Human Mannose Receptor 1 Attenuates HIV-1 Infectivity in a Virus Isolate-Specific Manner
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Plasmid and Viral Vectors
2.3. Transmitted/Founder (T/F) Viruses
2.4. Antibodies
2.5. Transient Transfection of HEK293T Cells
2.6. Quantitation of Extracellular Virus Using RT Assay
2.7. Immunoblot Analysis
2.8. Viral Infectivity Assay
2.9. Determination of Viral Tropism
2.10. OptiPrep Density Gradient Ultracentrifugation
2.11. Co-Immunoprecipitation Analysis of Env and hMRC1
2.12. Glycoprotein Analysis
3. Results
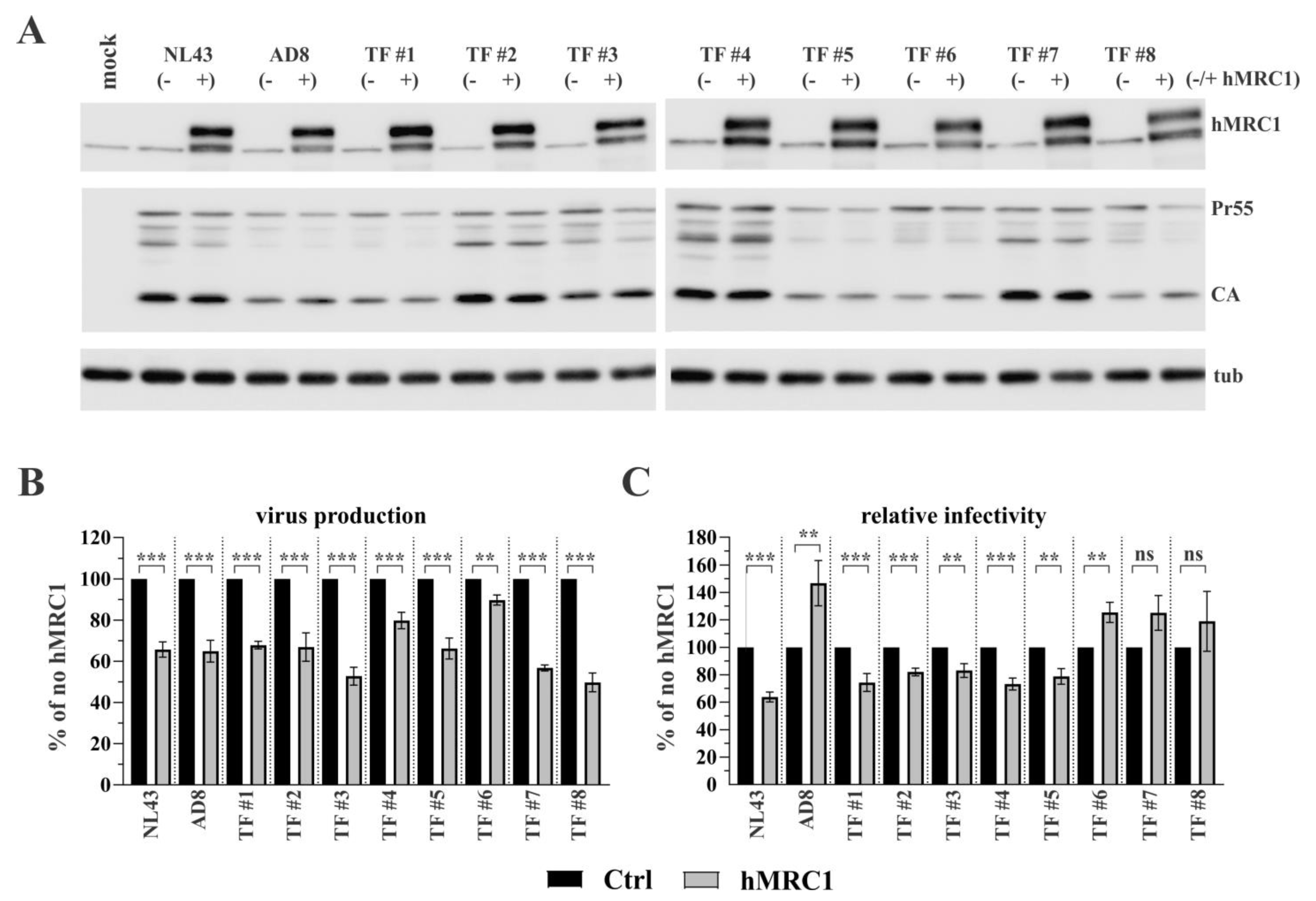
3.1. Exogenous Expression of hMRC1 Attenuates Virus Infectivity in an Isolate-Specific Manner
3.2. R5-Tropic T/F Viruses Vary in Their Sensitivity to Inhibition by hMRC1
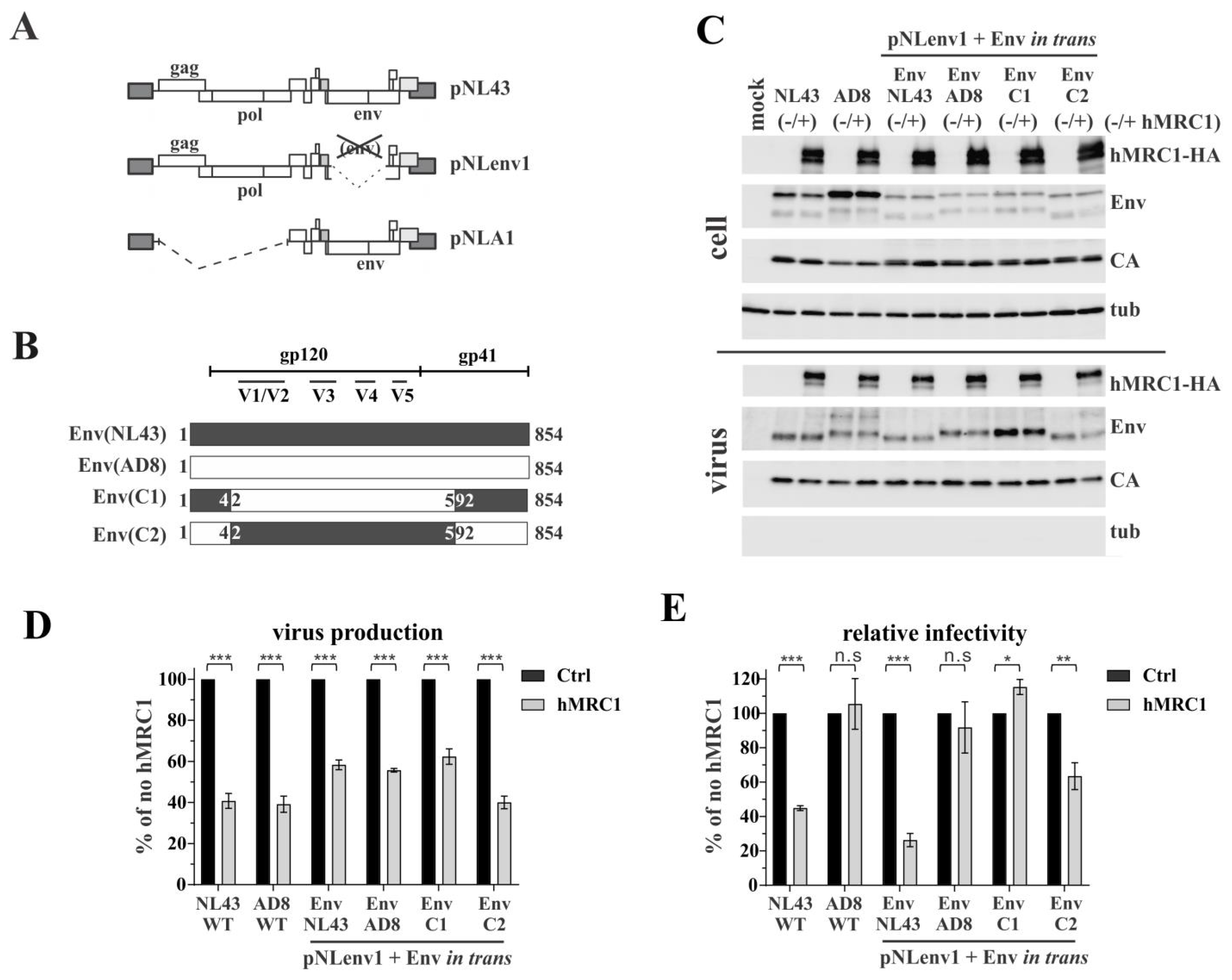
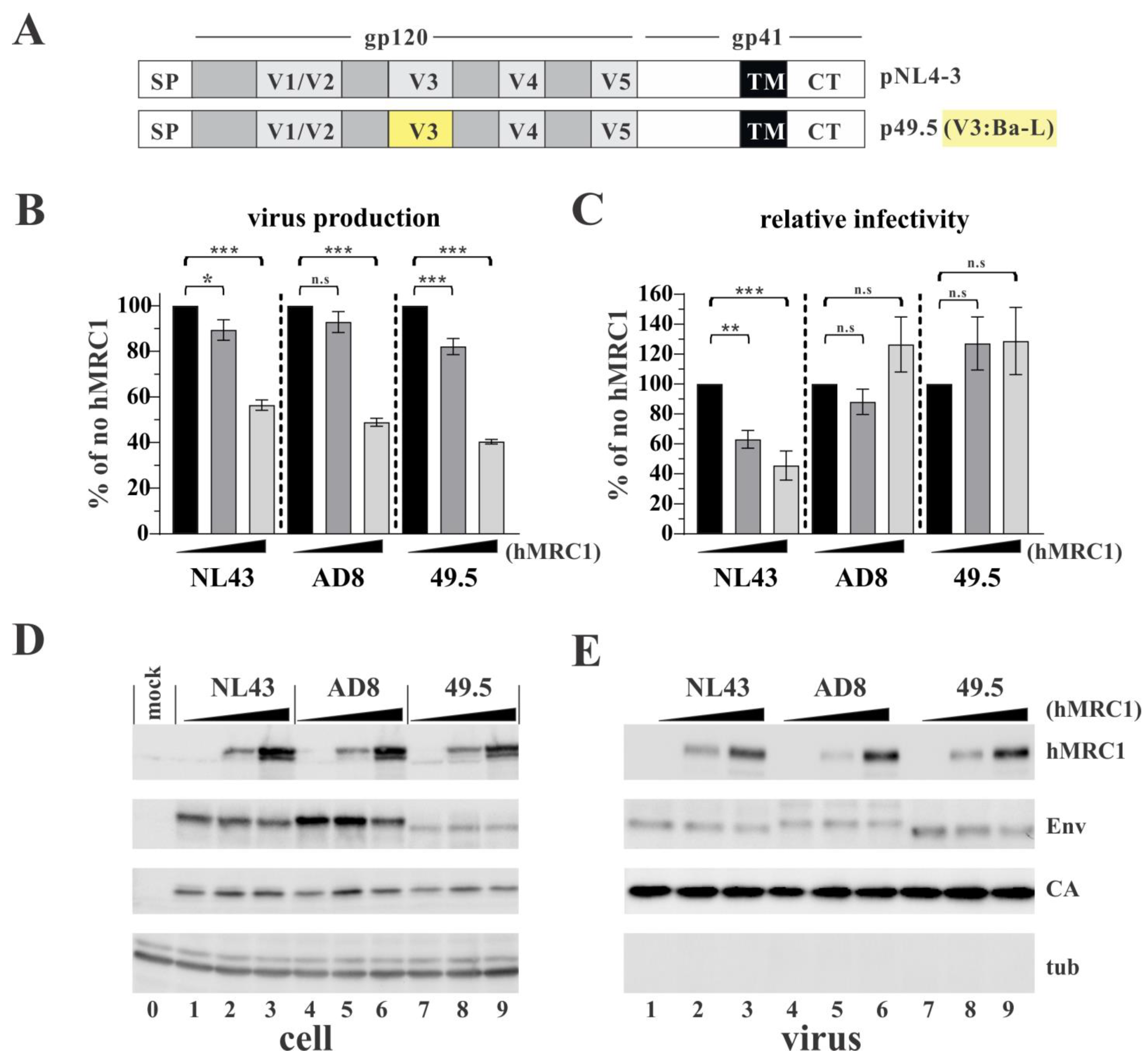
3.3. The Env V3 Region Is One of the Key Determinants for hMRC1-Mediated Attenuation of Virus Infectivity
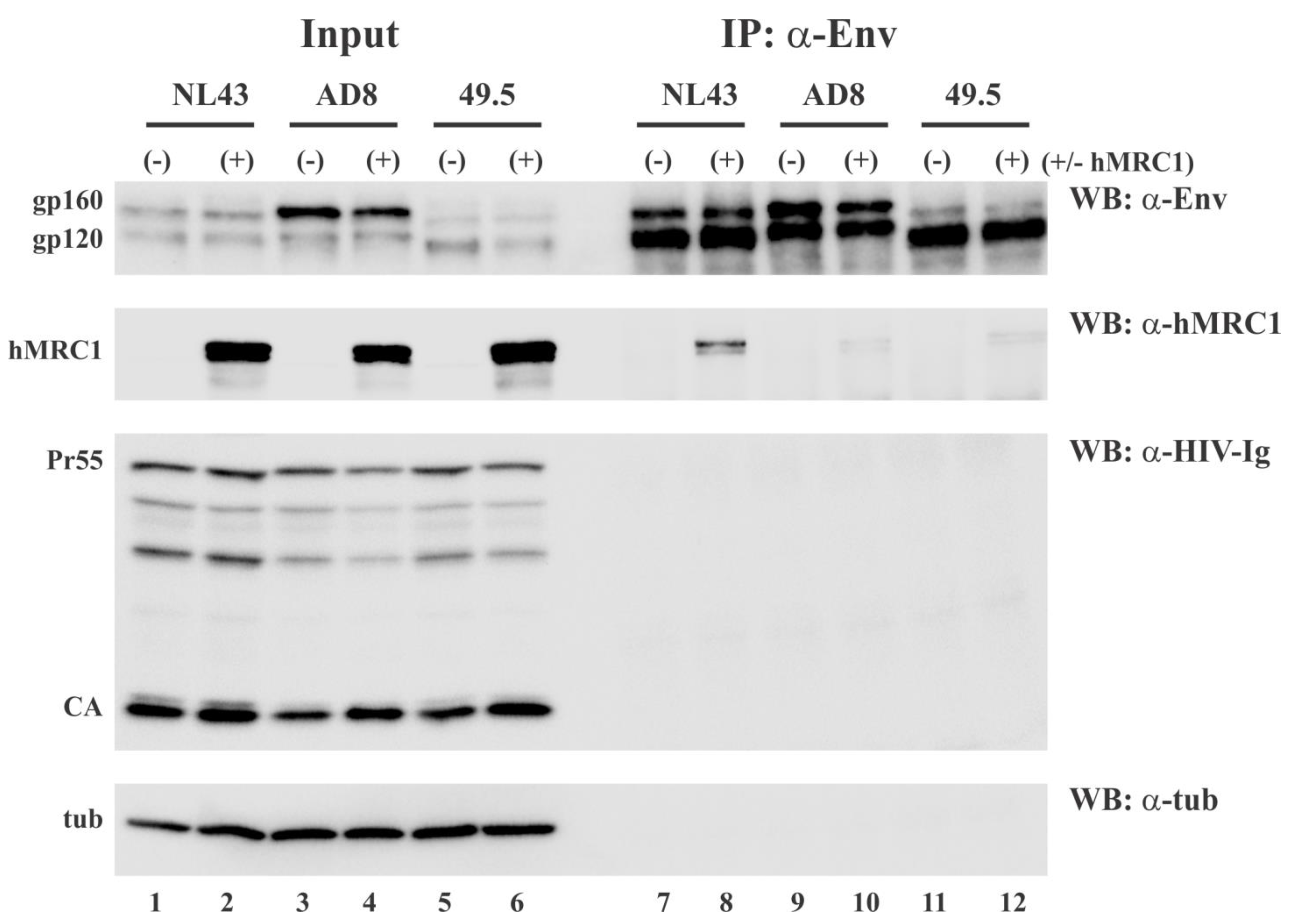
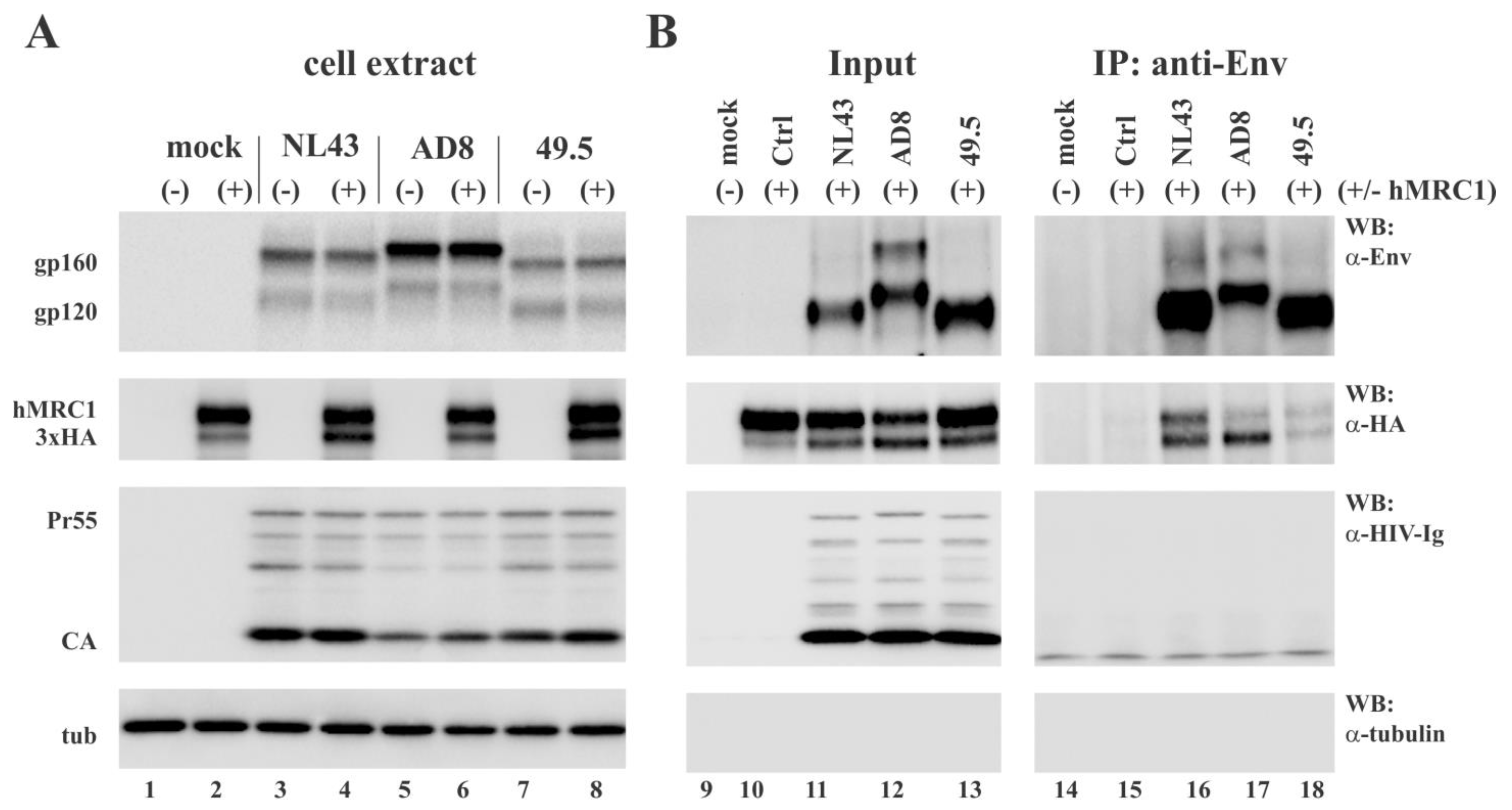
3.4. hMRC1 Interacts with NL43 Env but Not Env from AD8 and 49.5
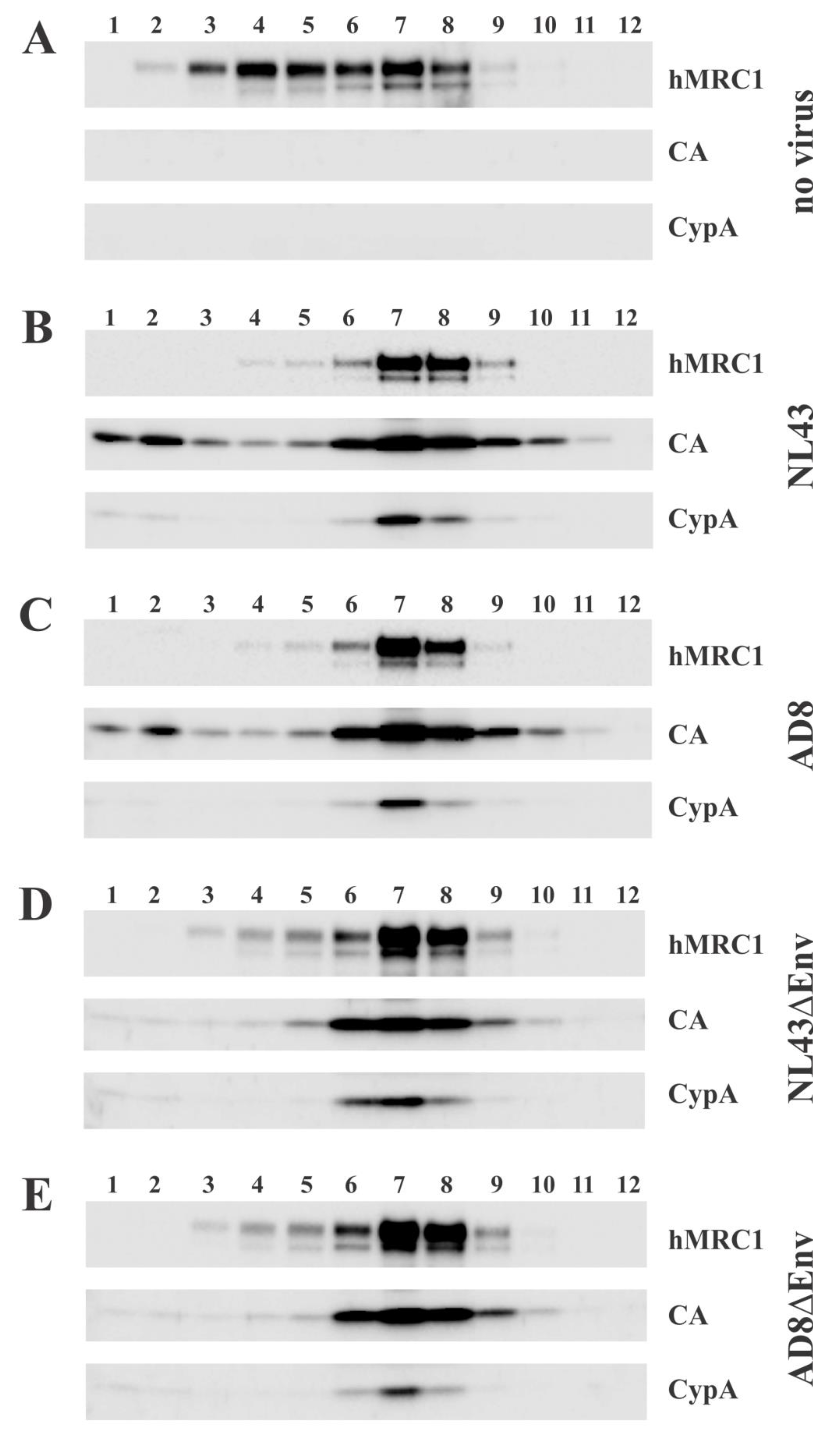
3.5. hMRC1 Is Packaged into Viral Particles in an Env Independent Manner
3.6. Virus-Incorporated hMRC1 Binds to NL43 Env but Only Weakly Interacts with the Env Proteins of AD8 and the Closely Related 49.5 Virus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Azad, A.K.; Rajaram, M.V.; Schlesinger, L.S. Exploitation of the Macrophage Mannose Receptor (CD206) in Infectious Disease Diagnostics and Therapeutics. J. Cytol. Mol. Biol. 2014, 1, 1000003. [Google Scholar]
- Stahl, P.D.; Ezekowitz, R.A. The mannose receptor is a pattern recognition receptor involved in host defense. Curr. Opin. Immunol. 1998, 10, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.R.; Gordon, S.; Martinez-Pomares, L. The mannose receptor: Linking homeostasis and immunity through sugar recognition. Trends Immunol. 2005, 26, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, J.R.; Rogers, R.; Molina, R.M.; Dangond, F.; McLane, M.F.; Essex, M.; Brain, J.D. Noninfectious entry of HIV-1 into peripheral and brain macrophages mediated by the mannose receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 5097–5102. [Google Scholar] [CrossRef]
- Pontow, S.E.; Kery, V.; Stahl, P.D. Mannose receptor. Int. Rev. Cytol. 1992, 137B, 221–244. [Google Scholar]
- Astarie-Dequeker, C.; N’Diaye, E.N.; Le Cabec, V.; Rittig, M.G.; Prandi, J.; Maridonneau-Parini, I. The mannose receptor mediates uptake of pathogenic and nonpathogenic mycobacteria and bypasses bactericidal responses in human macrophages. Infect. Immun. 1999, 67, 469–477. [Google Scholar] [CrossRef]
- Ezekowitz, R.A.; Williams, D.J.; Koziel, H.; Armstrong, M.Y.; Warner, A.; Richards, F.F.; Rose, R.M. Uptake of Pneumocystis carinii mediated by the macrophage mannose receptor. Nature 1991, 351, 155–158. [Google Scholar] [CrossRef]
- Allavena, P.; Chieppa, M.; Monti, P.; Piemonti, L. From pattern recognition receptor to regulator of homeostasis: The double-faced macrophage mannose receptor. Crit. Rev. Immunol. 2004, 24, 179–192. [Google Scholar] [CrossRef]
- Ezekowitz, R.A.; Austyn, J.; Stahl, P.D.; Gordon, S. Surface properties of bacillus Calmette-Guerin-activated mouse macrophages. Reduced expression of mannose-specific endocytosis, Fc receptors, and antigen F4/80 accompanies induction of Ia. J. Exp. Med. 1981, 154, 60–76. [Google Scholar] [CrossRef]
- Basu, N.; Sett, R.; Das, P.K. Down-regulation of mannose receptors on macrophages after infection with Leishmania donovani. Biochem. J. 1991, 277, 451–456. [Google Scholar] [CrossRef]
- Shepherd, V.L.; Lane, K.B.; Abdolrasulnia, R. Ingestion of Candida albicans down-regulates mannose receptor expression on rat macrophages. Arch. Biochem. Biophys. 1997, 344, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Koziel, H.; Eichbaum, Q.; Kruskal, B.A.; Pinkston, P.; Rogers, R.A.; Armstrong, M.Y.; Richards, F.F.; Rose, R.M.; Ezekowitz, R.A. Reduced binding and phagocytosis of Pneumocystis carinii by alveolar macrophages from persons infected with HIV-1 correlates with mannose receptor downregulation. J. Clin. Investig. 1998, 102, 1332–1344. [Google Scholar] [CrossRef] [PubMed]
- Lubow, J.; Collins, D.R.; Mashiba, M.; Peterson, B.; Virgilio, M.; Collins, K.L. Mannose receptor is a restriction factor of HIV in macrophages and is counteracted by the accessory protein Vpr. BioRxiv 2019. [Google Scholar] [CrossRef]
- Caldwell, R.L.; Egan, B.S.; Shepherd, V.L. HIV-1 Tat represses transcription from the mannose receptor promoter. J. Immunol. 2000, 165, 7035–7041. [Google Scholar] [CrossRef]
- Vigerust, D.J.; Egan, B.S.; Shepherd, V.L. HIV-1 Nef mediates post-translational down-regulation and redistribution of the mannose receptor. J. Leukoc. Biol. 2005, 77, 522–534. [Google Scholar] [CrossRef]
- Lubow, J.; Virgilio, M.C.; Merlino, M.; Collins, D.R.; Mashiba, M.; Peterson, B.G.; Lukic, Z.; Painter, M.M.; Gomez-Rivera, F.; Terry, V.; et al. Mannose receptor is an HIV restriction factor counteracted by Vpr in macrophages. eLife 2020, 9, e51035. [Google Scholar] [CrossRef]
- Kao, S.; Miyagi, E.; Mallorson, R.; Saito, H.; Sukegawa, S.; Mukherji, A.; Mateja, A.; Ferhadian, D.; Fabryova, H.; Clouse, K.; et al. The Myeloid-Specific Transcription Factor PU.1 Upregulates Mannose Receptor Expression but Represses Basal Activity of the HIV-LTR Promoter. J. Virol. 2022, 96, e00652-22. [Google Scholar] [CrossRef]
- Sukegawa, S.; Miyagi, E.; Bouamr, F.; Farkasova, H.; Strebel, K. Mannose Receptor 1 Restricts HIV Particle Release from Infected Macrophages. Cell Rep. 2018, 22, 786–795. [Google Scholar] [CrossRef]
- Hoffman, T.L.; Doms, R.W. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 1999, 16, 57–65. [Google Scholar] [CrossRef]
- Strebel, K.; Klimkait, T.; Martin, M.A. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 1988, 241, 1221–1223. [Google Scholar] [CrossRef]
- Adachi, A.; Gendelman, H.E.; Koenig, S.; Folks, T.; Willey, R.; Rabson, A.; Martin, M.A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986, 59, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Theodore, T.S.; Englund, G.; Buckler-White, A.; Buckler, C.E.; Martin, M.A.; Peden, K.W. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 1996, 12, 191–194. [Google Scholar] [CrossRef]
- Clavel, F.; Guyader, M.; Guetard, D.; Salle, M.; Montagnier, L.; Alizon, M. Molecular cloning and polymorphism of the human immunodeficiency virus type 2. Nature 1986, 324, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Van Heuverswyn, F.; Li, Y.; Bailes, E.; Neel, C.; Lafay, B.; Keele, B.F.; Shaw, K.S.; Takehisa, J.; Kraus, M.H.; Loul, S.; et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 2007, 368, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Willey, R.L.; Smith, D.H.; Lasky, L.A.; Theodore, T.S.; Earl, P.L.; Moss, B.; Capon, D.J.; Martin, M.A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 1988, 62, 139–147. [Google Scholar] [CrossRef]
- Fabryova, H.; Kao, S.; Sukegawa, S.; Miyagi, E.; Taylor, L.; Ferhadian, D.; Saito, H.; Schaal, H.; Hillebrand, F.; Strebel, K. HIV-1 Vpr Induces Degradation of Gelsolin, a Myeloid Cell-Specific Host Factor That Reduces Viral Infectivity by Inhibiting the Expression and Packaging of the HIV-1 Env Glycoprotein. Mbio 2023, 14, e02973-22. [Google Scholar] [CrossRef]
- Doring, M.; Borrego, P.; Buch, J.; Martins, A.; Friedrich, G.; Camacho, R.J.; Eberle, J.; Kaiser, R.; Lengauer, T.; Taveira, N.; et al. A genotypic method for determining HIV-2 coreceptor usage enables epidemiological studies and clinical decision support. Retrovirology 2016, 13, 85. [Google Scholar] [CrossRef]
- Polzer, S.; Dittmar, M.T.; Schmitz, H.; Schreiber, M. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology 2002, 304, 70–80. [Google Scholar] [CrossRef][Green Version]
- Ochsenbauer, C.; Edmonds, T.G.; Ding, H.; Keele, B.F.; Decker, J.; Salazar, M.G.; Salazar-Gonzalez, J.F.; Shattock, R.; Haynes, B.F.; Shaw, G.M.; et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 2012, 86, 2715–2728. [Google Scholar] [CrossRef]
- Toohey, K.; Wehrly, K.; Nishio, J.; Perryman, S.; Chesebro, B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 1995, 213, 70–79. [Google Scholar] [CrossRef]
- Strebel, K.; Daugherty, D.; Clouse, K.; Cohen, D.; Folks, T.; Martin, M.A. The HIV ‘A’(sor) gene product is essential for virus infectivity. Nature 1987, 328, 728–730. [Google Scholar] [CrossRef] [PubMed]
- Franke, E.K.; Yuan, H.E.; Luban, J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 1994, 372, 359–362. [Google Scholar] [CrossRef]
- Kornfeld, R.; Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar] [CrossRef]
- Van Damme, N.; Goff, D.; Katsura, C.; Jorgenson, R.L.; Mitchell, R.; Johnson, M.C.; Stephens, E.B.; Guatelli, J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 2008, 3, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Neil, S.J.; Eastman, S.W.; Jouvenet, N.; Bieniasz, P.D. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006, 2, e39. [Google Scholar] [CrossRef] [PubMed]
- Strebel, K. HIV accessory proteins versus host restriction factors. Curr. Opin. Virol. 2013, 3, 692–699. [Google Scholar] [CrossRef]
- Andrew, A.; Strebel, K. The interferon-inducible host factor bone marrow stromal antigen 2/tetherin restricts virion release, but is it actually a viral restriction factor? J. Interferon Cytokine Res. 2011, 31, 137–144. [Google Scholar] [CrossRef]
- Fouchier, R.A.; Groenink, M.; Kootstra, N.A.; Tersmette, M.; Huisman, H.G.; Miedema, F.; Schuitemaker, H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 1992, 66, 3183–3187. [Google Scholar] [CrossRef]
- van Rij, R.P.; Blaak, H.; Visser, J.A.; Brouwer, M.; Rientsma, R.; Broersen, S.; de Roda Husman, A.M.; Schuitemaker, H. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 2000, 106, 1569. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, H.; Sukegawa, S.; Kao, S.; Strebel, K. Human Mannose Receptor 1 Attenuates HIV-1 Infectivity in a Virus Isolate-Specific Manner. Viruses 2023, 15, 2057. https://doi.org/10.3390/v15102057
Saito H, Sukegawa S, Kao S, Strebel K. Human Mannose Receptor 1 Attenuates HIV-1 Infectivity in a Virus Isolate-Specific Manner. Viruses. 2023; 15(10):2057. https://doi.org/10.3390/v15102057
Chicago/Turabian StyleSaito, Hideki, Sayaka Sukegawa, Sandra Kao, and Klaus Strebel. 2023. "Human Mannose Receptor 1 Attenuates HIV-1 Infectivity in a Virus Isolate-Specific Manner" Viruses 15, no. 10: 2057. https://doi.org/10.3390/v15102057
APA StyleSaito, H., Sukegawa, S., Kao, S., & Strebel, K. (2023). Human Mannose Receptor 1 Attenuates HIV-1 Infectivity in a Virus Isolate-Specific Manner. Viruses, 15(10), 2057. https://doi.org/10.3390/v15102057





