1. Introduction
The coronavirus disease 2019 (COVID-19) has posed tremendous socioeconomic and scientific challenges to the global healthcare system and currently remains a threat as a result of the constant emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mutations. As a single-stranded positive-sense RNA virus, SARS-CoV-2 tends to more rapidly accumulate more pronounced genomic mutations than its DNA counterparts, primarily ascribing to the intrinsically error-prone RNA-dependent RNA polymerase (RdRp) [
1]. In addition, it has been reported that the host RNA-editing systems such as apolipoprotein B mRNA editing enzyme, catalytic polypeptide (APOBEC), and adenosine deaminase RNA-specific binding protein (ADAR) [
2], as well as the recombination events involving co-infection of two viral lineages, may further fuel the diversification of a particular viral strain [
3]. The enlarged mutation pool potentially creates new traits and offers selective advantages to the virus.
SARS-CoV-2 encodes four structural proteins, of which spike (S) serves as the main determinant that mediates viral entry and confers high immunogenicity. Spike proteins of coronaviruses are synthesized in the rough endoplasmic reticulum as a precursor SFL, where it is co-translationally glycosylated, trimerized, and subsequently transported to the Golgi apparatus where the proteolytic processing occurs [
4]. In the trans-Golgi network, the SFL precursor is cleaved by cellular proteases such as furin into S1 and S2 subunits. The receptor binding domain (RBD) in the S1 subunit, as the name refers, is responsible for the engagement with the receptor angiotensin-converting enzyme 2 (ACE2), while the S2 subunit harbors the viral fusion machinery and, thus, governs the subsequent viral–host membrane fusion. Of note, the viral entry route of SARS-CoV-2 can be divergent, depending on the accessibility of co-factors for a second proteolytic cleavage [
5]. The serine protease transmembrane protease, serine 2 (TMPRSS2), is required for SARS-CoV-2 entry via plasma membrane fusion, while endosomal cysteine proteases cathepsin B and L facilitate the viral entry through the endocytic pathway. The spike covers the external surface of the SARS-CoV-2 virion and, thus, the first and the most predominant encounter of the host immune cells. As a result, the generated immune responses concentrate on the spike protein. For instance, the protective neutralizing antibodies are virtually confined to the spike region, blocking the above-mentioned viral entry cascade. In addition, multiple dominant T cell epitopes have been identified in spike and suggested to confer protection against COVID-19 severe disease [
6,
7,
8]. Therefore, the spike itself is central to the fulfillment of the viral life cycle and, most importantly, serves as the key immunogen that the host immune system targets. Consequently, the alterations in the spike domains may influence the virological features, including the capacity of the viruses to enter the cell, the transmissibility that the strains spread among the hosts, and the sensitivity to the prophylaxis and treatments; moreover, it could concomitantly lead to a change in the ability to provoke an immune response, occasionally ensuing an exacerbated viral pathogenicity.
The genetic alterations archived during the early or late phase of the pandemic can be different, depending on the selection pressure by natural immunity or vaccine-induced immunity. There is a plethora of evidence showing that a swarm of SARS-CoV-2 variants, in particular, the variants of concern (VOCs) that emerged after the massive vaccine deployment, showed enhanced immune evasion, accelerated transmissibility, and even aggravated pathogenicity [
9,
10,
11,
12]. The features of the early emerging mutants are less well-characterized, and their biological relevance remains largely elusive. The genotyping and phenotyping of the early mutants provide an efficient means to study the epidemiology of COVID-19 and develop diagnostic approaches. Moreover, it is crucial for assessing viral drug resistance and immune escape pathogenesis-related mechanisms. Computation simulations-based evolutionary and structural analysis predicted the features of early mutants, mainly in the RBD region. Hot spots such as V367F have been implicated by a few groups to possess a stronger binding affinity to the receptor ACE2 than the ancestral strain, while others, such as V483A, displayed diminished binding [
13,
14,
15]. Interestingly, a few mutations, such as G476S, were predicted but with contradicting results. Regardless, these in silico models can be valuable, but a few traits need to be clarified, and more importantly, their biological relevance warrants further investigation.
In this study, we analyzed a panel of early SARS-CoV-2 spike mutants, characterized the virologic features, including protein expression, viral entry, and sensitivity to antivirals, and assessed their immunogenicity in eliciting antibody and T cell immune responses. The mixed traits provide us with an informative snapshot of the early phase of viral evolution, which may be conducive to COVID-19 origin tracing and future pandemic preparedness.
2. Materials and Methods
2.1. Plasmids, Cells, and Reagents
Sequences of a full-length spike (S) or 13 amino acids in cytoplasmic tail deleted S (SΔCT13) of SARS-CoV-2 (WA01/2020) were codon optimized and commercially synthesized (General Bio, Chuzhou, China). Synthetic genes were cloned into the mammalian expression plasmid pcDNA3.1+ (Invitrogen, Waltham, MA, USA). SARS-CoV-2 S D614G, V483A, S943P, D839Y, G476S, Q675H, V367F, W436R, N354D/D364Y variants were generated by site-directed mutagenesis based on SARS-CoV-2 SΔCT. Human ACE2 genes were commercially synthesized (General Bio, Chuzhou, China) and further cloned into a pQCXIP retroviral vector (Cat# 631516, Takara, San Jose, CA, USA). pHEF-VSVG expressing Vesicular Stomatitis Virus (VSV) glycoprotein was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH (Cat# 4693). PsPAX2 (Cat#12260), pLenti-CMV-Puro-Luc (Cat#17447), and pBS-CMV-gag-pol (Cat#35614) were obtained from Addgene (Boston, MA, USA). IFNα2b (Cat# HY-P7023) was purchased from MedChemExpress, Monmouth Junction, NJ, USA.
HEK293T (ATCC® CRL-11268™), Calu-3 (ATCC® HTB-55™), TZM-bl (AIDS Reagent, Manassas, VA, USA, Cat# 8129) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Cat# C11995500BT, Gibco, Waltham, MA, USA), supplemented with 0.5% penicillin/streptomycin (Cat# 15070063, Gibco, Waltham, MA, USA) and 10% FBS (F8318, Sigma, St. Louis, MO, USA). HEK293T-hACE2 was maintained in DMEM, supplemented with 0.5% penicillin/streptomycin, 10% FBS, and 1 μg/mL puromycin (Cat# 540222, Sigma, St. Louis, MO, USA).
Mouse monoclonal antibody against SARS-CoV-2 RBD (Cat# 40592-MM117), rabbit polyclonal antibody against SARS-CoV-2 S2 (Cat# 40590-T62), and hACE2 conjugated with mouse IgG Fc (hACE2-mFc, Cat# 10108) was purchased from Sino Biological (Beijing, China). HRP-conjugated β-actin antibody (Cat# HRP-60008) was purchased from Proteintech (Rosemont, IL, USA). Rabbit polyclonal antibody against HIV1 p55 + p24 + p17 (Cat# ab63917) was purchased from Abcam (Cambridge, MA, USA).
Bafilomycin A1 (BafA1) was purchased from Cell Signaling Technology (Cas# 88899-55-2), E64d was kindly provided by Dr. Shaobo Wang (Guangzhou Laboratory, Guangzhou, China), while teicoplanin was provided by Dr. Xiancai Ma [
16] (Guangzhou Laboratory, Guangzhou, China). A set of 15-mer peptides overlapping by 11 amino acids encompassing the spike (S) glycoprotein was synthesized by GL Biochem Ltd. (Shanghai, China) and used for the stimulation of T cells.
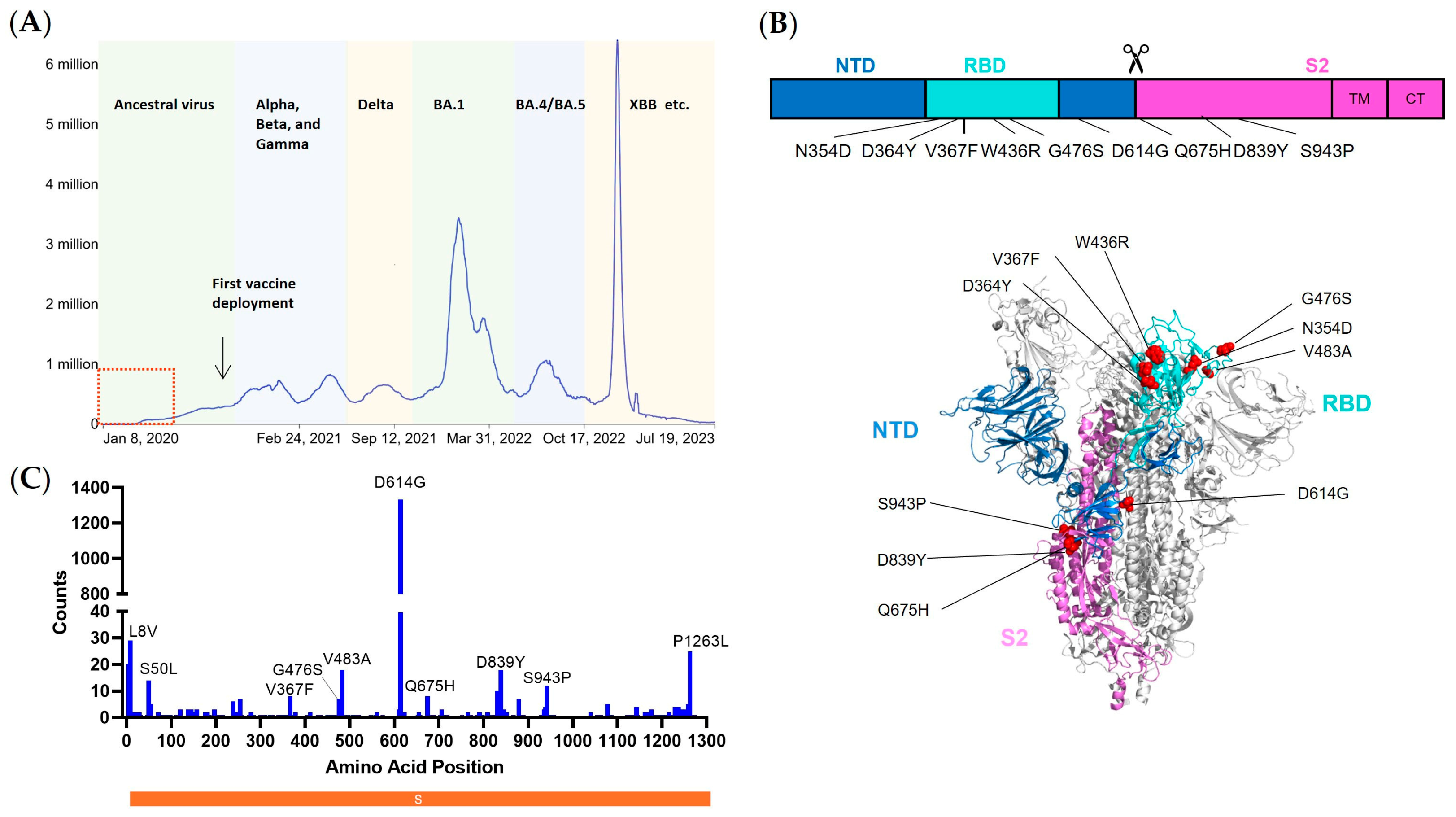
2.2. Selection of Early Emerging Spike Mutants
A total of 3165 whole genome sequences of SARS-CoV-2 sampled before April 2020 were downloaded from GISAID, and the workflow designed for SARS-CoV-2 was run using Nextstrain software, version 7.1.0 [
17,
18] (
https://docs.nextstrain.org/projects/ncov/en/latest/ (accessed on 2 September 2023)). After multiple-sequence alignments were completed by Mafft [
19] through the built-in program Augur, 2764 DNA sequences were aligned using WA01/2020 as the reference strain. Moreover, 2969 DNA sequences aligned to the spike gene, and the matching protein sequences were extracted from the intermediate file. The frequency of point mutations (non-synonymous mutations) was then measured by scanning each of the spike protein sequences’ amino acid positions one at a time. Nine spike mutations were filtered out for research based on the frequency of mutations after the N-terminal signal peptide area (the first 20 amino acids) and the C-terminal cytoplasmic tail region were excluded.
2.3. Generation of HEK293T-hACE2 Stable Cell
The retroviral pseudotypes expressing human ACE2 (hACE2) were generated by co-transfecting HEK293T cells with pHEF-VSVG, pBS-CMV-gag-pol, and pQCXIP-hACE2 at a ratio of 0.5:1:1. Viruses in the supernatants were collected every 12 h. The collected viruses were used to transduce HEK293T cells, and positive cell populations were selected by using 1 μg/mL puromycin.
2.4. Production of Pseudotyped Lentiviral Particles
The SARS-CoV-2 pseudoviruses expressing a luciferase reporter gene were generated in an approach similar to as described previously [
20,
21]. Moreover, 10 µg packaging construct psPAX2, 10 µg luciferase reporter plasmid pLenti-CMV Puro-Luc, and 5 µg spike protein expressing pcDNA3.1-SARS-CoV-2 SΔCT were co-transfected into 5 × 10
6 HEK293T cells in T-75 flask with lipofectamine 2000 (Sigma, St. Louis, MO, USA). Six hours post-transfection, the supernatants were replaced with fresh DMEM (plus 5% FBS). The supernatants containing the pseudotype viruses were collected 48 h post-transfection; pseudotype viruses were purified by filtration with a 0.45 µm filter.
2.5. Cell–Cell Fusion Assay
HEK293T cells were transfected with spike-encoding plasmids, along with HIV-1 Tat optimized expression vector (AIDS Reagent, Manassas, VA, USA, Cat# 827). Meanwhile, TZM-bl cells were transfected with human ACE2 plasmid. Twenty-four hours after transfection, cells were cocultured with TZM-bl at a 1:1 ratio for 12 h, lysed, and measured for firefly luciferase activity. All samples were tested in duplicates, and the results were averaged.
2.6. Virus Entry and Kinetics
In a flat 96-well plate, 1.5 × 104 HEK293T-ACE2 cells/well were seeded at a density of 1.5 × 105 cells per mL. On the second day, 50 µL of equal amount of p24 virus (approximately 20 ng) was added into each well and spun at 1680 g, 30 min, 4 °C. The cell culture plates were transferred into a 37 °C incubator immediately after spin to initiate viral internalization and infection. Moreover, 15 µL DMEM medium containing 50 nM BafA1 was added into each well immediately after spinoculation (0 h) and 10 min, 1, 2, 3, 4, 6, and 8 h post-temperature shift. Furthermore, 12 h after culturing, cells were replenished with fresh DMEM. Additionally, 48 h post spinoculation, cells were lysed for luciferase activity quantification.
2.7. Western Blot
Western blot was performed as previously described [
22]. Briefly, transfected HEK293T cells were collected, washed once with icy 1 × PBS (Cat# BL302A, Biosharp, Hefei, China), and lysed in Radioimmunoprecipitation assay buffer (RIPA, Cat# P0013B, Beyotime, Nantong, China) for 20 min on ice. The pseudoviral particles were obtained from DMEM culture medium by centrifugation (4000 rpm, 10 min, 4 °C). The cell lysates and viral particles were dissolved in sample buffer (Cat# P1041, Solarbio, Beijing, China), separated on 20% gradient gel (Cat# M00657, GenScript, Benelux, The Netherlands), and detected by anti-HIV-1 p24, anti-SARS-CoV-2 RBD, anti-SARS-CoV-2 S2, and anti-β-actin antibodies.
2.8. Cell Surface Staining
Cells were washed twice with cold 1× DPBS plus 2% FBS, detached with 1× DPBS containing 5 mM EDTA, and incubated on ice with the appropriate primary antibodies for 1 h. After three washes with PBS plus 2% FBS, cells were further incubated with FITC-conjugated secondary antibodies for 45 min. After two washes, cells were fixed with 2% formaldehyde and analyzed in a BD LSRII flow cytometer.
2.9. Animals and Study Design
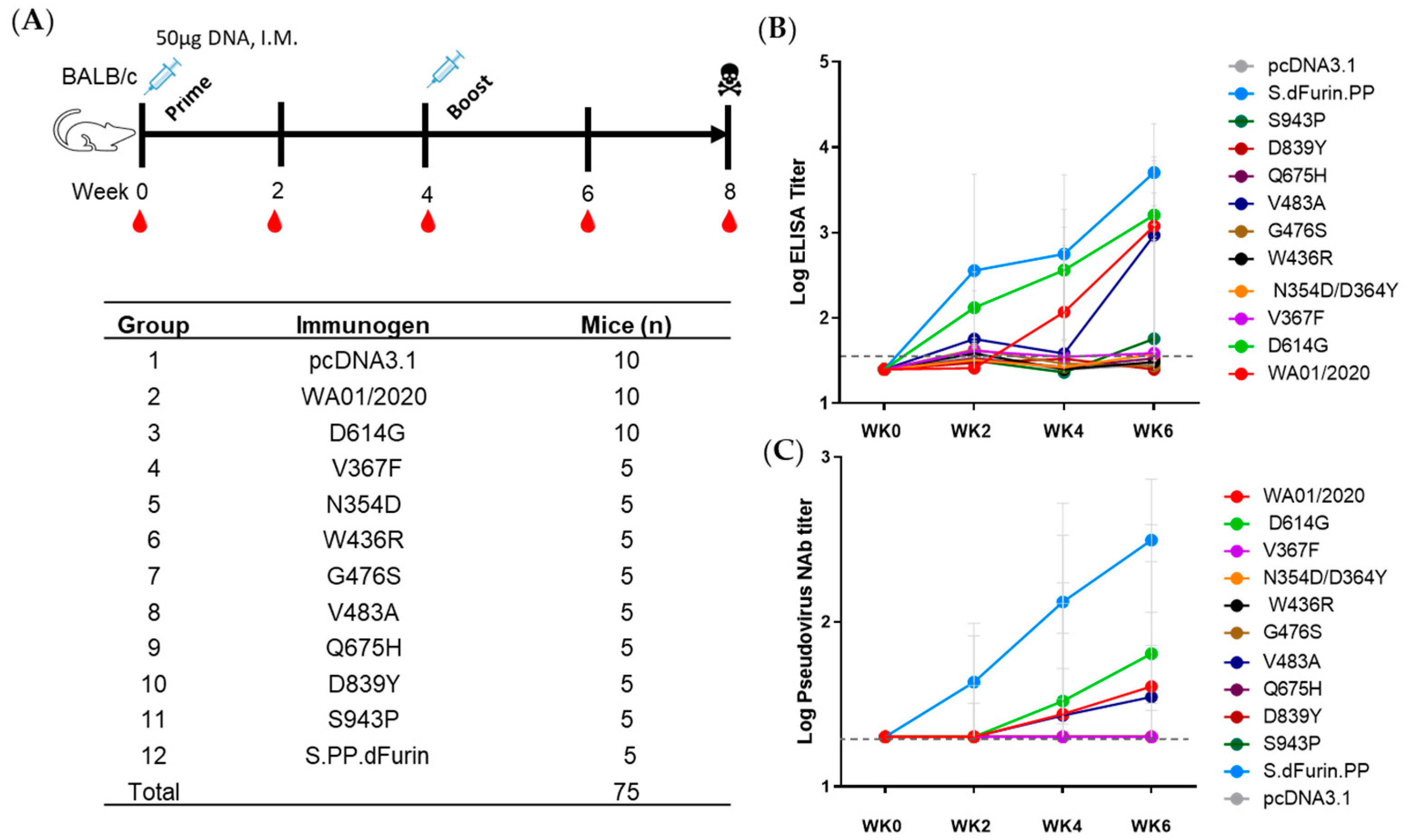
Additionally, 75 specific pathogen-free 6- to 8-week-old BALB/c mice were purchased from Hunan SJA Laboratory Animal Co., Hunan, China, and maintained in the Animal Care Facilities at the Guangzhou Laboratory and were randomly allocated to 12 groups. Mice received pcDNA3.1 vectors expressing different SARS-CoV-2 S protein or sham controls (n = 5 or 10 per group). Animals received a single immunization of 50 μg of plasmid by the intramuscular route without adjuvant. Then, 4 weeks after prime, the mice were boosted with another shot of 50 μg of plasmid. At indicated time points, peripheral blood was collected via the submandibular route to isolate serum for immunologic assays.
To test sensitivity of early spike mutants to antiserum, eight ICR mice (Gempharmatech Co., Ltd., Nanjing, China) were intramuscularly immunized with 0.5 µg (n = 2), 5 µg (n = 3), or 50 µg (n = 3) of spike ferritin nanoparticle vaccine adjuvanted with 100 µg of Alhydrogel (Invivogen, San Diego, CA, USA) at weeks 0 and 4. At week 6, peripheral blood was collected via the submandibular route to isolate serum for immunologic assays.
All animal studies were conducted in compliance with all relevant local and national regulations and were approved by the Institutional Animal Care and Use Committees of Guangzhou Laboratory.
2.10. Lentiviral Luciferase-Based Neutralization Assay
The SARS-CoV-2 pseudoviruses neutralization assay was generated in an approach similar to that described previously [
20,
21]. To determine the neutralization activity of the serum, plasma, or IgG samples from cohorts, HEK293T-hACE2 cells were seeded in 96-well tissue culture plates at a density of 1.75 × 10
4 cells/well overnight. Three-fold serial dilutions of heat-inactivated plasma samples were prepared and mixed with 50 µL of pseudovirus. The mixture was incubated at 37 °C for 1 h before being added to HEK293T-hACE2 cells. Moreover, 48 h after infection, cells were lysed in Steady-Glo Luciferase Assay (Promega) according to the manufacturer’s instructions. SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction in the relative light unit (RLU) was observed relative to the average of the virus control wells.
2.11. Enzyme-Linked Immunosorbent Assay (ELISA)
SARS-CoV-2 receptor-binding domain (RBD)-specific binding antibodies in serum were assessed by ELISA. Additionally, 96-well plates were coated with 1 μg/mL of similarly produced SARS-CoV-2 WA1/2020, RBD protein in 1× Dulbecco phosphate-buffered saline (DPBS) and incubated at 4 °C overnight. After incubation, plates were washed once with wash buffer (0.05% Tween 20 in 1× DPBS) and blocked with 350 μL of casein block solution per well for 2 to 3 h at room temperature. Following incubation, the block solution was discarded, and the plates were blotted dry. Serial dilutions of heat-inactivated serum diluted in casein block were added to wells, and plates were incubated for 1 h at room temperature prior to 3 more washes and a 1 h incubation with a 1 μg/mL dilution of anti-mouse IgG horseradish peroxidase (HRP) (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) at room temperature in the dark. Plates were washed 3 times, and 100 μL of SeraCare KPL TMB SureBlue Start solution was added to each well; plate development was halted by adding 100 μL of SeraCare KPL TMB Stop solution per well. The absorbance at 450 nm was recorded with a VersaMax microplate reader (Molecular Devices, San Jose, CA, USA). For each sample, the ELISA end-point titer was calculated using a 4-parameter logistic curve fit to calculate the reciprocal serum dilution that yields an absorbance value of 0.2 at 450 nm. Interpolated end-point titers were reported.
2.12. Intracellular Cytokine Staining (ICS) Assay
CD4+ and CD8+ T cell responses were quantitated by pooled peptide-stimulated intracellular cytokine staining (ICS) assays. Peptide pools were 15 amino acid peptides overlapping by 11 amino acids spanning the SARS-CoV-2 WA1/2020. Moreover, 2 × 105 peripheral blood mononuclear cells well were re-suspended in 100 µL of R10 media supplemented with CD49d monoclonal antibody (1 µg/mL) and CD28 monoclonal antibody (1 µg/mL). Each sample was assessed with mock (100 µL of R10 plus 0.5% DMSO; background control), peptides (2 µg/mL), and/or 10 ng/mL phorbol myristate acetate (PMA) and 1 µg/mL ionomycin (Sigma-Aldrich, St. Louis, MO, USA) (100 µL; positive control) and incubated at 37 °C for 1 h. After incubation, 0.25 µL of GolgiStop and 0.25 µL of GolgiPlug in 50 µL of R10 was added to each well and incubated at 37 °C for 8 h and then held at 4 °C overnight. The next day, the cells were washed twice with DPBS, stained with aqua live/dead dye for 10 min and then stained with predetermined titers of monoclonal against CD8a (clone 53–6.7; BUV395), CD3 (clone 17A2; BUV737), CD103 (clone 2E7; BV605), CD44 (clone IM7; BV711), CD62L (clone MEL-14; Alexa Fluor 488), CD4 (clone RM4–5; PE/Dazzle 594), and CD69 (clone H1.2F3; PE-Cy7) (all antibodies diluted 1:100) for 1 h. Cells were then washed twice with MACS buffer and incubated with 100 µL of BD Cytofix/CytoPerm Fixation/Permeabilization solution for 15 min. Cells were then washed twice with 1x Perm Wash buffer (BD Perm/WashTM Buffer 10× in CytoFix/CytoPerm Fixation/Permeabilization kit diluted with MilliQ water and passed through 0.22 µm filter) and stained intracellularly with mAbs against IFN-γ (clone XMG1.2; BV421), IL-2 (clone JES6-5H4; PE), IL-4 (clone 11B11; APC), and TNFα (clone MP6-XT22; Alexa Fluor 700) (all antibodies diluted 1:100) for 1 h. Cells were then washed twice with 1× Perm Wash buffer and washed once with MACS buffer, then fixed with 250 µL of freshly prepared 2% formaldehyde. Fixed cells were analyzed by the BD LSRII system. Data were analyzed using FlowJo v9.9.
2.13. ELISPOT
ELISPOT plates were coated with rat anti-mouse IFN-γ monoclonal antibody (BD Biosciences, Franklin Lakes, NJ, USA, 554410) diluted 1:100 at a final concentration of 500 ng per well overnight at 4 °C. Plates were washed with DPBS containing 0.25% Tween 20, and blocked with R10 media (RPMI with 10% FBS and 1% penicillin–streptomycin) for 1 h at 37 °C. The Spike peptide pools contain 15 amino acid peptides overlapping by 11 amino acids that span the protein sequence. Spike was prepared at a concentration of 2 µg per well, with 500,000 cells per well added. The peptides and cells were incubated for 18–24 h at 37 °C. All steps following this incubation were performed at room temperature. The plates were washed with coulter buffer and incubated for 2 h with Rat anti-mouse IFN-γ Biotin (diluted 1:200) from BD Biosciences, Franklin Lakes, NJ, USA (375 ng/well). Plates were washed again and then incubated with Streptavidin–alkaline phosphatase antibody from Southern Biotechnology, Birmingham, AL, USA (7105–04) (20 µL/well). The third wash was followed by the addition of Nitor-blue Tetrazolium Chloride/5-bromo-4-chloro 3′ indolyl phosphate p-toluidine salt (NBT/BCIP chromagen) from Thermo Fisher Scientific, Waltham, MA, USA for 7 min. Chromagen was discarded, and plates were washed with water and dried in a dark place for 24 h. Plates were scanned and counted on a Cellular Technologies Limited Immunospot Analyzer.
2.14. Statistical Analyses
Descriptive statistics and logistic regression were performed using GraphPad Prism 9.0.0 (GraphPad Software, San Diego, CA, USA). Immunologic data were generated in duplicate and were compared by two-sided Mann–Whitney tests. Correlations were assessed by two-sided Spearman rank-correlation tests. p values less than 0.05 were considered significant.
4. Discussion
The genomic surveillance of SARS-CoV-2 is crucial for the timely detection and characterization of circulating variants, as well as monitoring the emergence of new strains [
34]. Moreover, functional validation of the emerging and accumulating mutations can be equally important. In this study, we snapshot spike substitutions that occurred in the first four months of the pandemic and analyzed the virologic and immunologic characteristics of nine representative mutants, revealing that the virologic features are rather diversified while the immunologic traits are largely maintained.
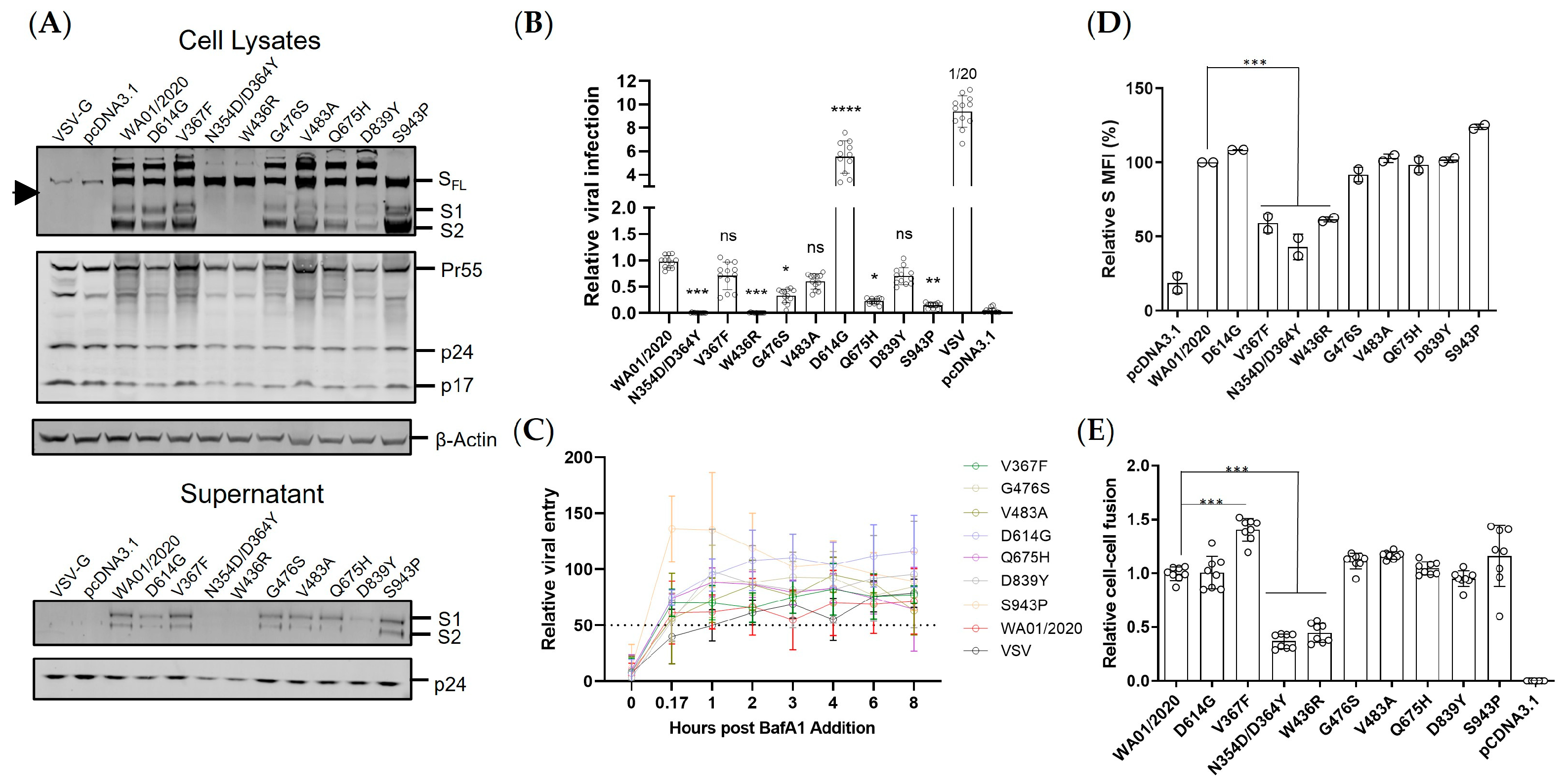
The spike expression level and the viral entry efficiency are two determinants of viral fitness. Under the presumable low immune pressure, the early mutants demonstrated surprisingly virologic heterogeneity. First, the protein expression either in total or on the cell surface varied even with only a single amino acid substitution, with 1 out of 9 (11%) up-regulated, 2 out of 9 (22%) down-regulated, and 6 out of 9 (67%) neutral. The mutations in the spike were blueprinted on the spike from the ancestral strain WA01/2020, which were codon optimized to allow optimal expression in human cells. Therefore, this heterogenous expression pattern of spike mutants may reflect the protein folding constraints and/or accelerated turnover instead of translation blockage [
35]. Second, the early mutants also showed an altered S1 to SFL ratio, implicating potentially compromised spike processing in the ER or Golgi apparatus. Third, functional characterization of viral entry by a lentiviral vectored pseudovirus demonstrated that the majority of the mutants mediated comparable viral entry to the ancestral spike, while the mutants N354D/D364Y and W436R manifested severe reduction, likely due to the compromised protein expression and/or processing. It remains to be determined how the N354D/D364Y and W436R substitution influenced the spike expression/processing. It could be due to a change of structural conformation that leads to altered accessibility to the furin cleavage site, the perturbed glycosylation that leads to reduced protein thermostability [
36], or other reasons. Conversely, the S943P showed greater S2 expression but lower infectivity. We reasoned that the S943P mutation likely leads to a conformation change on the spike that disfavors the pre-fusion process of viral entry, which warrants further validation studies. Interestingly, prior reports demonstrated that the V367F RBD showed elevated expression compared with the ancestral strain [
35]. However, we observed a much-diminished pattern. The full-length spike likely confers more constraints than the smaller RBD during the protein folding process. If that is the case, the previously built model for protein expression prediction may need to be optimized based on a full-length spike instead of RBD. We also noted that the well-characterized D614G showed much elevated viral entry than the parental and mutants, which may help to explain its rapid replacement of the ancestral strain and was considered the key mutation that makes the virus more transmissible worldwide. Cell–cell fusion represents an essential means of virus transmission, particularly in in vivo settings [
32]. The mutants N354D/D364Y and W436R again showed lower fusogenicity than the other mutants, likely resulting from a lower spike presence on the plasma membrane (
Figure 2C). Interestingly, the D614G showed comparable fusion to the WA01/2020 despite an approximate six-fold elevated viral infectivity. Moreover, the V367F, even though it showed lower protein expression and viral entry, induced 50% greater cell–cell fusion. Therefore, not only the spike protein itself but also the microenvironment where it resides can significantly impact its biological functioning.
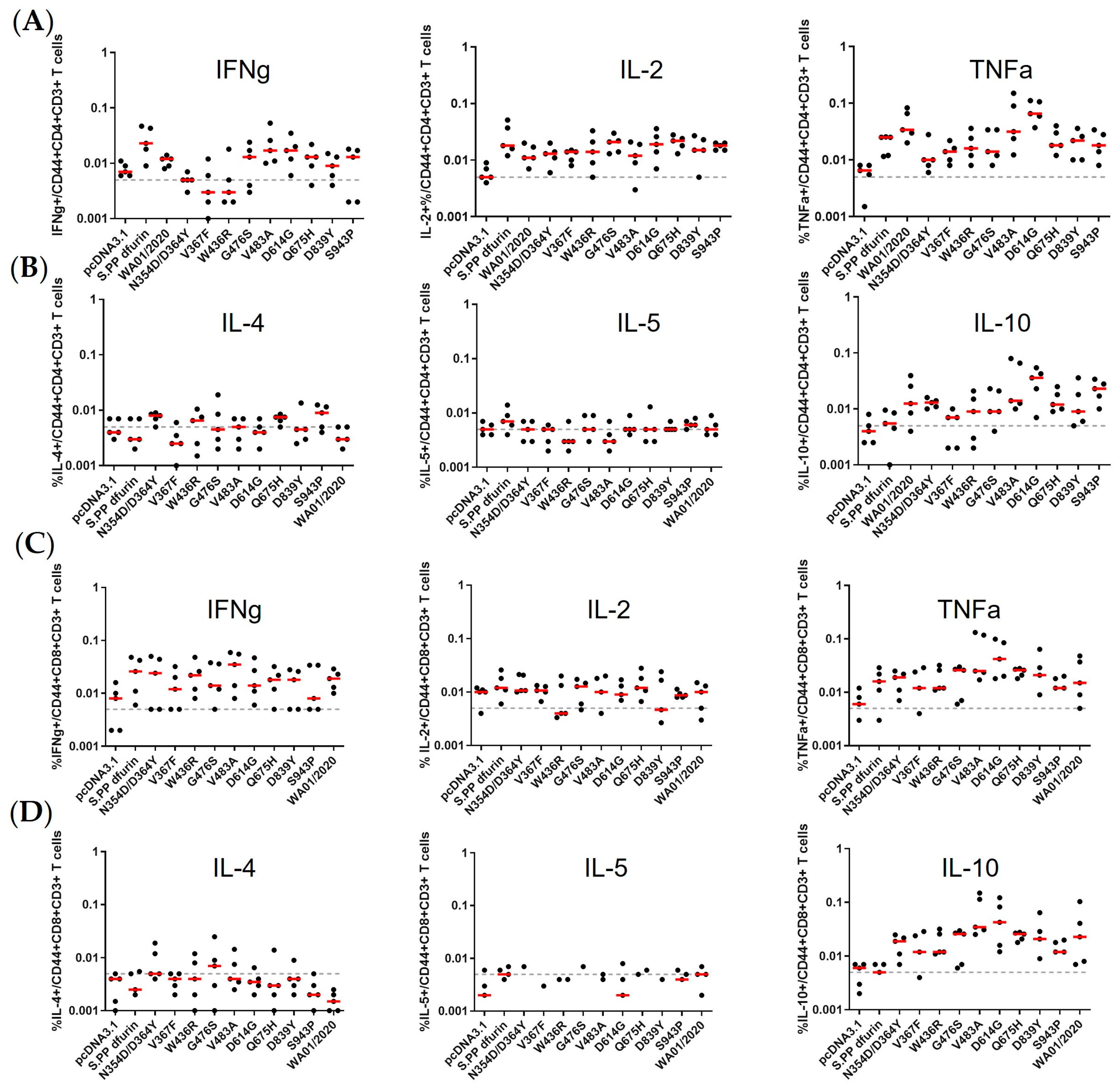
The immunogenicity of spikes was assessed via a prime-boost DNA immunization regimen. We observed only WA01/2020, D614G, and, to a lesser extent, V483A elicited binding and neutralizing antibody responses, leaving other mutants inert at least 6 weeks post-prime immunization. The mutants N354D/D364Y and W436R may be less immunogenic since their expression level are lower (
Figure 2A). However, these suppressed antibody responses by mutants with steady or elevated expression are unexpected. The apparent induction of T cell immune responses argues against the possibility of compromised in vivo expression as well (
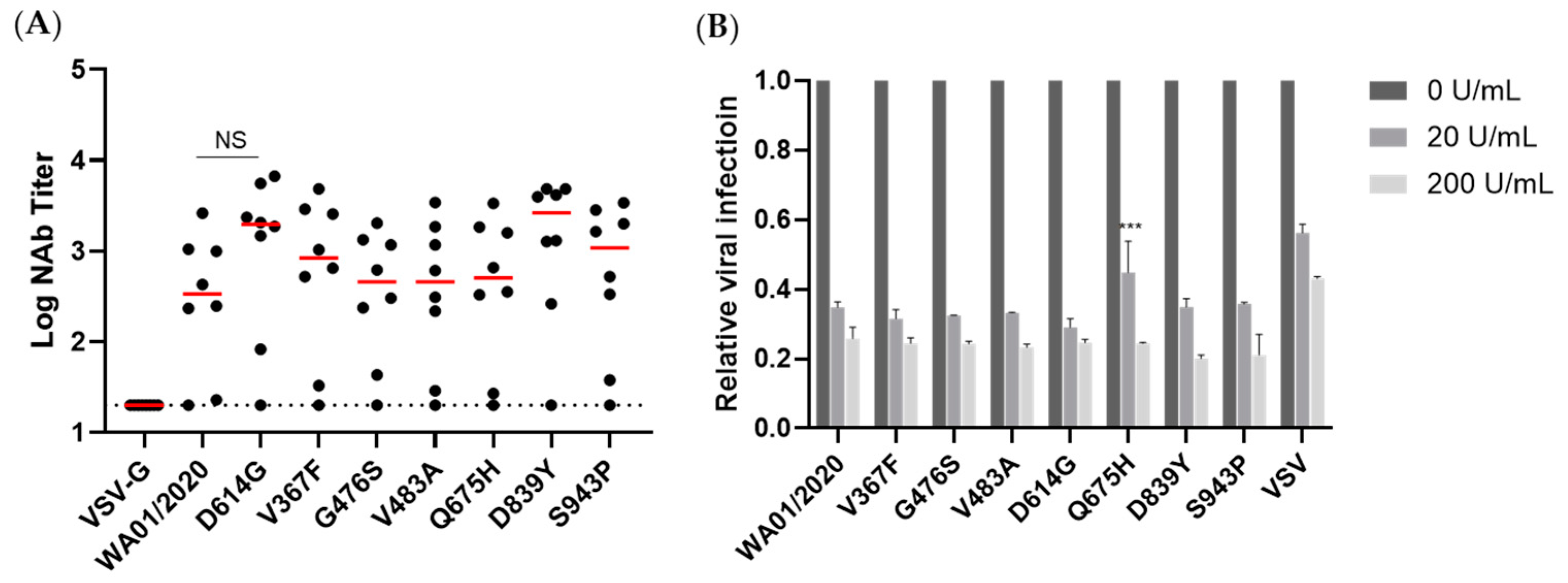
Figure 5). Whether this reflects a genuine difference between in vivo and in vitro expressions or is confined to a murine model warrants future characterization. This observation, on the other hand, may implicate that the early mutants may shield themselves from recognition by the host immune system and, thus, emerge. In addition, we take particular interest in the D614G mutation since it entirely replaced the ancestral strains shortly after its emergence [
23]. We noted that D614G induced more rapid and pronounced antibody titers than the ancestral strain. In addition, this single amino acid change is enough for the host immune system to differentiate. This piece of data suggests that this single amino acid change may cause a change of conformation [
31,
37], leading to favorable T and B cell activation. Regarding T cell responses, we noted generally comparable CD4+ T and CD8+ T cell responses, supporting recent reports that T cell epitopes are relatively conserved [
6,
38].
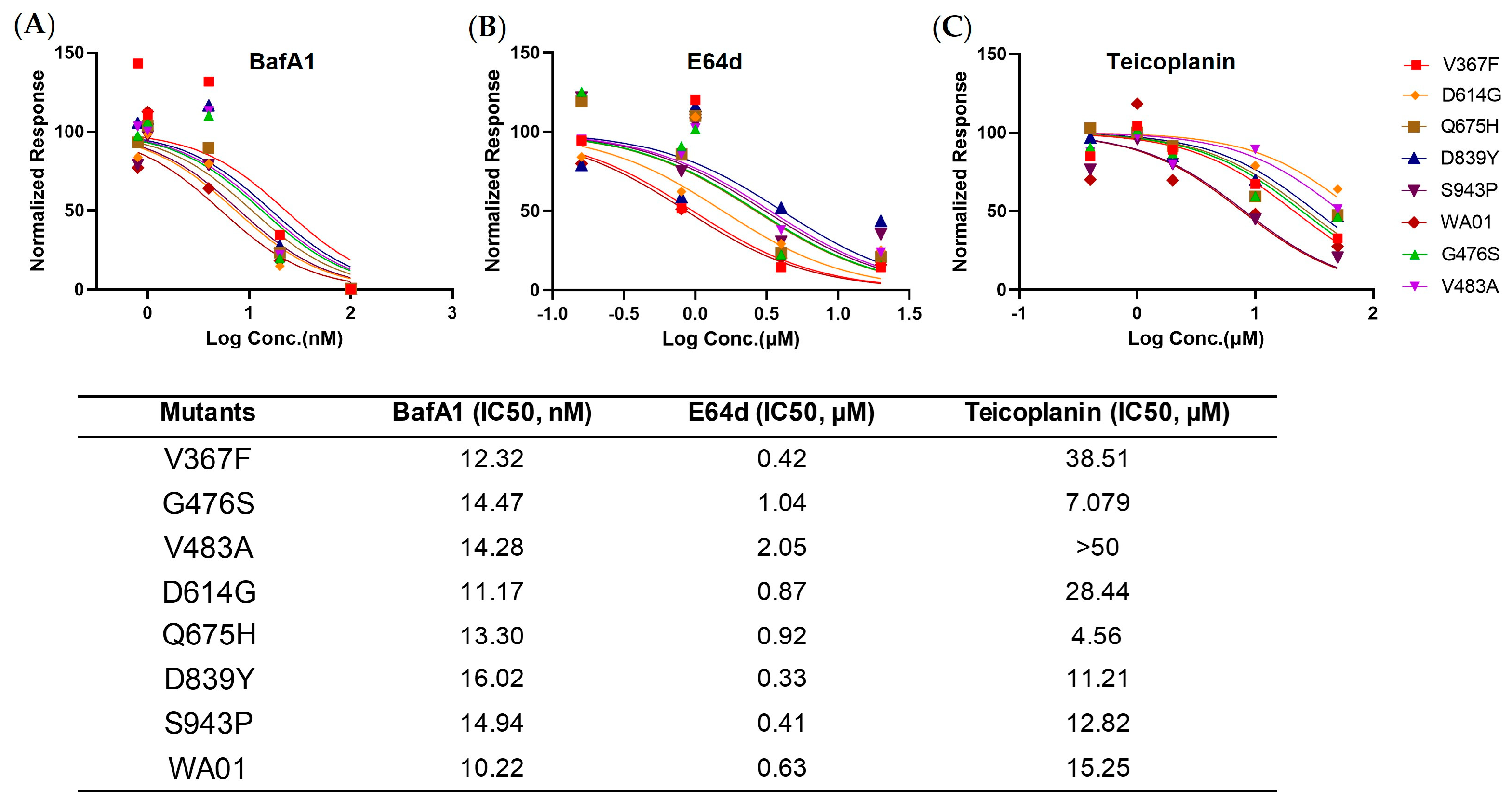
To gain a more comprehensive picture of the spike, we determined the viral entry kinetics and its sensitivity to viral inhibitors that block ACE2, Cathepsin B, or Cathepsin L, respectively. In general, most mutants tested demonstrated a similar half-maximal inhibitory concentration (IC50) and entry half-life to the ancestral WA01/2020, indicating limited selection pressure on these early mutants. In line with the virologic data, a panel of antisera from spike protein-immunized mice showed comparable neutralizing activity against all mutants, implicating a low chance of immune evasion. Similarly, the mutants showed a comparable type I IFN sensitivity. Therefore, these early mutants likely emerged sporadically and remain sensitive to antivirals and host immunity.
Our study has several limitations. First, our comparative fitness analysis was based on the sequence data released in GISAID and is, thus, subject to the selection bias of sequences being released to the public database. The proportion of SARS-CoV2 sequences varied substantially by sampling time and location, and therefore, we limited our analysis to the circulating period of the strains before 15 March 2020. In addition, the information based on spike mutation may not apply to authentic infectious viruses, since the mutations on other viral genes may deeply impact the viral fitness and shape the overall landscape collectively with spike mutations. Finally, all mutants except D614G were not present in the subsequent VOCs, again indicating limited immune pressure at the time of sampling. However, the early emerging mutations do formulate a pool, which supplied a more rapidly spreading infection, such as D614G and V367F in the case of viral infection and cell–cell fusion, respectively. Thus far, it would be necessary to keep surveilling the early emerging mutants and their viral fitness potential in future pandemics.
Together we snapshot a few mutants that emerged at the early phase of the pandemic and observed intriguingly divergent protein expression, viral entry, cell–cell fusion activity, and immunogenicity. This information can be useful in guiding vaccine design and future pandemic preparedness.