phylogenetic tree constructed with 50 amino acid sequences of GlcP protein. The NCBI accession number is shown to the right of the organism’s name. Gloeobacter violaceus PCC 7421 was used as an outgroup. The numbers near the tree branches indicate the TBE values. Bold numbers to the right of NCBI accession indicate the multiple instances of encoded proteins. The total number of bootstrap trees was 1000. The scale bar shows 1.0 estimated substitutions per site.
The genome of Tychonema sp. BBK16 encodes multiple amino acid ABC transporters. These are proteins that can participate in the import of amino acids, which can be a source of carbon and nitrogen [12]. A homology search revealed that most of the transporters discussed in meticulous work on mixotrophy [12], probably involved in the uptake of amino acids and other organic compounds, are encoded in the genome of BBK16 and other Tychonema strains. The genome contains the aapJQMP locus encoding the amino acid transport system (contig JAKJHX010000121.1), the phnECD locus encoding the phosphonate transport system (contig JAKJHX010000165.1), homologues of glnQ, the gene encoding glutamine transport system ATP-binding protein and homologues of proV and proW genes encoding the transporters participating in the uptake of dimethylsulfoniopropionate (DMSP).
3.6. Analysis of CRISPR Loci
Analysis of Tychonema sp. BBK16 and other cyanobacterial genomes revealed the presence of CRISPR-Cas phage defence systems (Figure 7). The analysed systems belong to class 1, which encodes multisubunit effector complexes. The organisation of loci indicated its belonging to type I and subtype I-D, typical of cyanobacteria [74,75]. The sequence search indicated that most Tychonema Cas proteins have close homologues in other cyanobacteria. Interestingly, the Tychonema sp. BBK16 CRISPR locus contains two hypothetical protein genes downstream of the cas3′ gene. Other analysed cyanobacterial genomes CRISPR loci may also contain additional inserted genes, including transposases. In some genomes, the order of the cas genes may be different, presumably due to recombinations. The size of the Tychonema sp. BBK16 CRISPR arrays (2906 bp) is similar to that of Tychonema bourrellyi FEM_GT703 (2978 bp).
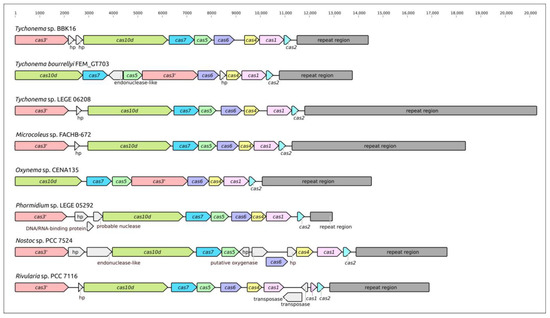
Figure 7.
Genetic map of the CRISPR loci of Tychonema sp. BBK16 and other cyanobacteria showing the location of cas gene, adjacent genes and spacer arrays (repeat regions). Sequences designated “hp” correspond to genes encoding hypothetical proteins. The scale above the sequences indicates the position in the locus.
The CRISPR array of Tychonema sp. BBK16 contains 39 spacers. The BLAST search using NCBI GenBank Phage database indicated similarities of spacer sequences with genomic sequences of several dozens of isolated cyanophages, but there were no exact matches found. According to the results of the search, 8 spacers demonstrated higher similarity to cyanophages, and 31 spacers were more similar to other phages (Table 3).

Table 3.
Cyanophages demonstrating sequence similarities with CRISPR spacers found with the BLAST search (the searches’ first hits).
3.7. Search for Prophage-Derived Regions
Search for prophages-derived regions (PDRs) in the genome of Tychonema sp. BBK16, performed with PHASTER, revealed two regions (Figure 8). Examination of genome content indicated the absence of structural proteins and the presence of transposases in both PDRs. Homologous genes were found in Nodularia phage vB_NspS-kac65v151 (NCBI accession NC_048756, genus Ravarandavirus), Synechococcus phage S-CBP2 (NC_025455, family Autographiviridae), Synechococcus phage S-CBS2 (NC_015463, unclassified siphovirus) and other phages. The presence of transposase can indicate genetic exchanges involving phage and host genomes.
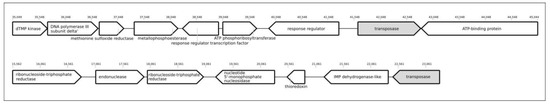
Figure 8.
Genetic map of suggested prophage-derived regions predicted by PHASTER in genome contigs of Tychonema sp. The scale above the sequence indicates the position in the corresponding contig.
3.8. Transporter Genes in Phage Genomes
To check the assumption of the involvement of bacteriophages in the transmission of transporter genes, including genes that may be useful for a mixotrophic lifestyle, the Tychonema sp. BBK16 mixotrophy-associated proteins were used for searching the homologous sequences in phages using the NCBI nt database. In addition, all GenBank phage database sequences (about 33 thousand sequences as of January 2023) were checked for the presence of transporter annotations.
The glcH and glcP genes were not found in the genomes of isolated cyanophage, but homologues GlcH and GlcP were found to be encoded in several phage human metagenomic sequences, including sequences MK231526.1 and BK032514.1. ABC transporter ATP-binding protein genes probably related to glnQ were found in the complete genome of isolated freshwater temperate cyanophage Planktothrix phage PaV-LD [27] and partial genomic sequences of another temperate freshwater cyanophage AS-1 [76]. Interestingly, sequences with a pairwise identity of 97% and above with the Planktothrix phage PaV-LD ATP-binding protein gene were found in several Planktothrix cyanobacterial genomes (Supplementary Figure S3), indicating that Planktothrix phage PaV-LD can represent a group of related temperate phages. Twenty other complete GenBank genomes of tailed cyanophages contain the genes annotated as ABC transporters, basically encoding the phosphate ABC transporter. A total of 14,353 complete genomes of Caudoviricetes phages deposited in the NCBI Genome database contain 603 genes annotated as different genes of the ABC transporter protein.
4. Discussion
Benthic biofilms are highly adapted to changing environmental conditions (dramatic changes in environmental parameters and nutrient concentrations) and are absorbers of excess nutrients from the environment [77]. The trichomes of the Baikal strain tightly intertwine and form a biofilm, characteristic of many species of Oscillatoriales cyanobacteria, in which the bacteria are held together by a polysaccharide mucus that is able to withstand harsh environmental conditions that might involve nutrient deficiencies and physical stress [78]. Various types of microscopy have been used to study the morphology of biofilms. SEM enables visualisation of the surface of the film, as well as its layers in cross-section, and confocal microscopy, which examines the film’s internal structure [79,80]. The structure of the Tychonema BBK16 biofilm was very similar to the 3D structures formed by its closest relatives, cyanobacteria of genera Microcoleus and Tychonema. Microcoleus and Tychonema form microbial mats in the benthos of lakes and rivers [77,81].
The genomic analysis showed a similar genome size and GC-content of Tychonema sp. BBK16 and Tychonema bourrellyi FEM_GT703. Calculations of ANI using all available Tychonema genomes and genomes of related cyanobacterial strains indicated that Tychonema sp. BBK16 may be classified as a representative of a new species. Phylogenetic analysis using concatenated alignments of orthologous protein sequences placed Tychonema sp. BBK16 and T. bourrellyi FEM_GT703 in a distinct clade and showed a complex picture of taxonomic relations between the strains classified as belonging to the Microcoleaceae family of the Oscillatoriales order. The proteome analysis also indicated close relatedness of Tychonema sp. BBK16 and T. bourrellyi FEM_GT703.
Previously, representatives of the genus Tychonema capable of forming biofilms were described in karst streams in a community with heterotrophic bacteria [82]. It has been shown that during the destruction of organic substances at the bottom of streams, a significant amount of organic matter is released, which is consumed by cyanobacteria in the lower layers of biofilms, where cells far from light are not able to photosynthesise. The bioinformatic analysis of the Tychonema sp. BBK16 genome indicated the presence of genes that can participate in the import of organic compounds. One of the sugar uptake genes, glcP, could be obtained by horizontal transfer. The Tychonema sp. BBK16 genome also contains clusters of genes of the aapJQMP amino acid transport system, the phnECD phosphonate transport system and other nutrient import genes. We hypothesise that cyanobacterial mixotrophy may be a factor for survival for Tychonema sp. BBK16 in benthic biofilms. It can be assumed that cyanobacteria of this genus are well adapted to the harsh conditions of Lake Baikal. The presence of protective polysaccharide sheaths, the ability to form dense biofilms and, probably, the capability of mixotrophy helped this species to occupy different ecological niches within the lake. We can also speculate that some genes associated with mixotrophy may be transferred by phages or other mobile elements.
It seems that there are no published isolated Tychonema phages at the moment. However, bioinformatic analysis indicated the presence of CRISPR-Cas and restriction modification phage defence systems in Tychonema sp. BBK16 genome. The Tychonema sp. BBK16 genome CRISPR-Cas locus belongs to characteristics for cyanobacteria subtype I-D of class 1 and includes a CRISPR array containing 39 spacers. Search for similar phages using the Tychonema sp. BBK16 spacers found no related cyanophages for about 80% of spacers, which can be related to the lack of genomic data on cyanophages [32]. Isolated cyanophages exhibiting similarities in genomic sequences with the CRISPR spacers of Tychonema sp. BBK16 belong to Kyanoviridae, Tamkungvirus and other phages.
Prophage predictions indicated the presence of prophage remnants in the Tychonema sp. BBK16 genome. These regions could be transferred in the genomes with the participation of transposases. Prophage-derived regions contain genes resembling cyanophage homologues in the genomes of Nodularia phage vB_NspS-kac65v151, Synechococcus phage S-CBP2 and Synechococcus phage S-CBS2.
5. Conclusions
Cyanobacterium Tychonema sp. BBK16 was isolated from the benthic zone of Lake Baikal. In culture, the strain formed biofilms with a felt-like interweaving of trichomes. Long fascicles of twisted trichomes were observed on the surface of the film, which caused folding. The ability to synthesise polysaccharide sheaths enabled the formation of more durable biofilms. Confocal microscopy showed the layered structure of the Tychonema biofilm. The genome of BBK16 was sequenced and analysed. The results of intergenomic comparisons and phylogenetic analyses indicate that Tychonema sp. BBK16 represent a new species related to Tychonema bourrellyi strain FEM_GT703. Genomic analysis indicated the presence of mixotrophy-associated genes. Analysis of CRISPR spacers and prophage-derived regions revealed cyanophages possibly related to hypothetical Tychonema phages.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v15020442/s1, Figure S1: ANI matrix constructed with cyanobacterial 50 genomes using ANI/AAI-Matrix Genome-based distance matrix calculator. The NCBI taxonomy is shown to the right of the organism’s name, Figure S2: best-scoring ML phylogenetic tree constructed with 90 amino acid sequences of GlcH protein. Bold numbers to the right of NCBI accession indicate the number of multiple instances of encoded proteins. The numbers near the tree branches indicate the TBE values. The total number of bootstrap trees was 1000. The scale bar shows 0.5 estimated substitutions per site, and Gloeobacter violaceus PCC 7421 was used as an outgroup, Figure S3: nucleotide alignment and ABC transporter ATP-binding protein genes of Planktothrix phage PaV-LD and Planktothrix agardhii NIES-204.
Author Contributions
Conceptualisation, P.E., I.T. and O.B.; investigation, I.T., A.K., E.S., A.G. and O.T.; writing—original draft preparation, P.E., I.T., A.K., E.S. and O.B.; writing—review and editing, E.S., P.E., K.M., I.T. and O.B.; funding acquisition, O.B.; supervision, O.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the national government and was carried out within the framework of State Project No. 0279-2021-0015, “Viral and bacterial communities as the basis for the stable functioning of freshwater ecosystems...”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This study was carried out in the Electron Microscopy Center of the Collective Instrumental Center “Ultramicroanalysis” (http://www.lin.irk.ru/copp/eng/accessed on 15 January 2023) and the Large-Scale Research Facilities “Experimental Freshwater Aquarium Complex for Baikal Hydrobionts” (http://www.lin.irk.ru/aqua (accessed on 15 January 2023) at the Limnological Institute SB RAS.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Paerl, H.W.; Otten, T.G.; Kudela, R. Mitigating the Expansion of Harmful Algal Blooms Across the Freshwater-to-Marine Continuum. Environ. Sci. Technol. 2018, 52, 5519–5529. [Google Scholar] [CrossRef] [PubMed]
- Plaas, H.E.; Paerl, H.W. Toxic Cyanobacteria: A Growing Threat to Water and Air Quality. Environ. Sci. Technol. 2021, 55, 44–64. [Google Scholar] [CrossRef] [PubMed]
- Timoshkin, O.A.; Samsonov, D.P.; Yamamuro, M.; Moore, M.V.; Belykh, O.I.; Malnik, V.V.; Sakirko, M.V.; Shirokaya, A.A.; Bondarenko, N.A.; Domysheva, V.M.; et al. Rapid Ecological Change in the Coastal Zone of Lake Baikal (East Siberia): Is the Site of the World’s Greatest Freshwater Biodiversity in Danger? J. Great Lakes Res. 2016, 42, 487–497. [Google Scholar] [CrossRef]
- Boyle, K.E.; Heilmann, S.; van Ditmarsch, D.; Xavier, J.B. Exploiting Social Evolution in Biofilms. Curr. Opin. Microbiol. 2013, 16, 207–212. [Google Scholar] [CrossRef]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Søgaard-Andersen, L.; van Wezel, G.P. Bacterial Solutions to Multicellularity: A Tale of Biofilms, Filaments and Fruiting Bodies. Nat. Rev. Microbiol. 2014, 12, 115–124. [Google Scholar] [CrossRef]
- Walsh, M.M.; Lowe, D.R. Filamentous Microfossils from the 3500-Myr-Old Onverwacht Group, Barberton Mountain Land, South Africa. Nature 1985, 314, 530–532. [Google Scholar] [CrossRef]
- Khanaev, I.V.; Kravtsova, L.S.; Maikova, O.O.; Bukshuk, N.A.; Sakirko, M.V.; Kulakova, N.V.; Butina, T.V.; Nebesnykh, I.A.; Belikov, S.I. Current State of the Sponge Fauna (Porifera: Lubomirskiidae) of Lake Baikal: Sponge Disease and the Problem of Conservation of Diversity. J. Great Lakes Res. 2018, 44, 77–85. [Google Scholar] [CrossRef]
- Bauer, F.; Fastner, J.; Bartha-Dima, B.; Breuer, W.; Falkenau, A.; Mayer, C.; Raeder, U. Mass Occurrence of Anatoxin-a- and Dihydroanatoxin-a-Producing Tychonema Sp. in Mesotrophic Reservoir Mandichosee (River Lech, Germany) as a Cause of Neurotoxicosis in Dogs. Toxins 2020, 12, 726. [Google Scholar] [CrossRef]
- Levasheva, M.V.; Timoshkin, O.A.; Vashukevich, N.V. A Landscape Approach to Ecological Monitoring in the Splash Zone of Bol’shye Koty Bay (Lake Baikal). Bull. Irkutsk State Univ. «Geoarchaeology Ethnol. Anthropol. Ser.» 2012, 3, 53–63. [Google Scholar]
- Sánchez-Baracaldo, P.; Cardona, T. On the Origin of Oxygenic Photosynthesis and Cyanobacteria. New Phytol. 2020, 225, 1440–1446. [Google Scholar] [CrossRef]
- Rippka, R. Photoheterotrophy and Chemoheterotrophy among Unicellular Blue-Green Algae. Archiv. Mikrobiol. 1972, 87, 93–98. [Google Scholar] [CrossRef]
- Muñoz-Marín, M.C.; Gómez-Baena, G.; López-Lozano, A.; Moreno-Cabezuelo, J.A.; Díez, J.; García-Fernández, J.M. Mixotrophy in Marine Picocyanobacteria: Use of Organic Compounds by Prochlorococcus and Synechococcus. ISME J. 2020, 14, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Stoecker, D.K.; Hansen, P.J.; Caron, D.A.; Mitra, A. Mixotrophy in the Marine Plankton. Ann. Rev. Mar. Sci. 2017, 9, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Eiler, A. Evidence for the Ubiquity of Mixotrophic Bacteria in the Upper Ocean: Implications and Consequences. Appl. Environ. Microbiol. 2006, 72, 7431–7437. [Google Scholar] [CrossRef]
- Khodzher, T.V.; Domysheva, V.M.; Sorokovikova, L.M.; Sakirko, M.V.; Tomberg, I.V. Current Chemical Composition of Lake Baikal Water. Inland Waters 2017, 7, 250–258. [Google Scholar] [CrossRef]
- Podlesnaya, G.V.; Krasnopeev, A.Y.; Potapov, S.A.; Tikhonova, I.V.; Shtykova, Y.R.; Suslova, M.Y.; Timoshkin, O.A.; Belykh, O.I. Diversity of Nitrifying Bacteria in Microbial Communities from Water and Epilithic Biofilms of the Lake Baikal Littoral Zone. Limnol. Freshw. Biol. 2020, 1008–1010. [Google Scholar] [CrossRef]
- Dufresne, A.; Salanoubat, M.; Partensky, F.; Artiguenave, F.; Axmann, I.M.; Barbe, V.; Duprat, S.; Galperin, M.Y.; Koonin, E.V.; Le Gall, F.; et al. Genome Sequence of the Cyanobacterium Prochlorococcus Marinus SS120, a Nearly Minimal Oxyphototrophic Genome. Proc. Natl. Acad. Sci. USA 2003, 100, 10020–10025. [Google Scholar] [CrossRef] [PubMed]
- Palenik, B.; Brahamsha, B.; Larimer, F.W.; Land, M.; Hauser, L.; Chain, P.; Lamerdin, J.; Regala, W.; Allen, E.E.; McCarren, J.; et al. The Genome of a Motile Marine Synechococcus. Nature 2003, 424, 1037–1042. [Google Scholar] [CrossRef]
- Rocap, G.; Larimer, F.W.; Lamerdin, J.; Malfatti, S.; Chain, P.; Ahlgren, N.A.; Arellano, A.; Coleman, M.; Hauser, L.; Hess, W.R.; et al. Genome Divergence in Two Prochlorococcus Ecotypes Reflects Oceanic Niche Differentiation. Nature 2003, 424, 1042–1047. [Google Scholar] [CrossRef]
- Casas, V.; Maloy, S. The Role of Phage in the Adaptation of Bacteria to New Environmental Niches. In Molecular Mechanisms of Microbial Evolution; Rampelotto, P.H., Ed.; Grand Challenges in Biology and Biotechnology; Springer International Publishing: Cham, Switzerland, 2018; pp. 267–306. ISBN 978-3-319-69078-0. [Google Scholar]
- Shestakov, S.V.; Karbysheva, E.A. The Role of Viruses in the Evolution of Cyanobacteria. Biol. Bull. Rev. 2015, 5, 527–537. [Google Scholar] [CrossRef]
- Bryan, M.J.; Burroughs, N.J.; Spence, E.M.; Clokie, M.R.J.; Mann, N.H.; Bryan, S.J. Evidence for the Intense Exchange of MazG in Marine Cyanophages by Horizontal Gene Transfer. PLoS ONE 2008, 3, e2048. [Google Scholar] [CrossRef]
- Liu, X.; Kong, S.; Shi, M.; Fu, L.; Gao, Y.; An, C. Genomic Analysis of Freshwater Cyanophage Pf-WMP3 Infecting Cyanobacterium Phormidium Foveolarum: The Conserved Elements for a Phage. Microb. Ecol. 2008, 56, 671–680. [Google Scholar] [CrossRef]
- Jacquet, S.; Zhong, X.; Parvathi, A.; Ram, A.S.P. First Description of a Cyanophage Infecting the Cyanobacterium Arthrospira Platensis (Spirulina). J. Appl. Phycol. 2013, 25, 195–203. [Google Scholar] [CrossRef]
- Zhang, D.; You, F.; He, Y.; Te, S.H.; Gin, K.Y.-H. Isolation and Characterization of the First Freshwater Cyanophage Infecting Pseudanabaena. J. Virol. 2020, 94, e00682-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shi, M.; Kong, S.; Gao, Y.; An, C. Cyanophage Pf-WMP4, a T7-like Phage Infecting the Freshwater Cyanobacterium Phormidium Foveolarum: Complete Genome Sequence and DNA Translocation. Virology 2007, 366, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Gao, E.-B.; Gui, J.-F.; Zhang, Q.-Y. A Novel Cyanophage with a Cyanobacterial Nonbleaching Protein A Gene in the Genome. J. Virol. 2012, 86, 236–245. [Google Scholar] [CrossRef]
- Zhou, Y.; Lin, J.; Li, N.; Hu, Z.; Deng, F. Characterization and Genomic Analysis of a Plaque Purified Strain of Cyanophage PP. Virol. Sin. 2013, 28, 272–279. [Google Scholar] [CrossRef]
- Du, K.; Yang, F.; Zhang, J.-T.; Yu, R.-C.; Deng, Z.; Li, W.-F.; Chen, Y.; Li, Q.; Zhou, C.-Z. Comparative Genomic Analysis of Five Freshwater Cyanophages and Reference-Guided Metagenomic Data Mining. Microbiome 2022, 10, 128. [Google Scholar] [CrossRef]
- Benler, S.; Yutin, N.; Antipov, D.; Rayko, M.; Shmakov, S.; Gussow, A.B.; Pevzner, P.; Koonin, E.V. Thousands of Previously Unknown Phages Discovered in Whole-Community Human Gut Metagenomes. Microbiome 2021, 9, 78. [Google Scholar] [CrossRef]
- Shmakov, S.A.; Wolf, Y.I.; Savitskaya, E.; Severinov, K.V.; Koonin, E.V. Mapping CRISPR Spaceromes Reveals Vast Host-Specific Viromes of Prokaryotes. Commun. Biol. 2020, 3, 1–9. [Google Scholar] [CrossRef]
- Nasko, D.J.; Ferrell, B.D.; Moore, R.M.; Bhavsar, J.D.; Polson, S.W.; Wommack, K.E. CRISPR Spacers Indicate Preferential Matching of Specific Virioplankton Genes. mBio 2019, 10, e02651-18. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brüssow, H. Prophage Genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and Bacterial Genomics: What Have We Learned so Far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef]
- Marques, A.T.; Tanoeiro, L.; Duarte, A.; Gonçalves, L.; Vítor, J.M.B.; Vale, F.F. Genomic Analysis of Prophages from Klebsiella Pneumoniae Clinical Isolates. Microorganisms 2021, 9, 2252. [Google Scholar] [CrossRef]
- Evseev, P.; Lukianova, A.; Tarakanov, R.; Tokmakova, A.; Popova, A.; Kulikov, E.; Shneider, M.; Ignatov, A.; Miroshnikov, K. Prophage-Derived Regions in Curtobacterium Genomes: Good Things, Small Packages. Int. J. Mol. Sci. 2023, 24, 1586. [Google Scholar] [CrossRef]
- Rippka, R. Isolation and Purification of Cyanobacteria. Methods Enzym. 1988, 167, 3–27. [Google Scholar] [CrossRef]
- Wood, E.J. Molecular Cloning. A Laboratory Manual by T Maniatis, E F Fritsch and J Sambrook. Pp 545. Cold Spring Harbor Laboratory, New York. 1982. $48. Biochem. Educ. 1983, 11, 82. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An Automated Binning Algorithm to Recover Genomes from Multiple Metagenomic Datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Download—NCBI. Available online: https://www.ncbi.nlm.nih.gov/home/download/ (accessed on 3 November 2022).
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline Reach with Protein Family Model Curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef] [PubMed]
- Geneious|Bioinformatics Software for Sequence Data Analysis. Available online: https://www.geneious.com/ (accessed on 11 November 2021).
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Gabler, F.; Nam, S.-Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef] [PubMed]
- Index of /Pub/Databases/Pfam/Tools. Available online: http://ftp.ebi.ac.uk/pub/databases/Pfam/Tools/ (accessed on 15 January 2023).
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An Improved Version of the NJ Algorithm Based on a Simple Model of Sequence Data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. RaxmlGUI 2.0: A Graphical Interface and Toolkit for Phylogenetic Analyses Using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Lemoine, F.; Domelevo Entfellner, J.-B.; Wilkinson, E.; Correia, D.; Dávila Felipe, M.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s Phylogenetic Bootstrap in the Era of Big Data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef]
- MinCED-Mining CRISPRs in Environmental Datasets 2022. Available online: https://github.com/ctSkennerton/minced (accessed on 15 January 2023).
- Arndt, D.; Marcu, A.; Liang, Y.; Wishart, D.S. PHAST, PHASTER and PHASTEST: Tools for Finding Prophage in Bacterial Genomes. Brief Bioinform. 2017, 20, 1560–1567. [Google Scholar] [CrossRef]
- Pinto, F.; Tett, A.; Armanini, F.; Asnicar, F.; Boscaini, A.; Pasolli, E.; Zolfo, M.; Donati, C.; Salmaso, N.; Segata, N. Draft Genome Sequence of the Planktic Cyanobacterium Tychonema Bourrellyi, Isolated from Alpine Lentic Freshwater. Genome Announc. 2017, 5, e01294-17. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.; Woodhouse, J.N. Defining Cyanobacterial Species: Diversity and Description Through Genomics. Crit. Rev. Plant Sci. 2020, 39, 101–124. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S RRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Barco, R.A.; Garrity, G.M.; Scott, J.J.; Amend, J.P.; Nealson, K.H.; Emerson, D. A Genus Definition for Bacteria and Archaea Based on a Standard Genome Relatedness Index. mBio 2020, 11, e02475-19. [Google Scholar] [CrossRef] [PubMed]
- Bouma-Gregson, K.; Olm, M.R.; Probst, A.J.; Anantharaman, K.; Power, M.E.; Banfield, J.F. Impacts of Microbial Assemblage and Environmental Conditions on the Distribution of Anatoxin-a Producing Cyanobacteria within a River Network. ISME J. 2019, 13, 1618–1634. [Google Scholar] [CrossRef]
- Moore, K.R.; Magnabosco, C.; Momper, L.; Gold, D.A.; Bosak, T.; Fournier, G.P. An Expanded Ribosomal Phylogeny of Cyanobacteria Supports a Deep Placement of Plastids. Front. Microbiol. 2019, 10, 1612. [Google Scholar] [CrossRef]
- Humbert, J.-F.; Barbe, V.; Latifi, A.; Gugger, M.; Calteau, A.; Coursin, T.; Lajus, A.; Castelli, V.; Oztas, S.; Samson, G.; et al. A Tribute to Disorder in the Genome of the Bloom-Forming Freshwater Cyanobacterium Microcystis Aeruginosa. PLoS ONE 2013, 8, e70747. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, X.; Liang, C.; Wu, J.; Bao, Q.; Qin, S. Genome-Wide Analysis of Restriction-Modification System in Unicellular and Filamentous Cyanobacteria. Physiol. Genom. 2006, 24, 181–190. [Google Scholar] [CrossRef]
- Dupuis, M.-È.; Villion, M.; Magadán, A.H.; Moineau, S. CRISPR-Cas and Restriction–Modification Systems Are Compatible and Increase Phage Resistance. Nat. Commun. 2013, 4, 2087. [Google Scholar] [CrossRef]
- Yelton, A.P.; Acinas, S.G.; Sunagawa, S.; Bork, P.; Pedrós-Alió, C.; Chisholm, S.W. Global Genetic Capacity for Mixotrophy in Marine Picocyanobacteria. ISME J. 2016, 10, 2946–2957. [Google Scholar] [CrossRef]
- Gómez-Baena, G.; López-Lozano, A.; Gil-Martínez, J.; Lucena, J.M.; Diez, J.; Candau, P.; García-Fernández, J.M. Glucose Uptake and Its Effect on Gene Expression in Prochlorococcus. PLoS ONE 2008, 3, e3416. [Google Scholar] [CrossRef]
- Muñoz-Marín, M.D.C.; Luque, I.; Zubkov, M.V.; Hill, P.G.; Diez, J.; García-Fernández, J.M. Prochlorococcus Can Use the Pro1404 Transporter to Take up Glucose at Nanomolar Concentrations in the Atlantic Ocean. Proc. Natl. Acad. Sci. USA 2013, 110, 8597–8602. [Google Scholar] [CrossRef]
- Moreno-Cabezuelo, J.Á.; López-Lozano, A.; Díez, J.; García-Fernández, J.M. Differential Expression of the Glucose Transporter Gene GlcH in Response to Glucose and Light in Marine Picocyanobacteria. PeerJ 2019, 6, e6248. [Google Scholar] [CrossRef]
- Zhang, C.C.; Durand, M.C.; Jeanjean, R.; Joset, F. Molecular and Genetical Analysis of the Fructose-Glucose Transport System in the Cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 1989, 3, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Picossi, S.; Flores, E.; Ekman, M. Diverse Roles of the GlcP Glucose Permease in Free-Living and Symbiotic Cyanobacteria. Plant Signal. Behav. 2013, 8, e27416. [Google Scholar] [CrossRef] [PubMed]
- Ekman, M.; Picossi, S.; Campbell, E.L.; Meeks, J.C.; Flores, E. A Nostoc Punctiforme Sugar Transporter Necessary to Establish a Cyanobacterium-Plant Symbiosis1[C][W]. Plant Physiol. 2013, 161, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Scholz, I.; Lange, S.J.; Hein, S.; Hess, W.R.; Backofen, R. CRISPR-Cas Systems in the Cyanobacterium Synechocystis Sp. PCC6803 Exhibit Distinct Processing Pathways Involving at Least Two Cas6 and a Cmr2 Protein. PLoS ONE 2013, 8, e56470. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, J.; Wang, B.; Han, J.; Hao, Y.; Wang, S.; Ma, X.; Yang, S.; Ma, L.; Yi, L.; et al. Endogenous Type I CRISPR-Cas: From Foreign DNA Defense to Prokaryotic Engineering. Front. Bioeng. Biotechnol. 2020, 8, 62. [Google Scholar] [CrossRef]
- Chu, T.-C.; Murray, S.R.; Hsu, S.-F.; Vega, Q.; Lee, L.H. Temperature-Induced Activation of Freshwater Cyanophage AS-1 Prophage. Acta Histochem. 2011, 113, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Quiblier, C.; Wood, S.; Echenique-Subiabre, I.; Heath, M.; Villeneuve, A.; Humbert, J.-F. A Review of Current Knowledge on Toxic Benthic Freshwater Cyanobacteria--Ecology, Toxin Production and Risk Management. Water Res. 2013, 47, 5464–5479. [Google Scholar] [CrossRef]
- Veerabadhran, M.; Chakraborty, S.; Mitra, S.; Karmakar, S.; Mukherjee, J. Effects of Flask Configuration on Biofilm Growth and Metabolites of Intertidal Cyanobacteria Isolated from a Mangrove Forest. J. Appl. Microbiol. 2018, 125, 190–202. [Google Scholar] [CrossRef]
- de los Ríos, A.; Ascaso, C.; Wierzchos, J.; Fernández-Valiente, E.; Quesada, A. Microstructural Characterization of Cyanobacterial Mats from the McMurdo Ice Shelf, Antarctica. Appl. Environ. Microbiol. 2004, 70, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Solé, A.; Gaju, N.; Méndez-Álvarez, S.; Esteve, I. Confocal Laser Scanning Microscopy as a Tool to Determine Cyanobacteria Biomass in Microbial Mats. J. Microsc. 2001, 204, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Burnat, M.; Diestra, E.; Esteve, I.; Solé, A. In Situ Determination of the Effects of Lead and Copper on Cyanobacterial Populations in Microcosms. PLoS ONE 2009, 4, e6204. [Google Scholar] [CrossRef]
- Arp, G.; Bissett, A.; Brinkmann, N.; Cousin, S.; Beer, D.D.; Friedl, T.; Mohr, K.I.; Neu, T.R.; Reimer, A.; Shiraishi, F.; et al. Tufa-Forming Biofilms of German Karstwater Streams: Microorganisms, Exopolymers, Hydrochemistry and Calcification. Geol. Soc. 2010, 336, 83. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).