Crude Extracts of Talaromyces Strains (Ascomycota) Affect Honey Bee (Apis mellifera) Resistance to Chronic Bee Paralysis Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Fungal Crude Extracts
2.2. Honey Bee Survival Assay
2.2.1. Honey Bee Feeding and Rearing in vitro
2.2.2. Virus Titration—End Point Replication Assay (BID50)
2.2.3. Infection of Adult Honey Bees
2.3. Antiviral Activity of the Fungal Extracts in Mammalian Cell-Based Assays
2.3.1. Virus Stock Solutions
2.3.2. Replication/Infection Inhibition Test with Fungal Crude Extracts
2.3.3. Virus Inactivation Test with Fungal Crude Extracts
2.4. Analysis of Fungal Compounds
2.4.1. UHPLC-HR-MS Analysis and Metabolite Annotation
2.4.2. Molecular Networking Analysis
3. Results
3.1. Survival Analysis
3.1.1. Preparation of Extracts and Feeding Assays
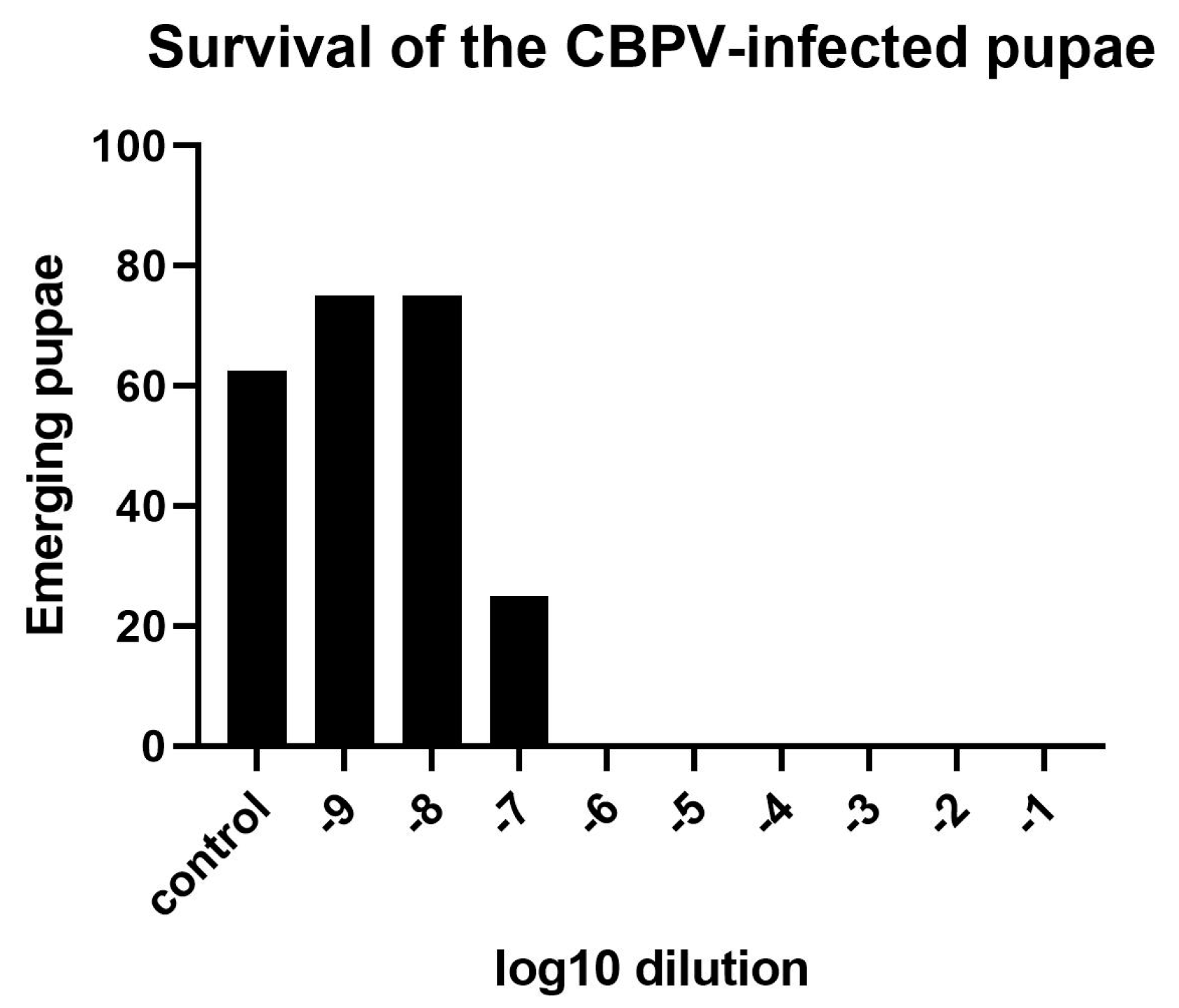
3.1.2. Determination of an Effective CBPV Infection Dose
3.2. Antiviral Activity of the Fungal Extracts in Mammalian Cell Culture
3.2.1. Pre-Treated Cells
3.2.2. Pretreated Virions
3.3. Metabolite Annotation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, A.M.; Vaissiere, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of Pollinators in Changing Landscapes for World Crops. Proc. R. Soc. B Biol. Sci. 2007, 274, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Ollerton, J.; Winfree, R.; Tarrant, S. How Many Flowering Plants Are Pollinated by Animals? Oikos 2011, 120, 321–326. [Google Scholar] [CrossRef]
- Wagner, D.L.; Grames, E.M.; Forister, M.L.; Berenbaum, M.R.; Stopak, D. Insect Decline in the Anthropocene: Death by a Thousand Cuts. Proc. Natl. Acad. Sci. USA 2021, 118, e2023989118. [Google Scholar] [CrossRef] [PubMed]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global Pollinator Declines: Trends, Impacts and Drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Klatt, B.K.; Holzschuh, A.; Westphal, C.; Clough, Y.; Smit, I.; Pawelzik, E.; Tscharntke, T. Bee Pollination Improves Crop Quality, Shelf Life and Commercial Value. Proc. R. Soc. B Biol. Sci. 2013, 281, 20132440. [Google Scholar] [CrossRef]
- Garibaldi, L.A.; Aizen, M.A.; Cunningham, S.A.; Klein, A.M. Pollinator Shortage and Global Crop Yield: Looking at the Whole Spectrum of Pollinator Dependency. Commun. Integr. Biol. 2009, 2, 37–39. [Google Scholar] [CrossRef]
- Steinhauer, N.; Kulhanek, K.; Antúnez, K.; Human, H.; Chantawannakul, P.; Chauzat, M.-P.; VanEngelsdorp, D. Drivers of Colony Losses. Curr. Opin. Insect Sci. 2018, 26, 142–148. [Google Scholar] [CrossRef]
- Ravoet, J.; Maharramov, J.; Meeus, I.; De Smet, L.; Wenseleers, T.; Smagghe, G.; de Graaf, D.C. Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia Mellificae as a New Contributory Factor to Winter Mortality. PLoS ONE 2013, 8, e72443. [Google Scholar] [CrossRef]
- Fünfhaus, A.; Ebeling, J.; Genersch, E. Bacterial Pathogens of Bees. Curr. Opin. Insect Sci. 2018, 26, 89–96. [Google Scholar] [CrossRef]
- Quintana, L. Fungal Infections In Honey Bees. Fungal Genom. Biol. 2015, 05, 1000118. [Google Scholar] [CrossRef]
- Genersch, E.; Aubert, M. Emerging and Re-Emerging Viruses of the Honey Bee (Apis Mellifera L.). Vet. Res. 2010, 41, 54. [Google Scholar] [CrossRef] [PubMed]
- Budge, G.E.; Simcock, N.K.; Holder, P.J.; Shirley, M.D.F.; Brown, M.A.; Van Weymers, P.S.M.; Evans, D.J.; Rushton, S.P. Chronic Bee Paralysis as a Serious Emerging Threat to Honey Bees. Nat. Commun. 2020, 11, 2164. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, E.W.; Chen, Y.; Message, D.; Pettis, J.; Evans, J.D. Virus Infections in Brazilian Honey Bees. J. Invertebr. Pathol. 2008, 99, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Noël, A.; Le Conte, Y.; Mondet, F. Varroa Destructor: How Does It Harm Apis Mellifera Honey Bees and What Can Be Done about It? Emerg. Top. Life Sci. 2020, 4, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa Destructor Feeds Primarily on Honey Bee Fat Body Tissue and Not Hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef]
- Forgách, P.; Bakonyi, T.; Tapaszti, Z.; Nowotny, N.; Rusvai, M. Prevalence of Pathogenic Bee Viruses in Hungarian Apiaries: Situation before Joining the European Union. J. Invertebr. Pathol. 2008, 98, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Yañez, O.; Fries, I.; De Miranda, J.R. Varroa Invasion and Virus Adaptation. Trends Parasitol. 2012, 28, 353–354. [Google Scholar] [CrossRef]
- Wilfert, L.; Long, G.; Leggett, H.C.; Schmid-Hempel, P.; Butlin, R.; Martin, S.J.M.; Boots, M. Honeybee Disease: Deformed Wing Virus Is a Recent Global Epidemic in Honeybees Driven by Varroa Mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef]
- Kulhanek, K.; Steinhauer, N.; Rennich, K.; Caron, D.M.; Sagili, R.R.; Pettis, J.S.; Ellis, J.D.; Wilson, M.E.; Wilkes, J.T.; Tarpy, D.R.; et al. Encuesta Nacional 2015–2016 Sobre Pérdidas Anuales de Colonias de La Abeja de La Miel Manejada En Los EE.UU. J. Apic. Res. 2017, 56, 328–340. [Google Scholar] [CrossRef]
- Lee, K.V.; Steinhauer, N.; Rennich, K.; Wilson, M.E.; Tarpy, D.R.; Caron, D.M.; Rose, R.; Delaplane, K.S.; Baylis, K.; Lengerich, E.J.; et al. A National Survey of Managed Honey Bee 2013-2014 Annual Colony Losses in the USA. Apidologie 2015, 46, 292–305. [Google Scholar] [CrossRef]
- Seitz, N.; Traynor, K.S.; Steinhauer, N.; Rennich, K.; Wilson, M.E.; Ellis, J.D.; Rose, R.; Tarpy, D.R.; Sagili, R.R.; Caron, D.M.; et al. Encuesta Nacional Sobre La Pérdida Anual de Colmenas de Abejas Manejadas Durante 2014–2015 En Los EEUU. J. Apic. Res. 2015, 54, 292–304. [Google Scholar] [CrossRef]
- Steinhauer, N.A.; Rennich, K.; Wilson, M.E.; Caron, D.M.; Lengerich, E.J.; Pettis, J.S.; Rose, R.; Skinner, J.A.; Tarpy, D.R.; Wilkes, J.T.; et al. A National Survey of Managed Honey Bee 2012-2013 Annual Colony Losses in the USA: Results from the Bee Informed Partnership. J. Apic. Res. 2014, 53, 1–18. [Google Scholar] [CrossRef]
- Jacques, A.; Laurent, M.; Ribière-Chabert, M.; Saussac, M.; Bougeard, S.; Budge, G.E.; Hendrikx, P.; Chauzat, M.P. A Pan-European Epidemiological Study Reveals Honey Bee Colony Survival Depends on Beekeeper Education and Disease Control. PLoS ONE 2017, 12, e0172591. [Google Scholar] [CrossRef] [PubMed]
- Brodschneider, R.; Gray, A.; Adjlane, N.; Ballis, A.; Brusbardis, V.; Charrière, J.D.; Chlebo, R.; Coffey, M.F.; Dahle, B.; de Graaf, D.C.; et al. Multi-Country Loss Rates of Honey Bee Colonies during Winter 2016/2017 from the COLOSS Survey. J. Apic. Res. 2018, 57, 452–457. [Google Scholar] [CrossRef]
- Brodschneider, R.; Gray, A.; van der Zee, R.; Adjlane, N.; Brusbardis, V.; Charrière, J.D.; Chlebo, R.; Coffey, M.F.; Crailsheim, K.; Dahle, B.; et al. Preliminary Analysis of Loss Rates of Honey Bee Colonies during Winter 2015/16 from the COLOSS Survey. J. Apic. Res. 2016, 55, 375–378. [Google Scholar] [CrossRef]
- Gisder, S.; Genersch, E. Special Issue: Honey Bee Viruses. Viruses 2015, 7, 5603–5608. [Google Scholar] [CrossRef] [PubMed]
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and Control of Varroa Destructor. J. Invertebr. Pathol. 2010, 103, S96–S119. [Google Scholar] [CrossRef]
- Traynor, K.S.; Mondet, F.; de Miranda, J.R.; Techer, M.; Kowallik, V.; Oddie, M.A.Y.; Chantawannakul, P.; McAfee, A. Varroa Destructor: A Complex Parasite, Crippling Honey Bees Worldwide. Trends Parasitol. 2020, 36, 592–606. [Google Scholar] [CrossRef]
- Yang, D.; Xu, X.; Zhao, H.; Yang, S.; Wang, X.; Zhao, D.; Diao, Q.; Hou, C. Diverse Factors Affecting Efficiency of RNAi in Honey Bee Viruses. Front. Genet. 2018, 9, 384. [Google Scholar] [CrossRef]
- Simone-Finstrom, M.; Aronstein, K.; Goblirsch, M.; Rinkevich, F.; de Guzman, L. Gamma Irradiation Inactivates Honey Bee Fungal, Microsporidian, and Viral Pathogens and Parasites. J. Invertebr. Pathol. 2018, 153, 57–64. [Google Scholar] [CrossRef]
- Sumpter, D.J.T.; Martin, S.J. The Dynamics of Virus Epidemics in Varroa-Infested Honey Bee Colonies. J. Anim. Ecol. 2004, 73, 51–63. [Google Scholar] [CrossRef]
- Le Conte, Y.; Ellis, M.; Ritter, W. Varroa Mites and Honey Bee Health: Can Varroa Explain Part of the Colony Losses? Apidologie 2010, 41, 353–363. [Google Scholar] [CrossRef]
- Locke, B.; Forsgren, E.; Fries, I.; de Miranda, J.R. Acaricide Treatment Affects Viral Dynamics in Varroa Destructor-Infested Honey Bee Colonies via Both Host Physiology and Mite Control. Appl. Environ. Microbiol. 2012, 78, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Eichberg, J.; Maiworm, E.; Oberpaul, M.; Czudai-matwich, V.; Lüddecke, T.; Vilcinskas, A.; Hardes, K. Virus Infections. Viruses 2022, 14, 2452. [Google Scholar] [CrossRef]
- Palmer-Young, E.C.; Tozkar, C.O.; Schwarz, R.S.; Chen, Y.; Irwin, R.E.; Adler, L.S.; Evans, J.D. Nectar and Pollen Phytochemicals Stimulate Honey Bee (Hymenoptera: Apidae) Immunity to Viral Infection. J. Econ. Entomol. 2017, 110, 1959–1972. [Google Scholar] [CrossRef]
- Hsieh, E.M.; Berenbaum, M.R.; Dolezal, A.G. Ameliorative Effects of Phytochemical Ingestion on Viral Infection in Honey Bees. Insects 2020, 11, 698. [Google Scholar] [CrossRef]
- Parekh, F.; Daughenbaugh, K.F.; Flenniken, M.L. Chemical Stimulants and Stressors Impact the Outcome of Virus Infection and Immune Gene Expression in Honey Bees (Apis Mellifera). Front. Immunol. 2021, 12, 747848. [Google Scholar] [CrossRef]
- Stamets, P.E.; Naeger, N.L.; Evans, J.D.; Han, J.O.; Hopkins, B.K.; Lopez, D.; Moershel, H.M.; Nally, R.; Sumerlin, D.; Taylor, A.W.; et al. Extracts of Polypore Mushroom Mycelia Reduce Viruses in Honey Bees. Sci. Rep. 2018, 8, 13936. [Google Scholar] [CrossRef]
- Gilliam, M.; Prest, D.B.; Lorenz, B.J. Microbiology of Pollen and Bee Bread: Taxonomy and Enzymology of Molds. Apidologie 1989, 20, 53–68. [Google Scholar] [CrossRef]
- Sinpoo, C.; Williams, G.R.; Chantawannakul, P. Dynamics of Fungal Communities in Corbicular Pollen and Bee Bread. Chiang Mai J. Sci. 2017, 44, 1235–1247. [Google Scholar]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.I.M.; Mott, B.M. Hive-Stored Pollen of Honey Bees: Many Lines of Evidence Are Consistent with Pollen Preservation, Not Nutrient Conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef] [PubMed]
- Gilliam, M.; Taber, S.; Lorenz, B.J.; Prest, D.B. Factors Affecting Development of Chalkbrood Disease in Colonies of Honey Bees, Apis Mellifera, Fed Pollen Contaminated with Ascosphaera Apis. J. Invertebr. Pathol. 1988, 52, 314–325. [Google Scholar] [CrossRef]
- Disayathanoowat, T.; Li, H.; Supapimon, N.; Suwannarach, N.; Lumyong, S.; Chantawannakul, P.; Guo, J. Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China. Microorganisms 2020, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Paludo, C.R.; Pishchany, G.; Andrade-Dominguez, A.; Silva-Junior, E.A.; Menezes, C.; Nascimento, F.S.; Currie, C.R.; Kolter, R.; Clardy, J.; Pupo, M.T. Microbial Community Modulates Growth of Symbiotic Fungus Required for Stingless Bee Metamorphosis. PLoS ONE 2019, 14, e0219696. [Google Scholar] [CrossRef] [PubMed]
- Zhai, M.M.; Li, J.; Jiang, C.X.; Shi, Y.P.; Di, D.L.; Crews, P.; Wu, Q.X. The Bioactive Secondary Metabolites from Talaromyces Species. Nat. Prod. Bioprospect 2016, 6, 1–24. [Google Scholar] [CrossRef]
- Rodríguez-Andrade, E.; Stchigel, A.M.; Terrab, A.; Guarro, J.; Cano-Lira, J.F. Diversity of Xerotolerant and Xerophilic Fungi in Honey. IMA Fungus 2019, 10, 20. [Google Scholar] [CrossRef]
- Sandeepani, H.P.; Ratnaweera, P.B. Antibacterial Activity of Entomopathogenic Fungi Isolated from Vespa Affinis and Apis Dorsata in Sri Lanka. In Proceedings of the International Conference on Frontiers in Chemical Technology 2020, Colombo, Sri Lanka, 20–22 July 2020. [Google Scholar]
- Barbosa, R.N.; Bezerra, J.D.P.; Souza-Motta, C.M.; Frisvad, J.C.; Samson, R.A.; Oliveira, N.T.; Houbraken, J. New Penicillium and Talaromyces Species from Honey, Pollen and Nests of Stingless Bees. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2018, 111, 1883–1912. [Google Scholar] [CrossRef]
- Lan, D.; Wu, B. Chemistry and Bioactivities of Secondary Metabolites from the Genus Talaromyces. Chem. Biodivers. 2020, 17, e2000229. [Google Scholar] [CrossRef]
- Yilmaz, N.; Houbraken, J.; Hoekstra, E.S.; Frisvad, J.C.; Visagie, C.M.; Samson, R.A. Delimitation and Characterisation of Talaromyces Purpurogenus and Related Species. Persoonia Mol. Phylogeny Evol. Fungi 2012, 29, 39–54. [Google Scholar] [CrossRef]
- Matsunaga, H.; Kamisuki, S.; Kaneko, M.; Yamaguchi, Y.; Takeuchi, T.; Watashi, K.; Sugawara, F. Isolation and Structure of Vanitaracin A, a Novel Anti-Hepatitis B Virus Compound from Talaromyces Sp. Bioorganic Med. Chem. Lett. 2015, 25, 4325–4328. [Google Scholar] [CrossRef]
- Olivier, V.; Blanchard, P.; Chaouch, S.; Lallemand, P.; Schurr, F.; Celle, O.; Dubois, E.; Tordo, N.; Thiéry, R.; Houlgatte, R.; et al. Molecular Characterisation and Phylogenetic Analysis of Chronic Bee Paralysis Virus, a Honey Bee Virus. Virus Res. 2008, 132, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.; Gibbs, A.J.; Woods, R.D. Two Viruses from Adult Honey Bees (Apis Mellifera Linnaeus). Virology 1963, 21, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L. The Occurrence of Chronic and Acute Bee Paralysis Viruses in Bees Outside Britain. J. Invertebr. Pathol. 1965, 7, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L. Paralysis of the Honey Bee, Apis Mellifera Linnaeus. J. Invertebr. Pathol. 1965, 7, 132–140. [Google Scholar] [CrossRef]
- Seitz, K.; Buczolich, K.; Dikunová, A.; Plevka, P.; Power, K.; Rümenapf, T.; Lamp, B. A Molecular Clone of Chronic Bee Paralysis Virus (CBPV) Causes Mortality in Honey Bee Pupae (Apis Mellifera). Sci. Rep. 2019, 9, 16274. [Google Scholar] [CrossRef]
- Lawson, J.S.; Syme, H.M.; Wheeler-Jones, C.P.D.; Elliott, J. Characterisation of Crandell-Rees Feline Kidney (CRFK) Cells as Mesenchymal in Phenotype. Res. Vet. Sci. 2019, 127, 99–102. [Google Scholar] [CrossRef]
- Marner, M.; Hartwig, C.; Patras, M.A.; Wodi, S.I.M.; Rieuwpassa, F.J.; Ijong, F.G.; Balansa, W. Sustainable Low-Volume Analysis of Environmental Samples by Semi-Automated Prioritization of Extracts for Natural Product Research (SeaPEPR). Mar. Drugs 2020, 18, 649. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Yang, B.; Peng, G.; Li, T.; Kadowaki, T. Molecular and Phylogenetic Characterization of Honey Bee Viruses, Nosema Microsporidia, Protozoan Parasites, and Parasitic Mites in China. Ecol. Evol. 2013, 3, 298–311. [Google Scholar] [CrossRef]
- Allen, F.; Greiner, R.; Wishart, D. Competitive Fragmentation Modeling of ESI-MS/MS Spectra for Putative Metabolite Identification. Metabolomics 2015, 11, 98–110. [Google Scholar] [CrossRef]
- Laatsch, H. AntiBase: The Natural Compound Identifier; Wiley: New York, NY, USA, 2017. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kraus, A.A.; Priemer, C.; Heider, H.; Krüger, D.H.; Ulrich, R. Inactivation of Hantaan Virus-Containing Samples for Subsequent Investigations Outside Biosafety Level 3 Facilities. Intervirology 2005, 48, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Hristov, P.; Shumkova, R.; Palova, N.; Neov, B. Factors Associated with Honey Bee Colony Losses: A Mini-Review. Vet. Sci. 2020, 7, 166. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee Declines Driven by Combined Stress from Parasites, Pesticides, and Lack of Flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef]
- Nicoletti, R. Talaromyces–Insect Relationships. Microorganisms 2022, 10, 45. [Google Scholar] [CrossRef]
- Nonaka, K.; Chiba, T.; Suga, T.; Asami, Y.; Iwatsuki, M.; Masuma, R.; Omura, S.; Shiomi, K. Coculnol, a New Penicillic Acid Produced by a Coculture of Fusarium Solani FKI-6853 and Talaromyces Sp. FKA-65. J. Antibiot. 2015, 68, 530–532. [Google Scholar] [CrossRef]
- Yue, Y.; Jiang, M.; Hu, H.; Wu, J.; Sun, H.; Jin, H.; Hou, T.; Tao, K. Isolation, Identification and Insecticidal Activity of the Secondary Metabolites of Talaromyces Purpureogenus BS5. J. Fungi 2022, 8, 288. [Google Scholar] [CrossRef]
- Kostić, A.; Milinčić, D.D.; Petrović, T.S.; Krnjaja, V.S.; Stanojević, S.P.; Barać, M.B.; Tešić, Ž.L.; Pešić, M.B. Mycotoxins and Mycotoxin Producing Fungi in Pollen: Review. Toxins 2019, 11, 64. [Google Scholar] [CrossRef]
- Beuchat, L.R. Influence of Water Activity on Growth, Metabolic Activities and Survival of Yeasts and Molds. J. Food Prot. 1983, 46, 135–141. [Google Scholar] [CrossRef]
- Mannaa, M.; Kim, K.D. Influence of Temperature and Water Activity on Deleterious Fungi and Mycotoxin Production during Grain Storage. Mycobiology 2017, 45, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Feofilova, E.P.; Ivashechkin, A.A.; Alekhin, A.I.; Sergeeva, Y.E. Fungal Spores: Dormancy, Germination, Chemical Composition, and Role in Biotechnology (Review). Appl. Biochem. Microbiol. 2012, 48, 1–11. [Google Scholar] [CrossRef]
- Moore, D.; Robson, G.D.; Trinci, A.P.J. 4.2 Spore Germination and Dormancy E. In 21st Century Guidebook to Fungi, 2nd ed.; Cambridge University Press: Cambridge, UK, 2020. [Google Scholar]
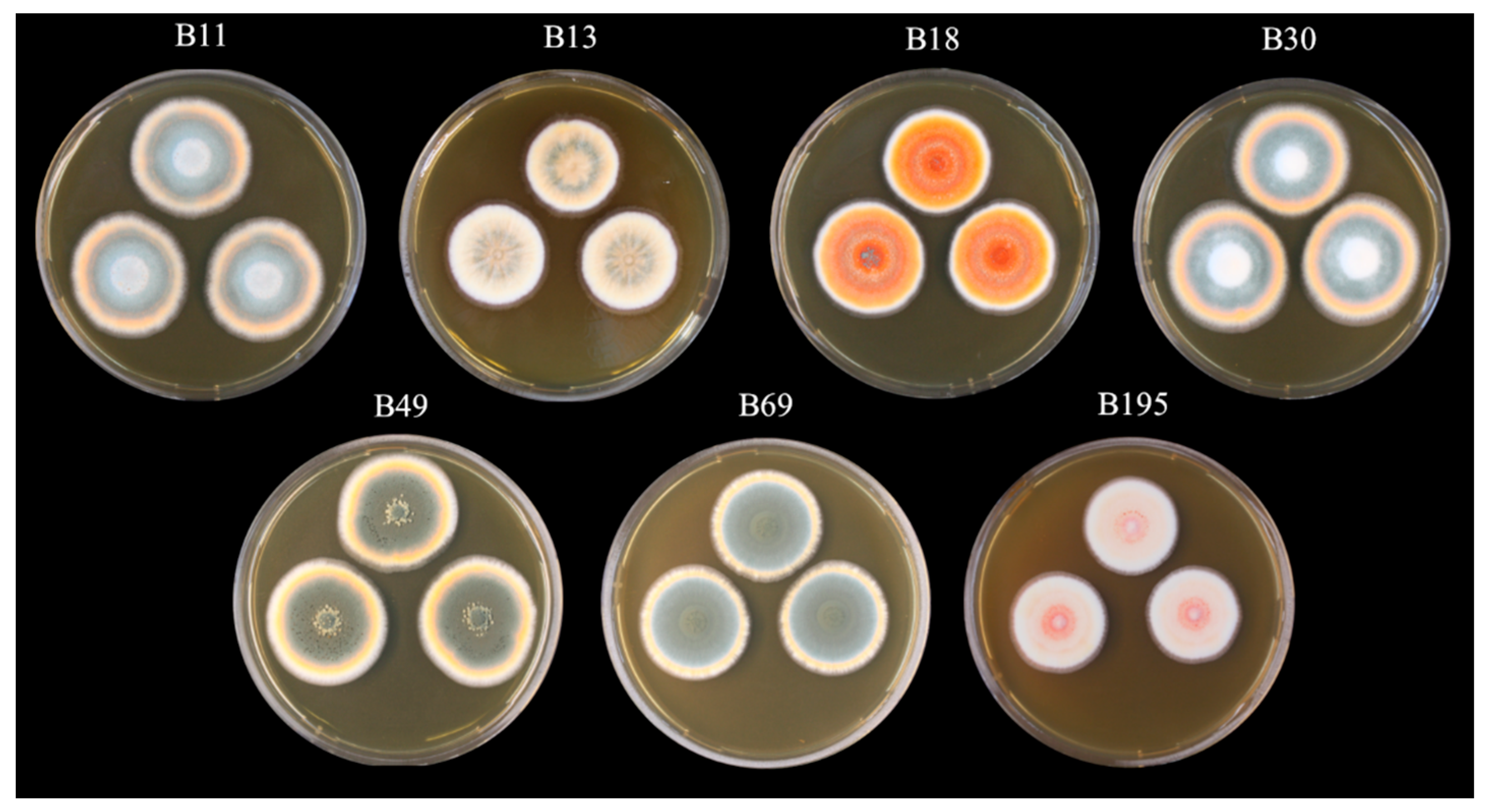




| Fungal Strain | Crude Extract Yield [g] |
|---|---|
| B11 | 5.52 |
| B13 | 3.47 |
| B18 | 1.05 |
| B30 | 4.42 |
| B49 | 5.33 |
| B69 | 3.54 |
| B195 | 1.24 |
| Experimental Group | Mortality [%] | No. of Injected Bees |
|---|---|---|
| B11 | 1.67 | 59 |
| B13 | 1.67 | 59 |
| B18 | 2.36 | 124 |
| B30 | 6.67 | 56 |
| B49 | 4.00 | 120 |
| B69 | 5.00 | 76 |
| B195 | 50.00 | 10 |
| no extract | 3.75 | 77 |
| mock acetone | 5.00 | 76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vocadlova, K.; Lamp, B.; Benes, K.; Matha, V.; Lee, K.-Z.; Vilcinskas, A. Crude Extracts of Talaromyces Strains (Ascomycota) Affect Honey Bee (Apis mellifera) Resistance to Chronic Bee Paralysis Virus. Viruses 2023, 15, 343. https://doi.org/10.3390/v15020343
Vocadlova K, Lamp B, Benes K, Matha V, Lee K-Z, Vilcinskas A. Crude Extracts of Talaromyces Strains (Ascomycota) Affect Honey Bee (Apis mellifera) Resistance to Chronic Bee Paralysis Virus. Viruses. 2023; 15(2):343. https://doi.org/10.3390/v15020343
Chicago/Turabian StyleVocadlova, Katerina, Benjamin Lamp, Karel Benes, Vladimir Matha, Kwang-Zin Lee, and Andreas Vilcinskas. 2023. "Crude Extracts of Talaromyces Strains (Ascomycota) Affect Honey Bee (Apis mellifera) Resistance to Chronic Bee Paralysis Virus" Viruses 15, no. 2: 343. https://doi.org/10.3390/v15020343
APA StyleVocadlova, K., Lamp, B., Benes, K., Matha, V., Lee, K.-Z., & Vilcinskas, A. (2023). Crude Extracts of Talaromyces Strains (Ascomycota) Affect Honey Bee (Apis mellifera) Resistance to Chronic Bee Paralysis Virus. Viruses, 15(2), 343. https://doi.org/10.3390/v15020343







