Transcriptomic and Translatomic Analyses Reveal Insights into the Signaling Pathways of the Innate Immune Response in the Spleens of SPF Chickens Infected with Avian Reovirus
Abstract
1. Introduction
2. Results
2.1. Overview of High-Throughput Sequencing Data between the Spleens of Control Group and ARV SPF Chickens
2.2. Global Translatome Characteristics
2.3. Translational Efficiency Significantly Decreased after ARV Infection
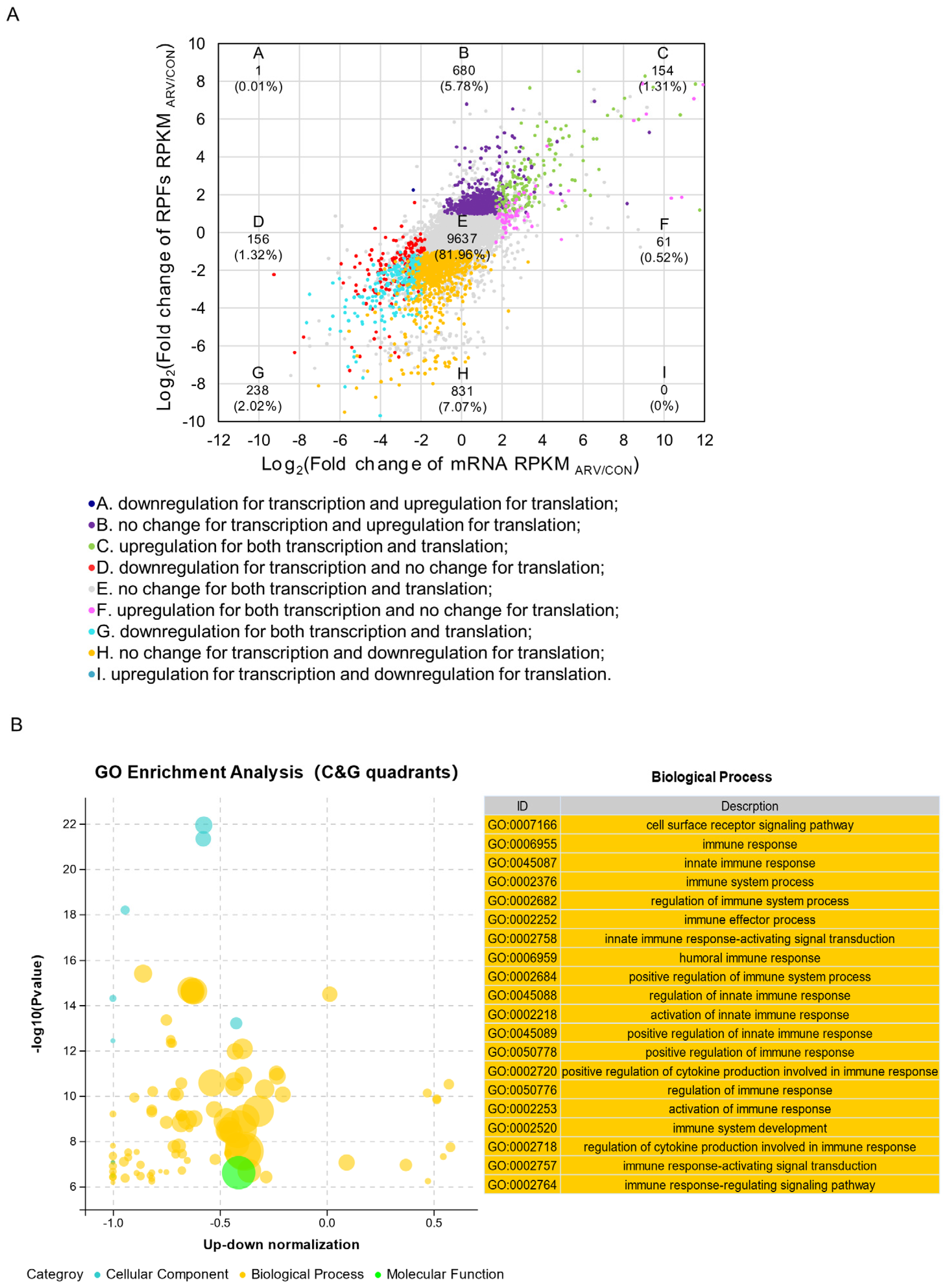
2.4. Regulation Patterns of the Transcriptome and Translatome
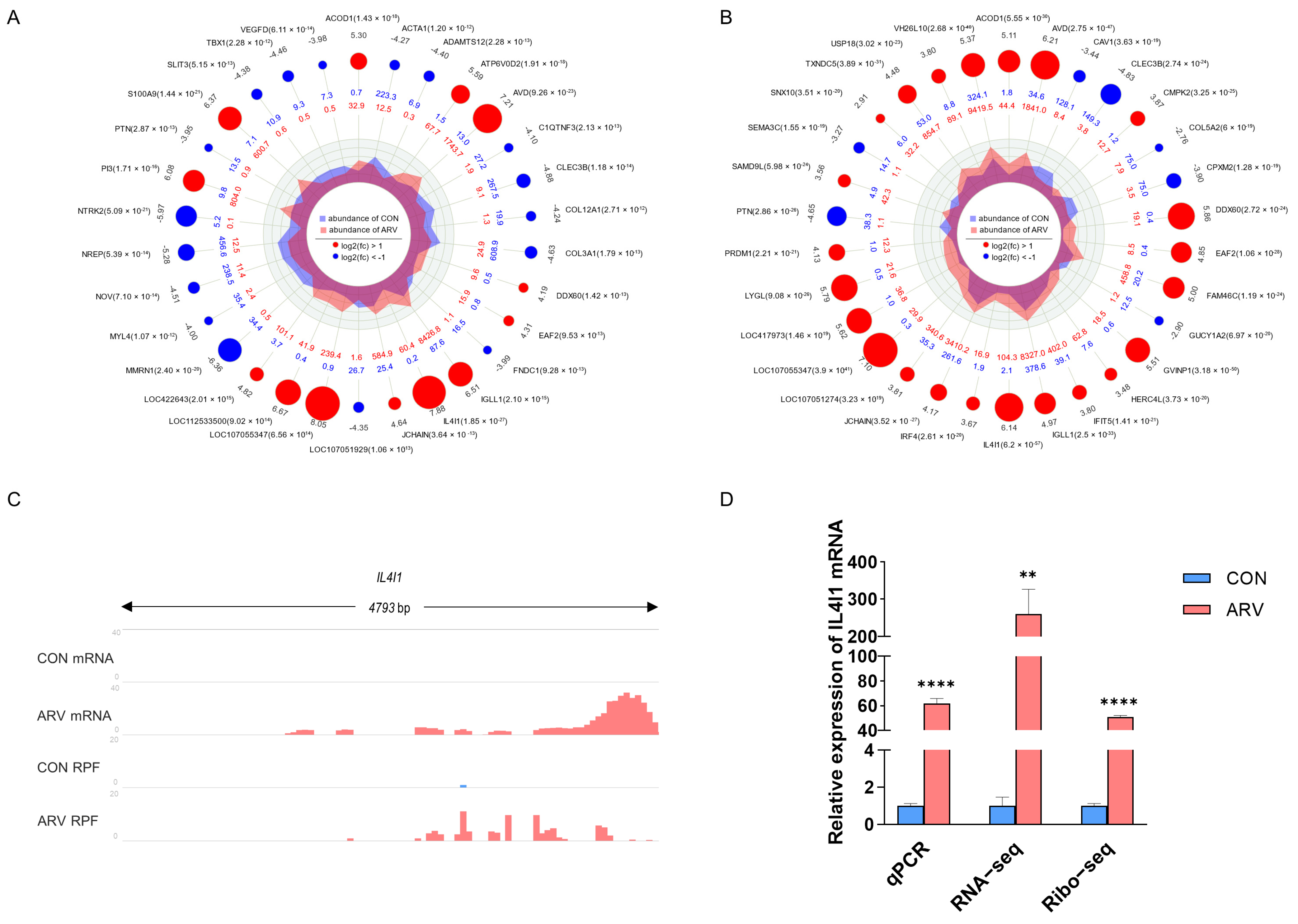
2.5. Screening Functional Genes after ARV Infection
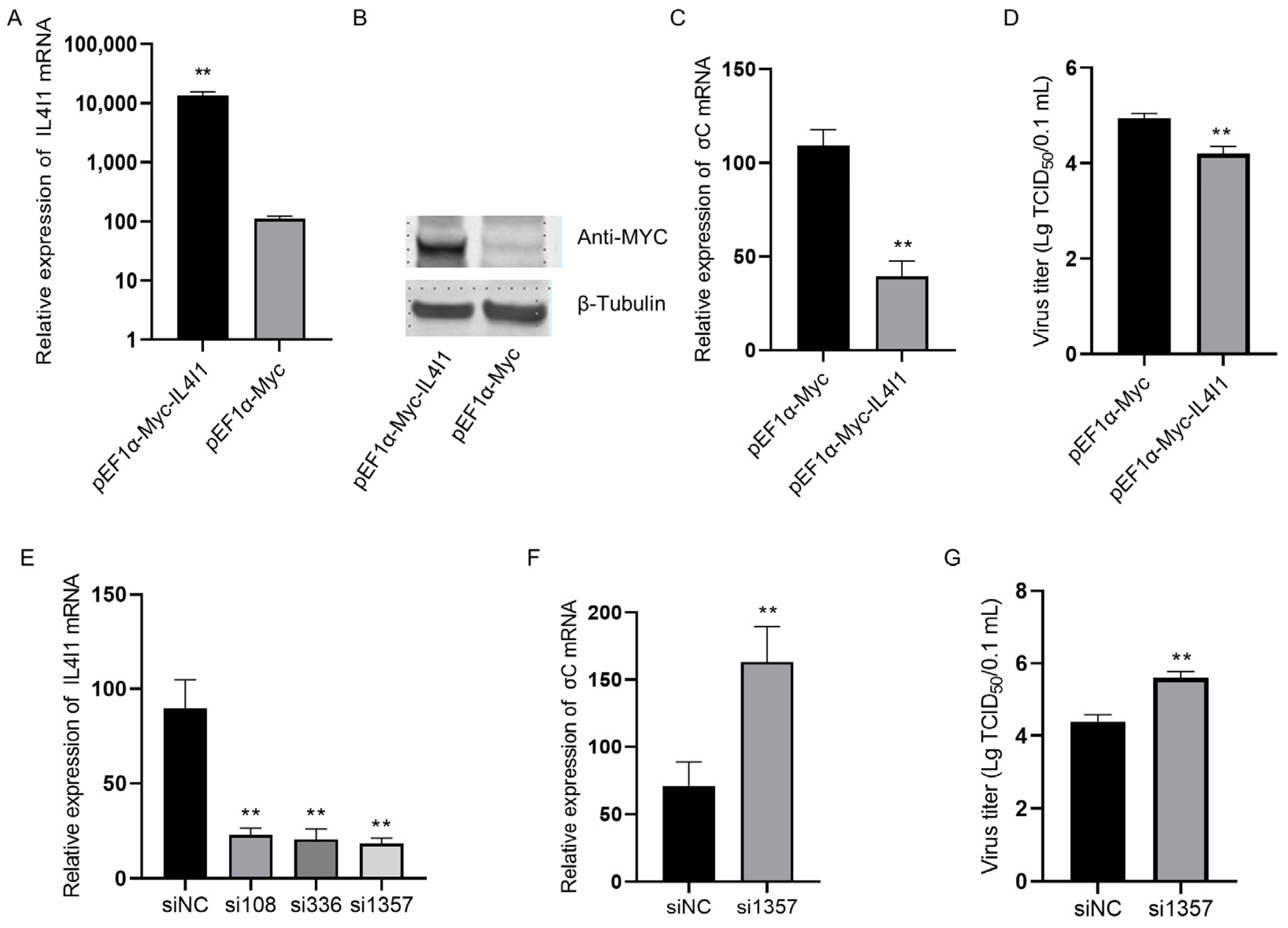
2.6. IL4I1 Expression Reduced ARV Replication
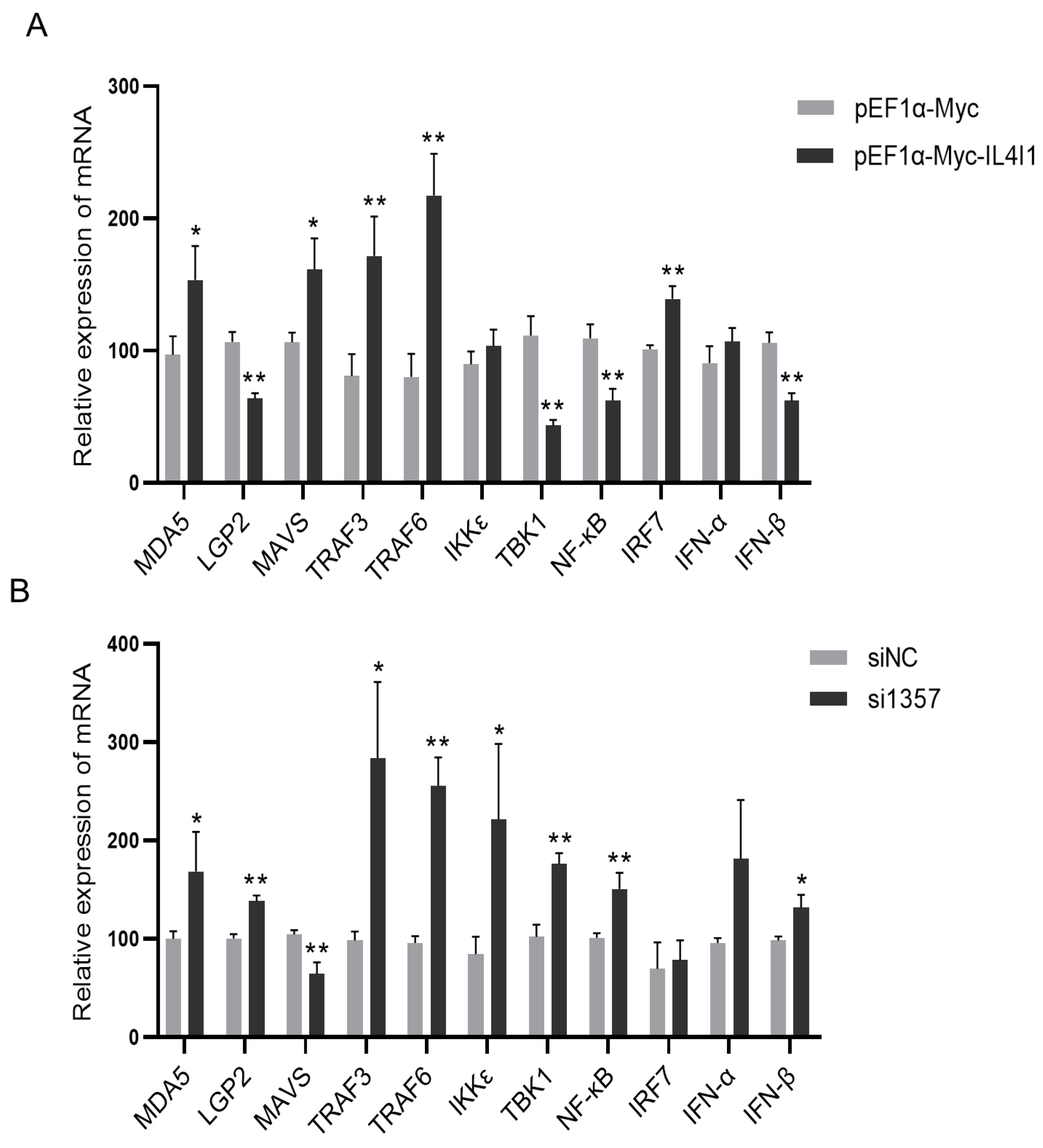
2.7. The Effect of IL4I1 Expression on the Innate Immune Response during ARV Infection
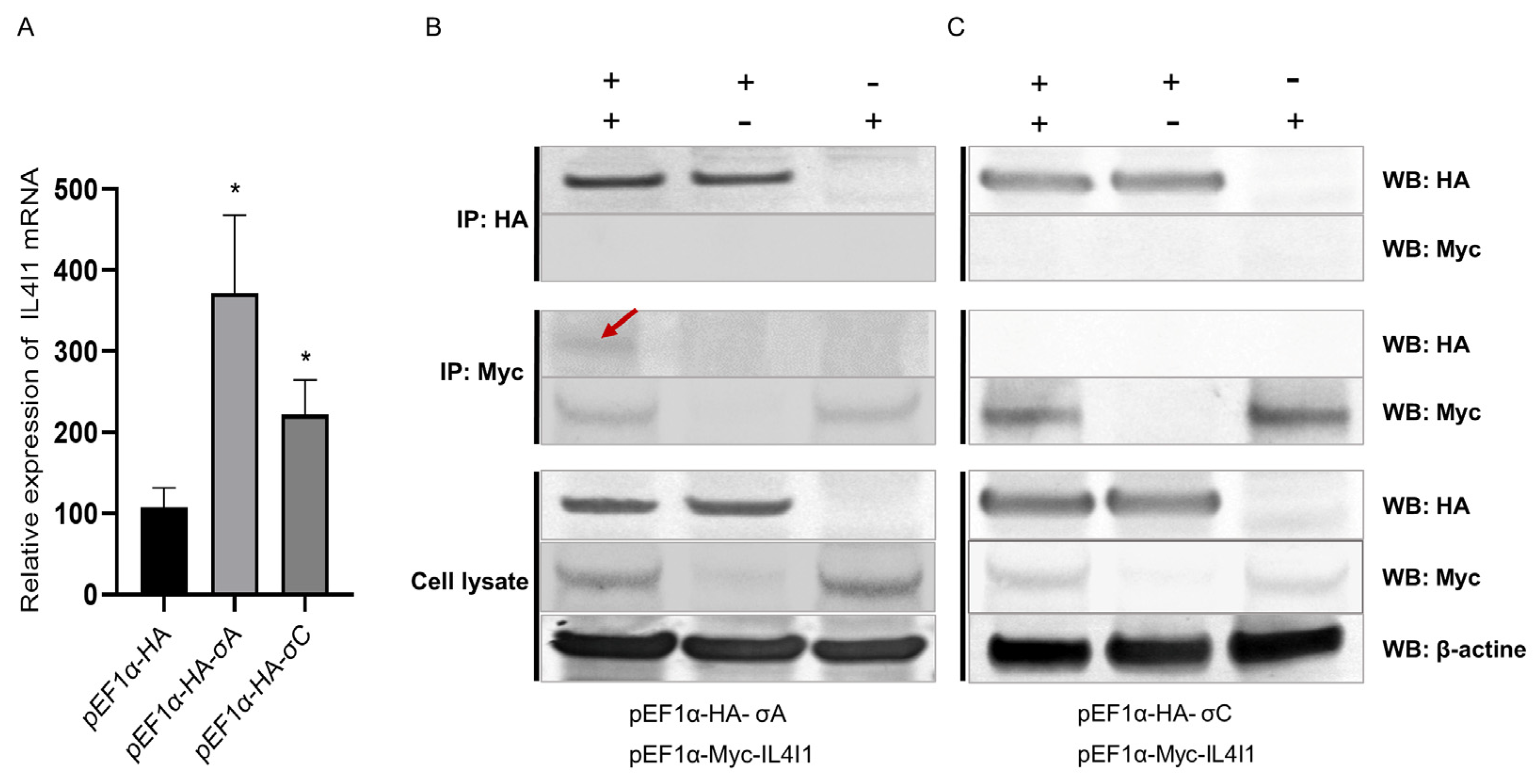
2.8. Interaction between IL4I1 and ARV σA/σC Proteins
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Viral Inoculations and Animal Experiments
4.3. RNA Extraction and Transcriptome Sequencing
4.4. Preparation of Ribosome-Protected Fragments and Ribosome Profiling
4.5. Sequence Analysis
4.6. Overexpression of IL4I1 Protein
4.7. RNA Interference Assay
4.8. Real-Time PCR
4.9. Co-immunoprecipitation (Co-IP) Assays
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van der Heide, L. The history of avian reovirus. Avian Dis. 2000, 44, 638–641. [Google Scholar] [CrossRef] [PubMed]
- Dandar, E.; Balint, A.; Kecskemeti, S.; Szentpali-Gavaller, K.; Kisfali, P.; Melegh, B.; Farkas, S.L.; Banyai, K. Detection and characterization of a divergent avian reovirus strain from a broiler chicken with central nervous system disease. Arch. Virol. 2013, 158, 2583–2588. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; Xie, Z.; Xie, L.; Liu, J.; Pang, Y.; Deng, X.; Xie, Z.; Fan, Q.; Luo, S.; Feng, J.; et al. Sequencing and phylogenetic analysis of an avian reovirus genome. Virus Genes 2014, 48, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Green, A.M.; Beatty, P.R.; Hadjilaou, A.; Harris, E. Innate immunity to dengue virus infection and subversion of antiviral responses. J. Mol. Biol. 2014, 426, 1148–1160. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Pillai, P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014, 14, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.Y.; Liu, H.J.; Liao, M.H.; Chang, C.D.; Chang, C.I.; Cheng, H.L.; Lee, J.W.; Shih, W.L. Activation of PI 3-kinase/Akt/NF-kappaB and Stat3 signaling by avian reovirus S1133 in the early stages of infection results in an inflammatory response and delayed apoptosis. Virology 2010, 400, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Neelima, S.; Ram, G.C.; Kataria, J.M.; Goswami, T.K. Avian reovirus induces an inhibitory effect on lymphoproliferation in chickens. Vet. Res. Commun. 2003, 27, 73–85. [Google Scholar] [CrossRef]
- Lostale-Seijo, I.; Martinez-Costas, J.; Benavente, J. Interferon induction by avian reovirus. Virology 2016, 487, 104–111. [Google Scholar] [CrossRef][Green Version]
- Xie, L.; Xie, Z.; Wang, S.; Huang, J.; Deng, X.; Xie, Z.; Luo, S.; Zeng, T.; Zhang, Y.; Zhang, M. Altered gene expression profiles of the MDA5 signaling pathway in peripheral blood lymphocytes of chickens infected with avian reovirus. Arch. Virol. 2019, 164, 2451–2458. [Google Scholar] [CrossRef]
- Wang, S.; Xie, L.; Xie, Z.; Wan, L.; Huang, J.; Deng, X.; Xie, Z.Q.; Luo, S.; Zeng, T.; Zhang, Y.; et al. Dynamic Changes in the Expression of Interferon-Stimulated Genes in Joints of SPF Chickens Infected with Avian Reovirus. Front. Vet. Sci. 2021, 8, 618124. [Google Scholar] [CrossRef]
- Wang, S.; Wan, L.; Ren, H.; Xie, Z.; Xie, L.; Huang, J.; Deng, X.; Xie, Z.; Luo, S.; Li, M.; et al. Screening of interferon-stimulated genes against avian reovirus infection and mechanistic exploration of the antiviral activity of IFIT5. Front. Microbiol. 2022, 13, 998505. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Yu, J.; Luo, Z.; Yu, L.; Liu, S.; Wang, P.; Jia, M.; Wu, T.; Miao, W.; Zhou, L.; et al. Translatome analysis reveals the regulatory role of betaine in high fat diet (HFD)-induced hepatic steatosis. Biochem. Biophys. Res. Commun. 2021, 575, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Yu, J.; Ma, Z.; Fu, Q.; Liu, S.; Luo, Z.; Liu, K.; Yu, L.; Miao, W.; Yu, D.; et al. Translatomics Probes into the Role of Lycopene on Improving Hepatic Steatosis Induced by High-Fat Diet. Front. Nutr. 2021, 8, 727785. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T. Ribosome profiling: New views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014, 15, 205–213. [Google Scholar] [CrossRef]
- Lian, X.; Guo, J.; Gu, W.; Cui, Y.; Zhong, J.; Jin, J.; He, Q.; Wang, T.; Zhang, G. Genome-Wide and Experimental Resolution of Relative Translation Elongation Speed at Individual Gene Level in Human Cells. PLoS Genet. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Huang, T.; Yu, L.; Pan, H.; Ma, Z.; Wu, T.; Zhang, L.; Liu, K.; Qi, Q.; Miao, W.; Song, Z.; et al. Integrated Transcriptomic and Translatomic Inquiry of the Role of Betaine on Lipid Metabolic Dysregulation Induced by a High-Fat Diet. Front. Nutr. 2021, 8, 751436. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Lee, M.T.; Giraldez, A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012, 336, 233–237. [Google Scholar] [CrossRef]
- Zhang, H.; Dou, S.; He, F.; Luo, J.; Wei, L.; Lu, J. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol. 2018, 16, e2003903. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhong, L.; Du, J.; Zhu, C.; Peng, X.; He, X.; Fu, J.; Ouyang, L.; Bian, J.; Hu, L.; et al. Ribosome profiling reveals the effects of nitrogen application translational regulation of yield recovery after abrupt drought-flood alternation in rice. Plant Physiol. Bioch. 2020, 155, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef]
- Lei, L.; Shi, J.; Chen, J.; Zhang, M.; Sun, S.; Xie, S.; Li, X.; Zeng, B.; Peng, L.; Hauck, A.; et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015, 84, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, J.K.; Sterner, F.J.; Botts, S.; Lee, K.P.; Margolin, A. In vitro and in vivo characterization of avian reoviruses. I. Pathogenicity and antigenic relatedness of several avian reovirus isolates. Avian Dis. 1989, 33, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Roessler, D.E.; Rosenberger, J.K. In vitro and in vivo characterization of avian reoviruses. III. Host factors affecting virulence and persistence. Avian Dis. 1989, 33, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Kurihara, Y.; Iwasaki, S. The Plant Translatome Surveyed by Ribosome Profiling. Plant Cell Physiol. 2019, 60, 1917–1926. [Google Scholar] [CrossRef]
- Dunn, J.G.; Foo, C.K.; Belletier, N.G.; Gavis, E.R.; Weissman, J.S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2013, 2, e1179. [Google Scholar] [CrossRef]
- Maier, T.; Guell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; McGeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012, 7, 1534–1550. [Google Scholar] [CrossRef]
- Schafer, S.; Adami, E.; Heinig, M.; Rodrigues, K.; Kreuchwig, F.; Silhavy, J.; van Heesch, S.; Simaite, D.; Rajewsky, N.; Cuppen, E.; et al. Translational regulation shapes the molecular landscape of complex disease phenotypes. Nat. Commun. 2015, 6, 7200. [Google Scholar] [CrossRef]
- Cabrera-Quio, L.E.; Herberg, S.; Pauli, A. Decoding sORF translation—From small proteins to gene regulation. RNA Biol. 2016, 13, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Araujo, P.R.; Yoon, K.; Ko, D.; Smith, A.D.; Qiao, M.; Suresh, U.; Burns, S.C.; Penalva, L.O. Before It Gets Started: Regulating Translation at the 5′ UTR. Comp. Funct. Genom. 2012, 2012, 475731. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, A.; Hussey, G.S.; Howe, P.H. 3′-UTR-mediated post-transcriptional regulation of cancer metastasis: Beginning at the end. RNA Biol. 2011, 8, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Chuang, S.T.; Chen, Y.T.; Shih, W.L.; Chang, C.D.; Liu, H.J. Avian reovirus-induced apoptosis related to tissue injury. Avian Pathol. 2007, 36, 155–159. [Google Scholar] [CrossRef]
- Mason, J.M.; Naidu, M.D.; Barcia, M.; Porti, D.; Chavan, S.S.; Chu, C.C. IL-4-induced gene-1 is a leukocyte L-amino acid oxidase with an unusual acidic pH preference and lysosomal localization. J. Immunol. 2004, 173, 4561–4567. [Google Scholar] [CrossRef] [PubMed]
- Sadik, A.; Somarribas, P.L.; Ozturk, S.; Mohapatra, S.R.; Panitz, V.; Secker, P.F.; Pfander, P.; Loth, S.; Salem, H.; Prentzell, M.T.; et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 2020, 182, 1252–1270. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, S.; Jia, C.; Xue, S.; Dou, C.; Dai, Z.; Xu, H.; Sun, Z.; Geng, T.; Cui, H. Gene expression profile and long non-coding RNA analysis, using RNA-Seq, in chicken embryonic fibroblast cells infected by avian leukosis virus J. Arch. Virol. 2018, 163, 639–647. [Google Scholar] [CrossRef]
- Feng, M.; Xie, T.; Li, Y.; Zhang, N.; Lu, Q.; Zhou, Y.; Shi, M.; Sun, J.; Zhang, X. A balanced game: Chicken macrophage response to ALV-J infection. Vet. Res. 2019, 50, 20. [Google Scholar] [CrossRef]
- Dong, K.; Chang, S.; Xie, Q.; Zhao, P.; Zhang, H. RNA Sequencing revealed differentially expressed genes functionally associated with immunity and tumor suppression during latent phase infection of a vv + MDV in chickens. Sci. Rep. 2019, 9, 14182. [Google Scholar] [CrossRef]
- Marquet, J.; Lasoudris, F.; Cousin, C.; Puiffe, M.L.; Martin-Garcia, N.; Baud, V.; Chereau, F.; Farcet, J.P.; Molinier-Frenkel, V.; Castellano, F. Dichotomy between factors inducing the immunosuppressive enzyme IL-4-induced gene 1 (IL4I1) in B lymphocytes and mononuclear phagocytes. Eur. J. Immunol. 2010, 40, 2557–2568. [Google Scholar] [CrossRef]
- Vazquez-Iglesias, L.; Lostale-Seijo, I.; Martinez-Costas, J.; Benavente, J. Avian reovirus σA localizes to the nucleolus and enters the nucleus by a nonclassical energy- and carrier-independent pathway. J. Virol. 2009, 83, 10163–10175. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Lopez, C.; Martinez-Costas, J.; Esteban, M.; Benavente, J. Evidence that avian reovirus σA protein is an inhibitor of the double-stranded RNA-dependent protein kinase. J. Gen. Virol. 2003, 84, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Xie, Z.; Huang, L.; Fan, Q.; Luo, S.; Huang, J.; Deng, X.; Xie, Z.; Zeng, T.; Zhang, Y.; et al. Avian reovirus σA and σNS proteins activate the phosphatidylinositol 3-kinase-dependent Akt signalling pathway. Arch. Virol. 2016, 161, 2243–2248. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Wang, S.; Xie, Z.; Wang, X.; Wan, L.; Deng, X.; Xie, Z.; Luo, S.; Zeng, T.; Zhang, M.; et al. Gallus NME/NM23 nucleoside diphosphate kinase 2 interacts with viral σA and affects the replication of avian reovirus. Vet. Microbiol. 2021, 252, 108926. [Google Scholar] [CrossRef]
- Shmulevitz, M.; Yameen, Z.; Dawe, S.; Shou, J.; O’Hara, D.; Holmes, I.; Duncan, R. Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: Implications for translation initiation. J. Virol. 2002, 76, 609–618. [Google Scholar] [CrossRef]
- Zhang, Z.; Lin, W.; Li, X.; Cao, H.; Wang, Y.; Zheng, S.J. Critical role of eukaryotic elongation factor 1 alpha 1 (EEF1A1) in avian reovirus sigma-C-induced apoptosis and inhibition of viral growth. Arch. Virol. 2015, 160, 1449–1461. [Google Scholar] [CrossRef]
- Castellano, F.; Molinier-Frenkel, V. An Overview of l-Amino Acid Oxidase Functions from Bacteria to Mammals: Focus on the Immunoregulatory Phenylalanine Oxidase IL4I1. Molecules 2017, 22, 2151. [Google Scholar] [CrossRef]
- Xiao, C.L.; Mai, Z.B.; Lian, X.L.; Zhong, J.Y.; Jin, J.J.; He, Q.Y.; Zhang, G. FANSe2: A robust and cost-efficient alignment tool for quantitative next-generation sequencing applications. PLoS ONE 2014, 9, e94250. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Wang, T.; Cui, Y.; Jin, J.; Guo, J.; Wang, G.; Yin, X.; He, Q.; Zhang, G. Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res. 2013, 41, 4743–4754. [Google Scholar] [CrossRef] [PubMed]
- Li, G.W.; Burkhardt, D.; Gross, C.; Weissman, J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 2014, 157, 624–635. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xie, Z.; Dai, J.; Cao, Y.; Hou, J.; Zheng, Y.; Wei, T.; Mo, M.; Wei, P. Responses of the Toll-like receptor and melanoma differentiation-associated protein 5 signaling pathways to avian infectious bronchitis virus infection in chicks. Virol. Sin. 2016, 31, 57–68. [Google Scholar] [CrossRef] [PubMed]


| Gene | Genbank Accession Number | Primer Sequences (5′-3′) |
|---|---|---|
| ARV σC | L39002.1 | F: CCACGGGAAATCTCACGGTCACT, R: TACGCACGGTCAAGGAACGAATGT |
| IL4I1 | NM_001099351.3 | F: CACGCCGTATCAGTTCACC, R: CCTCACCGCAGCCTTCAT |
| IFN-α | AB021154.1 | F: ATGCCACCTTCTCTCACGAC, R: AGGCGCTGTAATCGTTGTCT |
| IFN-β | X92479.1 | F: ACCAGGATGCCAACTTCT, R: TCACTGGGTGTTGAGACG |
| MDA5 | NM_001193638 | F: CAGCCAGTTGCCCTCGCCTCA, R: AACAGCTCCCTTGCACCGTCT |
| LGP2 | MF563595.1 | F: CCAGAATGAGCAGCAGGAC, R: AATGTTGCACTCAGGGATGT |
| MAVS | MF289560.1 | F: CCTGACTCAAACAAGGGAAG, R: AATCAGAGCGATGCCAACAG |
| TRAF3 | XM_040672281.1 | F: GGACGCACTTGTCGCTGTTT, R: CGGACCCTGATCCATTAGCAT |
| TRAF6 | XM_040673314.1 | F: GATGGAGACGCAAAACACTCAC, R: GCATCACAACAGGTCTCTCTTC |
| IKKε | XM_428036.4 | F: TGGATGGGATGGTGTCTGAAC, R: TGCGGAACTGCTTGTAGATG |
| TBK1 | MF159109.1 | F: AAGAAGGCACACATCCGAGA, R: GGTAGCGTGCAAATACAGC |
| IRF7 | NM_205372.1 | F: CAGTGCTTCTCCAGCACAAA, R: TGCATGTGGTATTGCTCGAT |
| NF-κB | NM_205129.1 | F: CATTGCCAGCATGGCTACTAT, R: TTCCAGTTCCCGTTTCTTCAC |
| GAPDH | NM_204305.1 | F: GCACTGTCAAGGCTGAGAACG, R: GATGATAACACGCTTAGCACCAC |
| siRNA | Sense | Antisense |
|---|---|---|
| si108 | GCUGCUGAGUAUUGUGAAATT | UUUCACAAUACUCAGCAGCTT |
| si336 | GCUGGUGCGUGAGUUUAUATT | UAUAAACUCACGCACCAGCTT |
| si1357 | CCGUAUCAGUUCACCGAUUTT | AAUCGGUGAACUGAUACGGTT |
| siNC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Huang, T.; Xie, Z.; Wan, L.; Ren, H.; Wu, T.; Xie, L.; Luo, S.; Li, M.; Xie, Z.; et al. Transcriptomic and Translatomic Analyses Reveal Insights into the Signaling Pathways of the Innate Immune Response in the Spleens of SPF Chickens Infected with Avian Reovirus. Viruses 2023, 15, 2346. https://doi.org/10.3390/v15122346
Wang S, Huang T, Xie Z, Wan L, Ren H, Wu T, Xie L, Luo S, Li M, Xie Z, et al. Transcriptomic and Translatomic Analyses Reveal Insights into the Signaling Pathways of the Innate Immune Response in the Spleens of SPF Chickens Infected with Avian Reovirus. Viruses. 2023; 15(12):2346. https://doi.org/10.3390/v15122346
Chicago/Turabian StyleWang, Sheng, Tengda Huang, Zhixun Xie, Lijun Wan, Hongyu Ren, Tian Wu, Liji Xie, Sisi Luo, Meng Li, Zhiqin Xie, and et al. 2023. "Transcriptomic and Translatomic Analyses Reveal Insights into the Signaling Pathways of the Innate Immune Response in the Spleens of SPF Chickens Infected with Avian Reovirus" Viruses 15, no. 12: 2346. https://doi.org/10.3390/v15122346
APA StyleWang, S., Huang, T., Xie, Z., Wan, L., Ren, H., Wu, T., Xie, L., Luo, S., Li, M., Xie, Z., Fan, Q., Huang, J., Zeng, T., Zhang, Y., Zhang, M., & Wei, Y. (2023). Transcriptomic and Translatomic Analyses Reveal Insights into the Signaling Pathways of the Innate Immune Response in the Spleens of SPF Chickens Infected with Avian Reovirus. Viruses, 15(12), 2346. https://doi.org/10.3390/v15122346






