Cellular Release of Infectious Hepatitis C Virus Particles via Endosomal Pathways
Abstract
1. Introduction
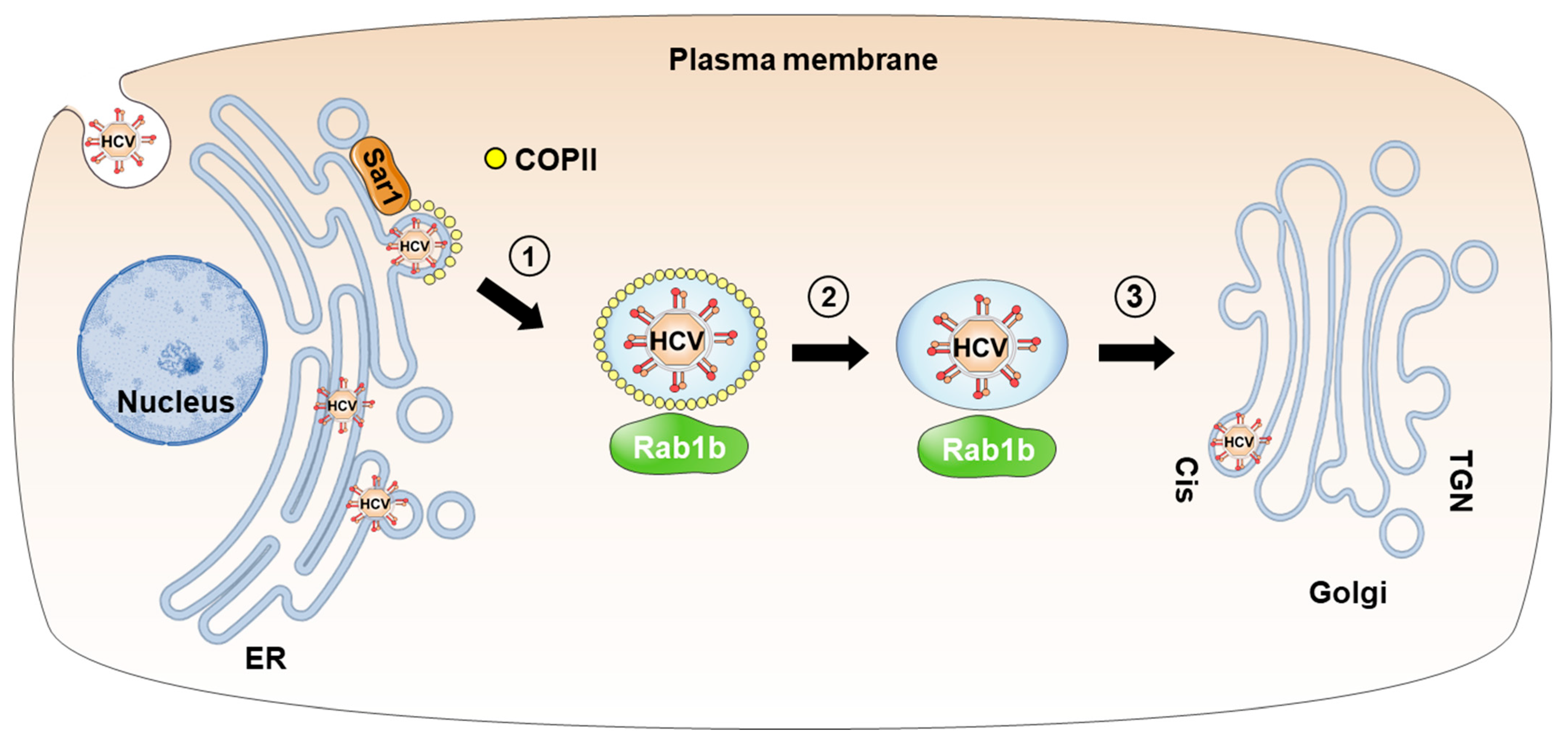
2. Transport of HCV Particles from the ER to the Golgi Apparatus in Coat Protein Complex II (COPII)-Coated Vesicles
2.1. COPII Vesicles and Rab1 GTPases
2.2. Transport of HCV Particles via ER-to-Golgi Trafficking
3. Transport of HCV Particles from the Golgi Apparatus to Recycling Endosomes
3.1. Golgi Apparatus and Cellular Proteins Required for Transport from Golgi to Recycling Endosomes
3.2. Transport of HCV Particle via TGN-to-Recycling Endosomes Trafficking
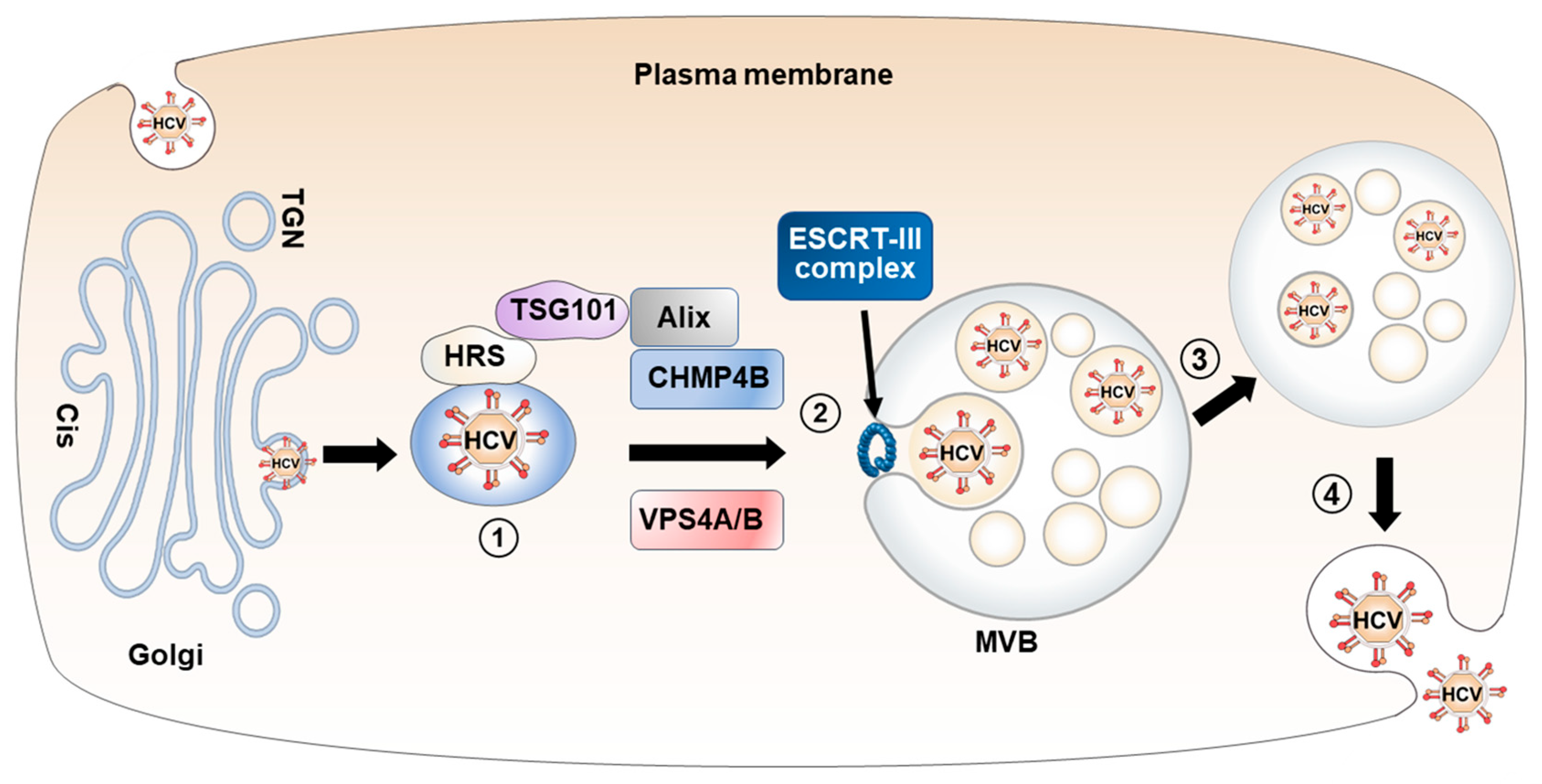
4. Transport of HCV Particles from the Golgi Apparatus to MVBs
4.1. MVB Biogenesis and ESCRT Machinery
4.2. Transport of HCV Particle via TGN-to-MVB Trafficking
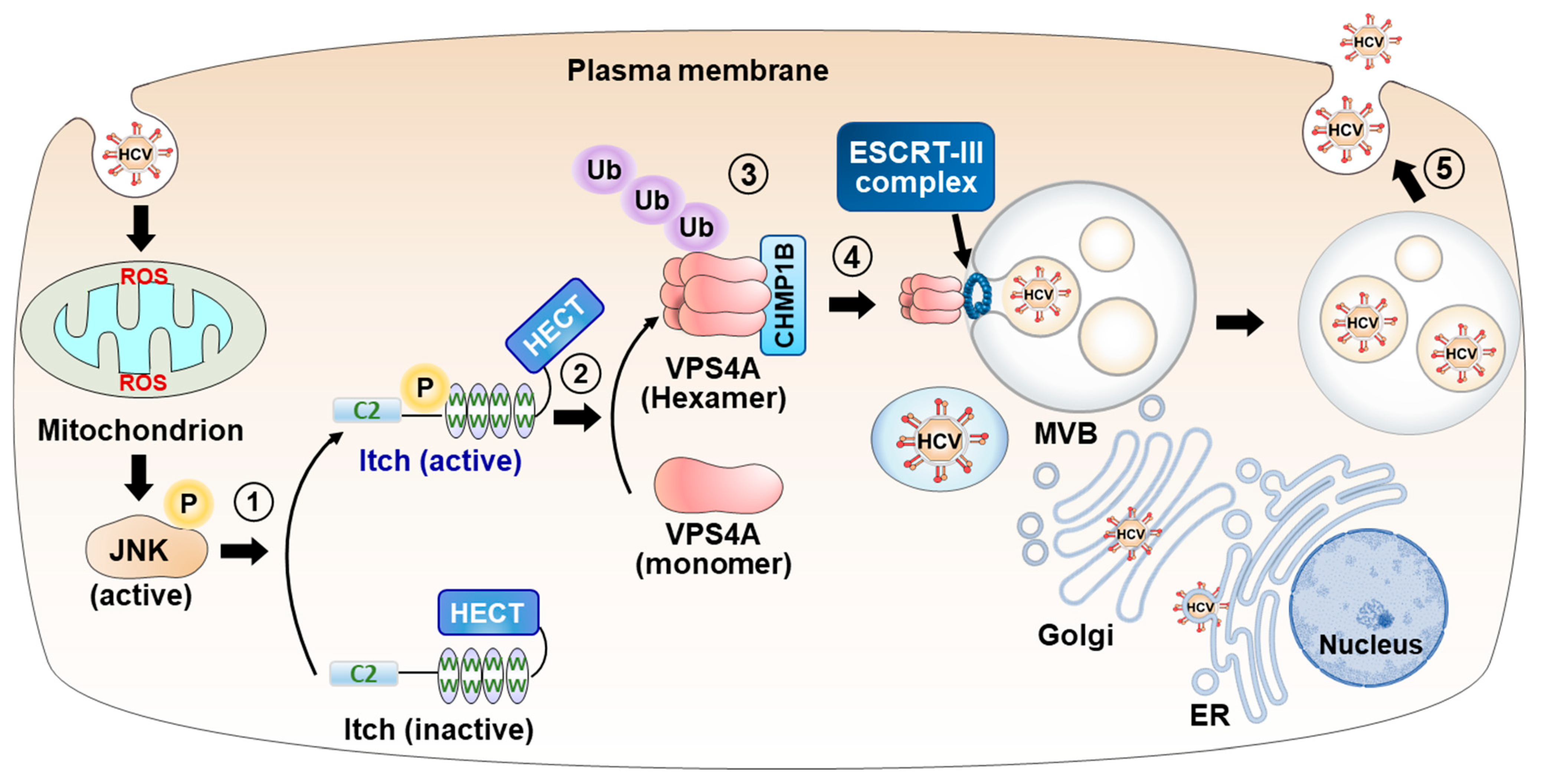
4.3. HCV-Induced ROS/JNK/Itch Signaling Pathway Promotes VSP4A Polyubiquitylation, Leading to Enhancement of VPS4A ATPase Activity, Thereby Upregulating the Release of HCV Particles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Hepatitis C. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 18 July 2023).
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef]
- Vermehren, J.; Park, J.S.; Jacobson, I.M.; Zeuzem, S. Challenges and perspectives of direct antivirals for the treatment of hepatitis C virus infection. J. Hepatol. 2018, 69, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, A.; Cooke, G.S.; Nayagam, S.; Thursz, M.; Hallett, T.B. Scaling up prevention and treatment towards the elimination of hepatitis C: A global mathematical model. Lancet 2019, 393, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Manns, M.P.; Maasoumy, B. Breakthroughs in hepatitis C research: From discovery to cure. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 533–550. [Google Scholar] [CrossRef] [PubMed]
- Scheel, T.K.; Rice, C.M. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 2013, 19, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Moradpour, D.; Penin, F.; Rice, C.M. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007, 5, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Unravelling hepatitis C virus replication from genome to function. Nature 2005, 436, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013, 11, 688–700. [Google Scholar] [CrossRef]
- Coller, K.E.; Berger, K.L.; Heaton, N.S.; Cooper, J.D.; Yoon, R.; Randall, G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 2009, 5, e1000702. [Google Scholar] [CrossRef]
- Gerold, G.; Moeller, R.; Pietschmann, T. Hepatitis C Virus Entry: Protein Interactions and Fusion Determinants Governing Productive Hepatocyte Invasion. Cold Spring Harb. Perspect. Med. 2020, 10, a036830. [Google Scholar] [CrossRef]
- Gosert, R.; Egger, D.; Lohmann, V.; Bartenschlager, R.; Blum, H.E.; Bienz, K.; Moradpour, D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 2003, 77, 5487–5492. [Google Scholar] [CrossRef] [PubMed]
- Egger, D.; Wolk, B.; Gosert, R.; Bianchi, L.; Blum, H.E.; Moradpour, D.; Bienz, K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002, 76, 5974–5984. [Google Scholar] [CrossRef]
- Huang, L.Y.; Hwang, J.; Sharma, S.D.; Hargittai, M.R.S.; Chen, Y.F.; Arnold, J.J.; Raney, K.D.; Cameron, C.E. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 2005, 280, 36417–36428. [Google Scholar] [CrossRef]
- Appel, N.; Zayas, M.; Miller, S.; Krijnse-Locker, J.; Schaller, T.; Friebe, P.; Kallis, S.; Engel, U.; Bartenschlager, R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008, 4, e1000035. [Google Scholar] [CrossRef] [PubMed]
- Pietschmann, T.; Zayas, M.; Meuleman, P.; Long, G.; Appel, N.; Koutsoudakis, G.; Kallis, S.; Leroux-Roels, G.; Lohmann, V.; Bartenschlager, R. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 2009, 5, e1000475. [Google Scholar] [CrossRef] [PubMed]
- Counihan, N.A.; Rawlinson, S.M.; Lindenbach, B.D. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog. 2011, 7, e1002302. [Google Scholar] [CrossRef] [PubMed]
- Shimotohno, K. HCV Assembly and Egress via Modifications in Host Lipid Metabolic Systems. Cold Spring Harb. Perspect. Med. 2021, 11, a036814. [Google Scholar] [CrossRef]
- Timpe, J.M.; Stamataki, Z.; Jennings, A.; Hu, K.; Farquhar, M.J.; Harris, H.J.; Schwarz, A.; Desombere, I.; Roels, G.L.; Bafe, P.; et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 2008, 47, 17–24. [Google Scholar] [CrossRef]
- Witteveldt, J.; Evans, M.J.; Bitzegeio, J.; Koutsoudakis, G.; Owsianka, A.M.; Angus, A.G.; Keck, Z.Y.; Foung, S.K.; Pietschmann, T.; Rice, C.M.; et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 2009, 90, 48–58. [Google Scholar] [CrossRef]
- Mankouri, J.; Walter, C.; Stewart, H.; Bentham, M.; Park, W.S.; Heo, W.D.; Fukuda, M.; Griffin, S.; Harris, M. Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion. J. Virol. 2016, 90, 7159–7170. [Google Scholar] [CrossRef] [PubMed]
- Coller, K.E.; Heaton, N.S.; Berger, K.L.; Cooper, J.D.; Saunders, J.L.; Randall, G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012, 8, e1002466. [Google Scholar] [CrossRef] [PubMed]
- Syed, G.H.; Khan, M.; Yang, S.; Siddiqui, A. Hepatitis C Virus Lipoviroparticles Assemble in the Endoplasmic Reticulum (ER) and Bud off from the ER to the Golgi Compartment in COPII Vesicles. J. Virol. 2017, 91, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Liang, Y.; Ariffianto, A.; Matsui, C.; Abe, T.; Muramatsu, M.; Wakita, T.; Maki, M.; Shibata, H.; Shoji, I. Hepatitis C Virus-Induced ROS/JNK Signaling Pathway Activates the E3 Ubiquitin Ligase Itch to Promote the Release of HCV Particles via Polyubiquitylation of VPS4A. J. Virol. 2022, 96, e0181121. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y.; Kuroki, M.; Maki, M.; Ikeda, M.; Dansako, H.; Wakita, T.; Kato, N. The ESCRT system is required for hepatitis C virus production. PLoS ONE 2011, 6, e14517. [Google Scholar] [CrossRef] [PubMed]
- Corless, L.; Crump, C.M.; Griffin, S.D.; Harris, M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J. Gen. Virol. 2010, 91, 362–372. [Google Scholar] [CrossRef]
- Shrivastava, S.; Devhare, P.; Sujijantarat, N.; Steele, R.; Kwon, Y.C.; Ray, R.; Ray, R.B. Knockdown of Autophagy Inhibits Infectious Hepatitis C Virus Release by the Exosomal Pathway. J. Virol. 2016, 90, 1387–1396. [Google Scholar] [CrossRef]
- Bunz, M.; Ritter, M.; Schindler, M. HCV egress—Unconventional secretion of assembled viral particles. Trends Microbiol. 2022, 30, 364–378. [Google Scholar] [CrossRef]
- Merquiol, E.; Uzi, D.; Mueller, T.; Goldenberg, D.; Nahmias, Y.; Xavier, R.J.; Tirosh, B.; Shibolet, O. HCV Causes Chronic Endoplasmic Reticulum Stress Leading to Adaptation and Interference with the Unfolded Protein Response. PLoS ONE 2011, 6, e24660. [Google Scholar] [CrossRef] [PubMed]
- Bayer, K.; Banning, C.; Bruss, V.; Wiltzer-Bach, L.; Schindler, M. Hepatitis C Virus Is Released via a Noncanonical Secretory Route. J. Virol. 2016, 90, 10558–10573. [Google Scholar] [CrossRef]
- Emr, S.; Glick, B.S.; Linstedt, A.D.; Lippincott-Schwartz, J.; Luini, A.; Malhotra, V.; Marsh, B.J.; Nakano, A.; Pfeffer, S.R.; Rabouille, C.; et al. Journeys through the Golgi--taking stock in a new era. J. Cell Biol. 2009, 187, 449–453. [Google Scholar] [CrossRef]
- Yarwood, R.; Hellicar, J.; Woodman, P.G.; Lowe, M. Membrane trafficking in health and disease. Dis. Models Mech. 2020, 13, dmm043448. [Google Scholar] [CrossRef] [PubMed]
- Jensen, D.; Schekman, R. COPII-mediated vesicle formation at a glance. J. Cell Sci. 2011, 124, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Barlowe, C.; Orci, L.; Yeung, T.; Hosobuchi, M.; Hamamoto, S.; Salama, N.; Rexach, M.F.; Ravazzola, M.; Amherdt, M.; Schekman, R. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 1994, 77, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Barlowe, C. Signals for COPII-dependent export from the ER: What’s the ticket out? Trends Cell Biol. 2003, 13, 295–300. [Google Scholar] [CrossRef]
- Allan, B.B.; Moyer, B.D.; Balch, W.E. Rab1 recruitment of p115 into a cis-SNARE complex: Programming budding COPII vesicles for fusion. Science 2000, 289, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Plutner, H.; Cox, A.D.; Pind, S.; Khosravi-Far, R.; Bourne, J.R.; Schwaninger, R.; Der, C.J.; Balch, W.E. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J. Cell Biol. 1991, 115, 31–43. [Google Scholar] [CrossRef]
- Saraste, J.; Lahtinen, U.; Goud, B. Localization of the small GTP-binding protein rab1p to early compartments of the secretory pathway. J. Cell Sci. 1995, 108 Pt 4, 1541–1552. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef]
- Slavin, I.; Garcia, I.A.; Monetta, P.; Martinez, H.; Romero, N.; Alvarez, C. Role of Rab1b in COPII dynamics and function. Eur. J. Cell Biol. 2011, 90, 301–311. [Google Scholar] [CrossRef]
- Stenmark, H.; Olkkonen, V.M. The Rab GTPase family. Genome Biol. 2001, 2, reviews3007.1. [Google Scholar] [CrossRef][Green Version]
- Takacs, C.N.; Andreo, U.; Dao Thi, V.L.; Wu, X.; Gleason, C.E.; Itano, M.S.; Spitz-Becker, G.S.; Belote, R.L.; Hedin, B.R.; Scull, M.A.; et al. Differential Regulation of Lipoprotein and Hepatitis C Virus Secretion by Rab1b. Cell Rep. 2017, 21, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, G.; Simons, K. The trans Golgi network: Sorting at the exit site of the Golgi complex. Science 1986, 234, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Klumperman, J. Architecture of the mammalian Golgi. Cold Spring Harb. Perspect. Biol. 2011, 3, a005181. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Boulan, E.; Musch, A. Protein sorting in the Golgi complex: Shifting paradigms. Biochim. Biophys. Acta 2005, 1744, 455–464. [Google Scholar] [CrossRef]
- Guo, Y.; Sirkis, D.W.; Schekman, R. Protein sorting at the trans-Golgi network. Annu. Rev. Cell Dev. Biol. 2014, 30, 169–206. [Google Scholar] [CrossRef] [PubMed]
- Progida, C.; Bakke, O. Bidirectional traffic between the Golgi and the endosomes—Machineries and regulation. J. Cell Sci. 2016, 129, 3971–3982. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.H.; Walsh, D. Microtubule Regulation and Function during Virus Infection. J. Virol. 2017, 91, 1110–1128. [Google Scholar] [CrossRef]
- Scott, C.C.; Vacca, F.; Gruenberg, J. Endosome maturation, transport and functions. Semin. Cell Dev. Biol. 2014, 31, 2–10. [Google Scholar] [CrossRef]
- Ang, A.L.; Taguchi, T.; Francis, S.; Folsch, H.; Murrells, L.J.; Pypaert, M.; Warren, G.; Mellman, I. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 2004, 167, 531–543. [Google Scholar] [CrossRef]
- Tanaka, N.; Kyuuma, M.; Sugamura, K. Endosomal sorting complex required for transport proteins in cancer pathogenesis, vesicular transport, and non-endosomal functions. Cancer Sci. 2008, 99, 1293–1303. [Google Scholar] [CrossRef]
- Keen, J.H. Clathrin and associated assembly and disassembly proteins. Annu. Rev. Biochem. 1990, 59, 415–438. [Google Scholar] [CrossRef]
- Brodsky, F.M. Diversity of clathrin function: New tricks for an old protein. Annu. Rev. Cell Dev. Biol. 2012, 28, 309–336. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.J.; Collins, B.M.; Evans, P.R. Adaptors for clathrin coats: Structure and function. Annu. Rev. Cell Dev. Biol. 2004, 20, 153–191. [Google Scholar] [CrossRef]
- Nakatsu, F.; Hase, K.; Ohno, H. The Role of the Clathrin Adaptor AP-1: Polarized Sorting and Beyond. Membranes 2014, 4, 747–763. [Google Scholar] [CrossRef] [PubMed]
- Gravotta, D.; Deora, A.; Perret, E.; Oyanadel, C.; Soza, A.; Schreiner, R.; Rodriguez-Boulan, E. AP1B sorts basolateral proteins in recycling and biosynthetic routes of MDCK cells. Proc. Natl. Acad. Sci. USA 2007, 104, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, R.C.; Boehm, M.; Gorshkova, I.; Crouch, R.J.; Tomita, K.; Saito, T.; Ohno, H.; Bonifacino, J.S. Signal-binding specificity of the μ4 subunit of the adaptor protein complex AP-4. J. Biol. Chem. 2001, 276, 13145–13152. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S. The GGA proteins: Adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004, 5, 23–32. [Google Scholar] [CrossRef]
- Nokes, R.L.; Fields, I.C.; Collins, R.N.; Folsch, H. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J. Cell Biol. 2008, 182, 845–853. [Google Scholar] [CrossRef]
- Ullrich, O.; Reinsch, S.; Urbe, S.; Zerial, M.; Parton, R.G. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 1996, 135, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Benedicto, I.; Gondar, V.; Molina-Jimenez, F.; Garcia-Buey, L.; Lopez-Cabrera, M.; Gastaminza, P.; Majano, P.L. Clathrin mediates infectious hepatitis C virus particle egress. J. Virol. 2015, 89, 4180–4190. [Google Scholar] [CrossRef]
- Xiao, F.; Wang, S.; Barouch-Bentov, R.; Neveu, G.; Pu, S.; Beer, M.; Schor, S.; Kumar, S.; Nicolaescu, V.; Lindenbach, B.D.; et al. Interactions between the Hepatitis C Virus Nonstructural 2 Protein and Host Adaptor Proteins 1 and 4 Orchestrate Virus Release. mBio 2018, 9, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Bekerman, E.; Neveu, G.; Shulla, A.; Brannan, J.; Pu, S.Y.; Wang, S.; Xiao, F.; Barouch-Bentov, R.; Bakken, R.R.; Mateo, R.; et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Investig. 2017, 127, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Hanson, P.I. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 2010, 11, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Christ, L.; Raiborg, C.; Wenzel, E.M.; Campsteijn, C.; Stenmark, H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 2017, 42, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Barouch-Bentov, R.; Neveu, G.; Xiao, F.; Beer, M.; Bekerman, E.; Schor, S.; Campbell, J.; Boonyaratanakornkit, J.; Lindenbach, B.; Lu, A.; et al. Hepatitis C Virus Proteins Interact with the Endosomal Sorting Complex Required for Transport (ESCRT) Machinery via Ubiquitination to Facilitate Viral Envelopment. mBio 2016, 7, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.K.; Jeng, K.S.; Machida, K.; Lai, M.M. Hepatitis C virus egress and release depend on endosomal trafficking of core protein. J. Virol. 2010, 84, 11590–11598. [Google Scholar] [CrossRef] [PubMed]
- Sandrin, V.; Boulanger, P.; Penin, F.; Granier, C.; Cosset, F.L.; Bartosch, B. Assembly of functional hepatitis C virus glycoproteins on infectious pseudoparticles occurs intracellularly and requires concomitant incorporation of E1 and E2 glycoproteins. J. Gen. Virol. 2005, 86, 3189–3199. [Google Scholar] [CrossRef]
- Tamai, K.; Shiina, M.; Tanaka, N.; Nakano, T.; Yamamoto, A.; Kondo, Y.; Kakazu, E.; Inoue, J.; Fukushima, K.; Sano, K.; et al. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology 2012, 422, 377–385. [Google Scholar] [CrossRef]
- Babst, M.; Wendland, B.; Estepa, E.J.; Emr, S.D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998, 17, 2982–2993. [Google Scholar] [CrossRef]
- Merrill, S.A.; Hanson, P.I. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem. 2010, 285, 35428–35438. [Google Scholar] [CrossRef]
- Norgan, A.P.; Davies, B.A.; Azmi, I.F.; Schroeder, A.S.; Payne, J.A.; Lynch, G.M.; Xu, Z.; Katzmann, D.J. Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J. Biol. Chem. 2013, 288, 26147–26156. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Adachi, T.; Kitayama, K.; Bungyoku, Y.; Kitazawa, S.; Ishido, S.; Shoji, I.; Hotta, H. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J. Virol. 2008, 82, 10375–10385. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Shoji, I.; Ogawa, W.; Kaneda, S.; Soga, T.; Jiang, D.P.; Ide, Y.H.; Hotta, H. Hepatitis C virus infection promotes hepatic gluconeogenesis through an NS5A-mediated, FoxO1-dependent pathway. J. Virol. 2011, 85, 8556–8568. [Google Scholar] [CrossRef]
- Lin, W.; Tsai, W.L.; Shao, R.X.; Wu, G.; Peng, L.F.; Barlow, L.L.; Chung, W.J.; Zhang, L.; Zhao, H.; Jang, J.Y.; et al. Hepatitis C virus regulates transforming growth factor beta1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gastroenterology 2010, 138, 2509–2518.e1. [Google Scholar] [CrossRef] [PubMed]
- Qadri, I.; Iwahashi, M.; Capasso, J.M.; Hopken, M.W.; Flores, S.; Schaack, J.; Simon, F.R. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: Role of JNK, p38 MAPK and AP-1. Biochem. J. 2004, 378, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Bernassola, F.; Karin, M.; Ciechanover, A.; Melino, G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell 2008, 14, 10–21. [Google Scholar] [CrossRef]
- Melino, G.; Gallagher, E.; Aqeilan, R.I.; Knight, R.; Peschiaroli, A.; Rossi, M.; Scialpi, F.; Malatesta, M.; Zocchi, L.; Browne, G.; et al. Itch: A HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008, 15, 1103–1112. [Google Scholar] [CrossRef]
- Zhu, K.; Shan, Z.; Chen, X.; Cai, Y.; Cui, L.; Yao, W.; Wang, Z.; Shi, P.; Tian, C.; Lou, J.; et al. Allosteric auto-inhibition and activation of the Nedd4 family E3 ligase Itch. EMBO Rep. 2017, 18, 1618–1630. [Google Scholar] [CrossRef]
- Gallagher, E.; Gao, M.; Liu, Y.C.; Karin, M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. USA 2006, 103, 1717–1722. [Google Scholar] [CrossRef]
- Su, W.C.; Chen, Y.C.; Tseng, C.H.; Hsu, P.W.; Tung, K.F.; Jeng, K.S.; Lai, M.M. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc. Natl. Acad. Sci. USA 2013, 110, 17516–17521. [Google Scholar] [CrossRef]
- Han, Z.; Sagum, C.A.; Bedford, M.T.; Sidhu, S.S.; Sudol, M.; Harty, R.N. ITCH E3 Ubiquitin Ligase Interacts with Ebola Virus VP40 to Regulate Budding. J. Virol. 2016, 90, 9163–9171. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Liu, G.T.; Kung, H.N.; Liu, P.T.; Liao, Y.T.; Chow, L.P.; Chang, L.S.; Chang, Y.H.; Chang, C.W.; Shu, W.C.; et al. The Ubiquitin Ligase Itch and Ubiquitination Regulate BFRF1-Mediated Nuclear Envelope Modification for Epstein-Barr Virus Maturation. J. Virol. 2016, 90, 8994–9007. [Google Scholar] [CrossRef] [PubMed]
- Dorjbal, B.; Derse, D.; Lloyd, P.; Soheilian, F.; Nagashima, K.; Heidecker, G. The role of ITCH protein in human T-cell leukemia virus type 1 release. J. Biol. Chem. 2011, 286, 31092–31104. [Google Scholar] [CrossRef] [PubMed]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Côté, M.; Rich, R.L.; et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.D.; Hensley, L.; McKnight, K.L.; Hu, F.Y.; Madden, V.; Ping, L.F.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Wirblich, C.; Bhattacharya, B.; Roy, P. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J. Virol. 2006, 80, 460–473. [Google Scholar] [CrossRef]
- Zheng, Y.C.; Wang, M.F.; Li, S.T.; Bu, Y.N.; Xu, Z.C.; Zhu, G.G.; Wu, C.J.; Zhao, K.T.; Li, A.X.; Chen, Q.; et al. Hepatitis B virus hijacks TSG101 to facilitate egress via multiple vesicle bodies. PLoS Pathog. 2023, 19, e1011382. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Matsuura, T.; Aoyagi, H.; Matsuda, M.; Hmwe, S.S.; Date, T.; Watanabe, N.; Watashi, K.; Suzuki, R.; Ichinose, S.; et al. Antiviral Activity of Glycyrrhizin against Hepatitis C Virus. PLoS ONE 2013, 8, e68992. [Google Scholar] [CrossRef]

| Protein Names | Role in the Cell | Role in HCV Infection | References |
|---|---|---|---|
| Sar1 | COPII-vesicle formation, ER-Golgi trafficking | KD leads to inhibition of release | [22] |
| Rab1b | COPII-dependent ER to Golgi trafficking | DN expression reduces release | [42] |
| COPII | Traffic from ER to Golgi | Budding from ER | [23] |
| Clathrin | Vesicle formation at TGN, endosomes, and plasma membrane | KD decreases extracellular infectivity and RNA amount | [61] |
| AP-1A | TGN to recycling endosome bidirectional transport | KD decreases extracellular infectivity and RNA amount | [22,61,62] |
| GGA2 | TGN to recycling endosome transport | KD decreases extracellular infectivity | [21] |
| Rab 11A | TGN to recycling endosome transport | KD results in accumulation of core at the Golgi | [22] |
| Rab 13 | TGN to recycling endosome transport | DN leads to inhibition of release | [21] |
| VPS4A | Late steps of MVB biogenesis, membrane fission | KD or DN expression reduces extracellular infectivity | [24,26] |
| VPS4B | Late steps of MVB biogenesis, membrane fission | KD or DN expression reduces extracellular infectivity | [25,26] |
| CHMP4B | Subunit of ESCRT-III complex, membrane fission | KD or DN expression reduces extracellular infectivity | [25,26] |
| TSG101 | Subunit of ESCRT-0, cargo sorting | KD reduces extracellular infectivity | [25] |
| Alix | MVB biogenesis | KD reduces extracellular infectivity | [25,66] |
| Itch | HECT-type E3 ubiquitin ligase | KD reduces extracellular infectivity | [24] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, L.; Solichin, M.R.; Adyaksa, D.N.M.; Septianastiti, M.A.; Fitri, R.A.; Suwardan, G.N.R.; Matsui, C.; Abe, T.; Shoji, I. Cellular Release of Infectious Hepatitis C Virus Particles via Endosomal Pathways. Viruses 2023, 15, 2430. https://doi.org/10.3390/v15122430
Deng L, Solichin MR, Adyaksa DNM, Septianastiti MA, Fitri RA, Suwardan GNR, Matsui C, Abe T, Shoji I. Cellular Release of Infectious Hepatitis C Virus Particles via Endosomal Pathways. Viruses. 2023; 15(12):2430. https://doi.org/10.3390/v15122430
Chicago/Turabian StyleDeng, Lin, Muchamad Ridotu Solichin, Dewa Nyoman Murti Adyaksa, Maria Alethea Septianastiti, Rhamadianti Aulia Fitri, Gede Ngurah Rsi Suwardan, Chieko Matsui, Takayuki Abe, and Ikuo Shoji. 2023. "Cellular Release of Infectious Hepatitis C Virus Particles via Endosomal Pathways" Viruses 15, no. 12: 2430. https://doi.org/10.3390/v15122430
APA StyleDeng, L., Solichin, M. R., Adyaksa, D. N. M., Septianastiti, M. A., Fitri, R. A., Suwardan, G. N. R., Matsui, C., Abe, T., & Shoji, I. (2023). Cellular Release of Infectious Hepatitis C Virus Particles via Endosomal Pathways. Viruses, 15(12), 2430. https://doi.org/10.3390/v15122430






