A Glimpse on the Evolution of RNA Viruses: Implications and Lessons from SARS-CoV-2
Abstract
1. Introduction
2. RNA Virus Classification
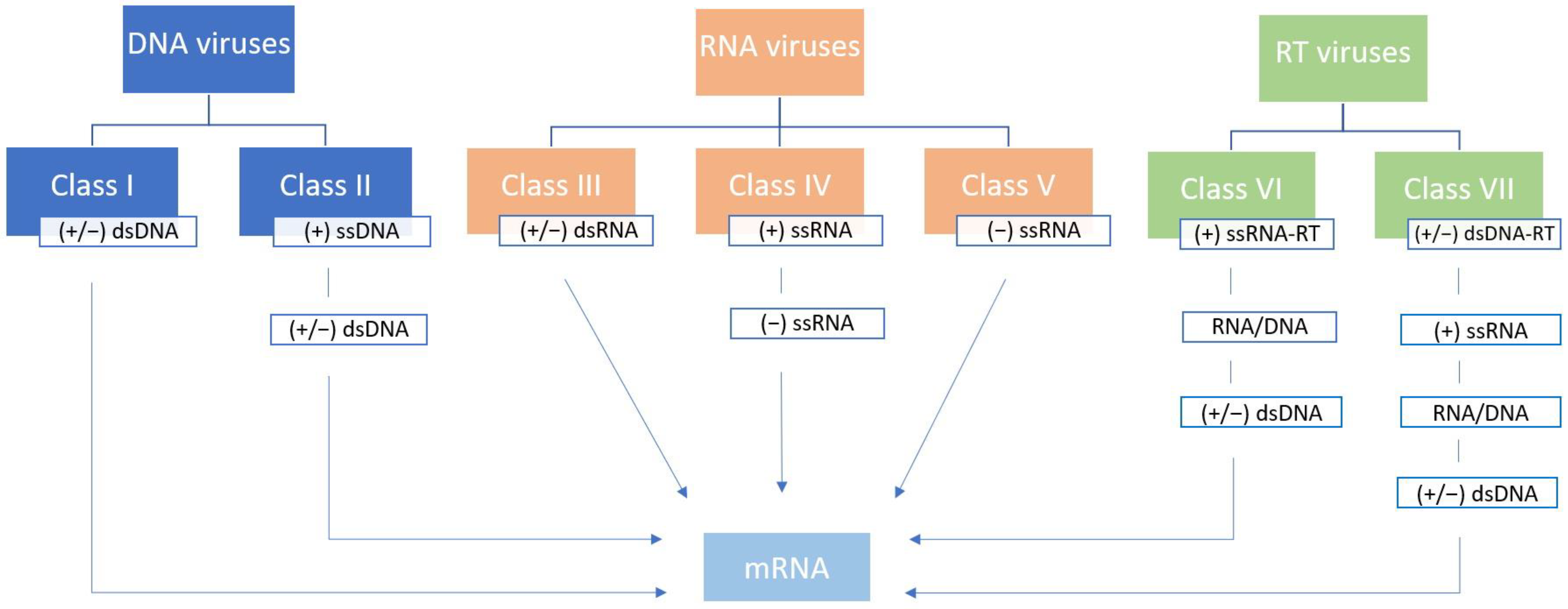
2.1. Baltimore Classification
2.2. The International Committee on Taxonomy of Viruses Classification
3. RNA Virus Characteristics
3.1. Host Range
3.2. Structural Genome Features
3.3. RNA Regulatory Processes
3.4. Quasispecies Concept
4. Mechanisms of RNA Virus Variation
4.1. Mutation
4.2. Recombination
4.3. Reassortment
5. Causes and Consequences of RNA Virus Variation
6. Evolution of Human Coronaviruses
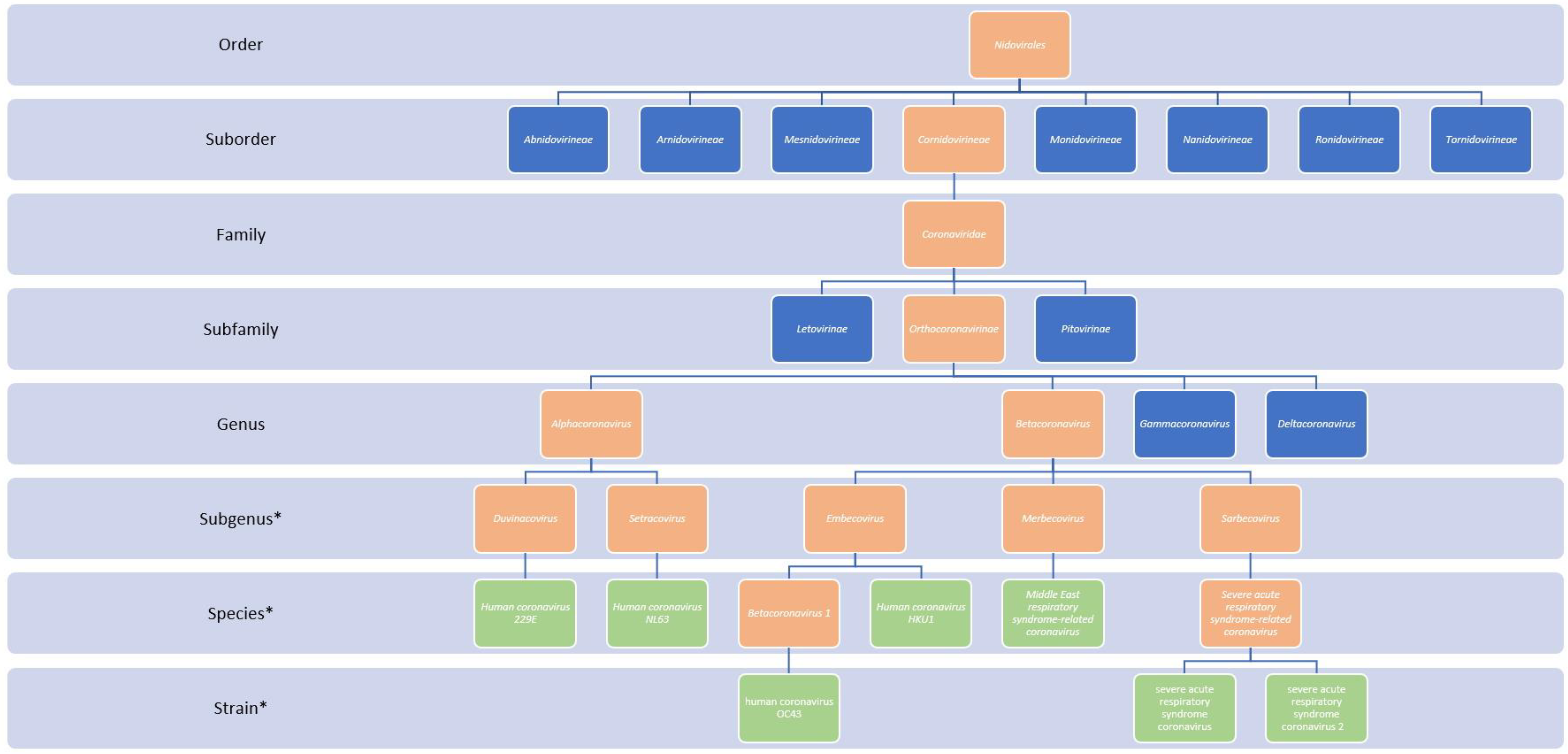
6.1. Classification of Coronaviruses
6.2. Structure and Genome of Coronaviruses
6.3. Mutation in Coronaviruses
6.4. Recombination in Coronaviruses
6.5. Origin of Human Coronaviruses
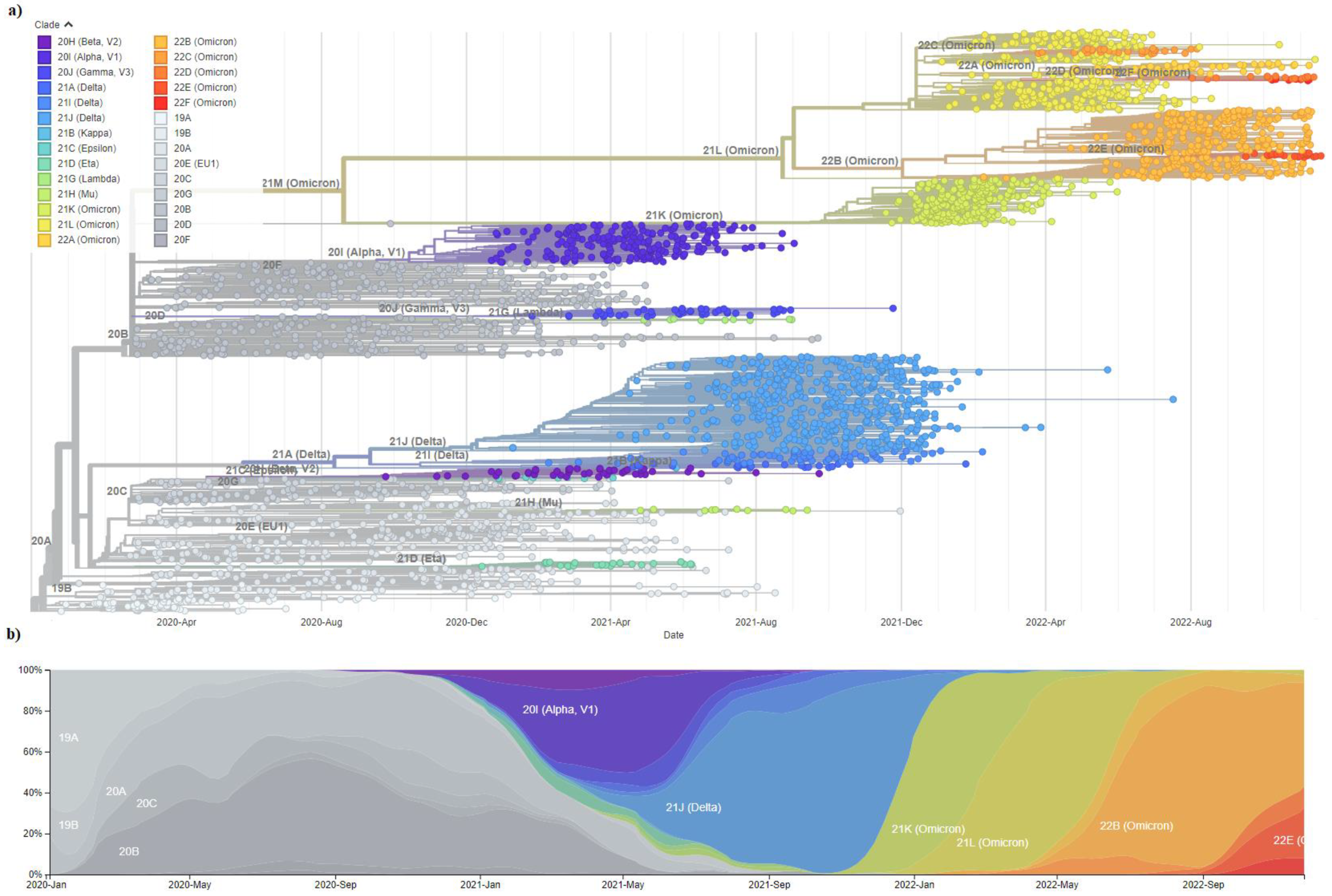
6.6. SARS-CoV-2 Variant Evolution
7. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bolduc, B.; Shaughnessy, D.P.; Wolf, Y.I.; Koonin, E.V.; Roberto, F.F.; Young, M. Identification of Novel Positive-Strand RNA Viruses by Metagenomic Analysis of Archaea-Dominated Yellowstone Hot Springs. J. Virol. 2012, 86, 5562–5573. [Google Scholar] [CrossRef] [PubMed]
- Callanan, J.; Stockdale, S.R.; Adriaenssens, E.M.; Kuhn, J.H.; Rumnieks, J.; Pallen, M.J.; Shkoporov, A.N.; Draper, L.A.; Ross, R.P.; Hill, C. Leviviricetes: Expanding and Restructuring the Taxonomy of Bacteria-Infecting Single-Stranded RNA Viruses. Microb. Genom. 2021, 7, 686. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. Expression of Animal Virus Genomes. Bacteriol. Rev. 1971, 35, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.; Mohr, I. Viral Subversion of the Host Protein Synthesis Machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef]
- Payne, S. Virus Interactions with the Cell. In Viruses: From Understanding to Investigation; Elsevier: Amsterdam, The Netherlands, 2017; pp. 23–35. [Google Scholar] [CrossRef]
- Becher, P.; Tautz, N. RNA Recombination in Pestiviruses: Cellular RNA Sequences in Viral Genomes Highlight the Role of Host Factors for Viral Persistence and Lethal Disease. RNA Biol. 2011, 8, 216–224. [Google Scholar] [CrossRef]
- Sanjuán, R.; Domingo-Calap, P. Mechanisms of Viral Mutation. Cell. Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef]
- Mattenberger, F.; Vila-Nistal, M.; Geller, R. Increased RNA Virus Population Diversity Improves Adaptability. Sci. Rep. 2021, 11, 6824. [Google Scholar] [CrossRef]
- Fleischmann, W.R., Jr. Viral Genetics. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 43. [Google Scholar]
- Holland, D.J. Emerging Viruses. Curr. Opin. Pediatr. 1998, 10, 34–40. [Google Scholar] [CrossRef]
- Graham, R.L.; Baric, R.S. Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission. J. Virol. 2009, 84, 3134–3146. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar]
- Haake, C.; Cook, S.; Pusterla, N.; Murphy, B. Coronavirus Infections in Companion Animals: Virology, Epidemiology, Clinical and Pathologic Features. Viruses 2020, 12, 1023. [Google Scholar] [CrossRef]
- The New Scope of Virus Taxonomy: Partitioning the Virosphere into 15 Hierarchical Ranks. Nat. Microbiol. 2020, 5, 668–674. [CrossRef]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the Largest RNA Virus Genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef]
- Eckerle, L.D.; Lu, X.; Sperry, S.M.; Choi, L.; Denison, M.R. High Fidelity of Murine Hepatitis Virus Replication Is Decreased in Nsp14 Exoribonuclease Mutants. J. Virol. 2007, 81, 12135–12144. [Google Scholar] [CrossRef]
- Koonin, E.V.; Krupovic, M.; Agol, V.I. The Baltimore Classification of Viruses 50 Years Later: How Does It Stand in the Light of Virus Evolution? Microbiol. Mol. Biol. Rev. 2021, 85, e0005321. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V. A Virocentric Perspective on the Evolution of Life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Senkevich, T.G.; Dolja, V.V. The Ancient Virus World and Evolution of Cells. Biol. Direct 2006, 1, 29. [Google Scholar]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and Evolution of the Global RNA Virome. mBio 2018, 9, e02329. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.M.; Nelson, M.I.; Turner, P.E.; Patton, J.T. Reassortment in Segmented RNA Viruses: Mechanisms and Outcomes. Nat. Rev. Microbiol. 2016, 14, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Newburn, L.R.; White, K.A. Trans-Acting RNA–RNA Interactions in Segmented RNA Viruses. Viruses 2019, 11, 751. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, e00061. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to Virus Taxonomy and the International Code of Virus Classification and Nomenclature Ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; et al. Changes to Virus Taxonomy and the Statutes Ratified by the International Committee on Taxonomy of Viruses (2020). Arch. Virol. 2020, 165, 2737–2748. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Curtis Hendrickson, R.; et al. Recent Changes to Virus Taxonomy Ratified by the International Committee on Taxonomy of Viruses (2022). Arch. Virol. 2022, 167, 2429–2440. [Google Scholar] [CrossRef]
- Current ICTV Taxonomy Release|ICTV. Available online: https://ictv.global/taxonomy (accessed on 6 October 2022).
- Krupovic, M.; Prangishvili, D.; Hendrix, R.W.; Bamford, D.H. Genomics of Bacterial and Archaeal Viruses: Dynamics within the Prokaryotic Virosphere. Microbiol. Mol. Biol. Rev. 2011, 75, 610–635. [Google Scholar] [CrossRef]
- Saberi, A.; Gulyaeva, A.A.; Brubacher, J.L.; Newmark, P.A.; Gorbalenya, A.E. A Planarian Nidovirus Expands the Limits of RNA Genome Size. PLoS Pathog. 2018, 14, e1007314. [Google Scholar] [CrossRef]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A New Wine Saccharomyces Cerevisiae Killer Toxin (Klus), Encoded by a Double-Stranded RNA Virus, with Broad Antifungal Activity Is Evolutionarily Related to a Chromosomal Host Gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef]
- Holmes, E.C. Error Thresholds and the Constraints to RNA Virus Evolution. Trends Microbiol. 2003, 11, 543–546. [Google Scholar] [CrossRef]
- Poole, A.M.; Logan, D.T. Modern MRNA Proofreading and Repair: Clues That the Last Universal Common Ancestor Possessed an RNA Genome? Mol. Biol. Evol. 2005, 22, 1444–1455. [Google Scholar] [CrossRef]
- Chirico, N.; Vianelli, A.; Belshaw, R. Why Genes Overlap in Viruses. Proc. R. Soc. B Biol. Sci. 2010, 277, 3809–3817. [Google Scholar] [CrossRef]
- Belshaw, R.; Pybus, O.G.; Rambaut, A. The Evolution of Genome Compression and Genomic Novelty in RNA Viruses. Genome Res. 2007, 17, 1496–1504. [Google Scholar] [CrossRef]
- Dreher, T.W. Functions of the 3’-untranslated regions of positive strand rna viral genomes. Annu. Rev. Phytopathol. 1999, 37, 151–174. [Google Scholar] [CrossRef]
- Zong, J.; Yao, X.; Yin, J.; Zhang, D.; Ma, H. Evolution of the RNA-Dependent RNA Polymerase (RdRP) Genes: Duplications and Possible Losses before and after the Divergence of Major Eukaryotic Groups. Gene 2009, 447, 29–39. [Google Scholar] [CrossRef]
- Rampersad, S.; Tennant, P. Replication and Expression Strategies of Viruses. In Viruses: From Understanding to Investigation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 55–82. [Google Scholar]
- Vanden Heuvel, J.P. Posttranscriptional Processing of Messenger RNA. In Regulation of Gene Expression; Perdew, G.H., Vanden Heuvel, J.P., Peters, J.M., Eds.; Humana Press: Totowa, NJ, USA, 2007; pp. 105–106. [Google Scholar]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and Unconventional Mechanisms for Capping Viral MRNA. Nat. Rev. Microbiol. 2012, 10, 51–65. [Google Scholar] [CrossRef]
- Li, X.; Palese, P. Characterization of the Polyadenylation Signal of Influenza Virus RNA. J. Virol. 1994, 68, 1245–1249. [Google Scholar] [CrossRef]
- Kempf, B.J.; Barton, D.J. Picornavirus RNA Polyadenylation by 3Dpol, the Viral RNA-Dependent RNA Polymerase. Virus Res. 2015, 206, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Piñeiro, D.; Martinez-Salas, E. RNA Structural Elements of Hepatitis c Virus Controlling Viral RNA Translation and the Implications for Viral Pathogenesis. Viruses 2012, 4, 2233–2250. [Google Scholar] [CrossRef] [PubMed]
- Chua, M.A.; Schmid, S.; Perez, J.T.; Langlois, R.A.; Oever, B.R. Influenza a Virus Utilizes Suboptimal Splicing to Coordinate the Timing of Infection. Cell Rep. 2013, 3, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, S.G.; Sawicki, D.L.; Siddell, S.G. A Contemporary View of Coronavirus Transcription. J. Virol. 2007, 81, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef] [PubMed]
- Jaafar, Z.A.; Kieft, J.S. Viral RNA Structure-Based Strategies to Manipulate Translation. Nat. Rev. Microbiol. 2018, 17, 110–123. [Google Scholar] [CrossRef]
- Jan, E.; Mohr, I.; Walsh, D. A Cap-To-Tail Guide to MRNA Translation Strategies in Virus-Infected Cells. Annu. Rev. Virol. 2016, 3, 283–307. [Google Scholar] [CrossRef]
- Lukavsky, P.J. Structure and Function of HCV IRES Domains. Virus Res. 2009, 139, 166–171. [Google Scholar] [CrossRef]
- Ishimaru, D.; Plant, E.P.; Sims, A.C.; Yount, B.L.; Roth, B.M.; Eldho, N.V.; Pérez-Alvarado, G.C.; Armbruster, D.W.; Baric, R.S.; Dinman, J.D.; et al. RNA Dimerization Plays a Role in Ribosomal Frameshifting of the SARS Coronavirus. Nucleic Acids Res. 2012, 41, 2594–2608. [Google Scholar] [CrossRef]
- Sender, R.; Bar-On, Y.M.; Gleizer, S.; Bernshtein, B.; Flamholz, A.; Phillips, R.; Milo, R. The Total Number and Mass of SARS-CoV-2 Virions. Proc. Natl. Acad. Sci. USA 2021, 118, e2024815118. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. A Principle of Natural Self-Organization. Naturwissenschaften 1977, 64, 541–565. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The Hypercycle. Naturwissenschaften 1978, 65, 7–41. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The Hypercycle; Springer: Berlin/Heidelberg, Germany, 1979. [Google Scholar]
- Eigen, M. On the Nature of Virus Quasispecies. Trends Microbiol. 1996, 4, 216–218. [Google Scholar] [CrossRef]
- Biebricher, C.K.; Eigen, M. The Error Threshold. Virus Res. 2005, 107, 117–127. [Google Scholar] [CrossRef]
- Eigen, M. The Origin of Genetic Information: Viruses as Models. Gene 1993, 135, 37–47. [Google Scholar] [CrossRef]
- Domingo, E.; Sabo, D.; Taniguchi, T.; Weissmann, C. Nucleotide Sequence Heterogeneity of an RNA Phage Population. Cell 1978, 13, 735–744. [Google Scholar] [CrossRef]
- Holland, J.; Spindler, K.; Horodyski, F.; Grabau, E.; Nichol, S.; VandePol, S. Rapid Evolution of RNA Genomes. Science 1982, 215, 1577–1585. [Google Scholar] [CrossRef]
- Duarte, E.A.; Novella, I.S.; Ledesma, S.; Clarke, D.K.; Moya, A.; Elena, S.F.; Domingo, E.; Holland, J.J. Subclonal Components of Consensus Fitness in an RNA Virus Clone. J. Virol. 1994, 68, 4295–4301. [Google Scholar] [CrossRef]
- Martell, M.; Esteban, J.I.; Quer, J.; Genescà, J.; Weiner, A.; Esteban, R.; Guardia, J.; Gómez, J. Hepatitis c Virus (HCV) Circulates as a Population of Different but Closely Related Genomes: Quasispecies Nature of HCV Genome Distribution. J. Virol. 1992, 66, 3225–3229. [Google Scholar] [CrossRef]
- Domingo, E. Quasispecies. Encycl. Virol. 1999, 1431–1436. [Google Scholar]
- Domingo, E.; Holland, J.J. RNA VIRUS MUTATIONS and FITNESS for SURVIVAL. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.M. RNA Recombination in Animal and Plant Viruses. Microbiol. Rev. 1992, 56, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Vijaykrishna, D.; Mukerji, R.; Smith, G.J.D. RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion. PLOS Pathog. 2015, 11, e1004902. [Google Scholar] [CrossRef] [PubMed]
- Jackowiak, P.; Kuls, K.; Budzko, L.; Mania, A.; Figlerowicz, M.; Figlerowicz, M. Phylogeny and Molecular Evolution of the Hepatitis c Virus. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 21, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Bentley, K.; Evans, D.J. Mechanisms and Consequences of Positive-Strand RNA Virus Recombination. J. Gen. Virol. 2018, 99, 1345–1356. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Holmes, E.C. Why Do RNA Viruses Recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Holmes, E.C. Gene Duplication Is Infrequent in the Recent Evolutionary History of RNA Viruses. Mol. Biol. Evol. 2013, 30, 1263–1269. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Morris, T.J. Evolution and Taxonomy of Positive-Strand RNA Viruses: Implications of Comparative Analysis of Amino Acid Sequences. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 375–430. [Google Scholar] [CrossRef]
- Iranzo, J.; Krupovic, M.; Koonin, E.V. The Double-Stranded DNA Virosphere as a Modular Hierarchical Network of Gene Sharing. mBio 2016, 7, e00978. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of Life: The RNA World. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origin of Viruses: Primordial Replicators Recruiting Capsids from Hosts. Nat. Rev. Microbiol. 2019, 17, 449–458. [Google Scholar] [CrossRef]
- Iyer, L.M.; Koonin, E.V.; Aravind, L. Evolutionary Connection between the Catalytic Subunits of DNA-Dependent RNA Polymerases and Eukaryotic RNA-Dependent RNA Polymerases and the Origin of RNA Polymerases. BMC Struct. Biol. 2003, 3, 1. [Google Scholar] [CrossRef]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef]
- Hirst, G.K. Genetic Recombination with Newcastle Disease Virus, Polioviruses, and Influenza. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 303–309. [Google Scholar] [CrossRef]
- Ledinko, N. Genetic Recombination with Poliovirus Type 1. Virology 1963, 20, 107–119. [Google Scholar] [CrossRef]
- Cooper, P.D.; Steiner-Pryor, A.; Scotti, P.D.; Delong, D. On the Nature of Poliovirus Genetic Recombinants. J. Gen. Virol. 1974, 23, 41–49. [Google Scholar] [CrossRef]
- Savolainen-Kopra, C.; Blomqvist, S. Mechanisms of Genetic Variation in Polioviruses. Rev. Med. Virol. 2010, 20, 358–371. [Google Scholar] [CrossRef]
- Banner, L.R.; Mc Lai, M. Random Nature of Coronavirus RNA Recombination in the Absence of Selection Pressure. Virology 1991, 185, 441–445. [Google Scholar] [CrossRef]
- Gmyl, A.P.; Belousov, E.V.; Maslova, S.V.; Khitrina, E.V.; Chetverin, A.B.; Agol, V.I. Nonreplicative RNA Recombination in Poliovirus. J. Virol. 1999, 73, 8958–8965. [Google Scholar] [CrossRef]
- Scheel, T.K.H.; Galli, A.; Li, Y.-P.; Mikkelsen, L.S.; Gottwein, J.M.; Bukh, J. Productive Homologous and Non-Homologous Recombination of Hepatitis c Virus in Cell Culture. PLoS Pathog. 2013, 9, e1003228. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Mounce, B.C.; Rozen-Gagnon, K.; Hooikaas, P.J.; Stapleford, K.A.; Moratorio, G.; Vignuzzi, M. Low-Fidelity Polymerases of Alphaviruses Recombine at Higher Rates to Overproduce Defective Interfering Particles. J. Virol. 2016, 90, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Woodman, A.; Arnold, J.J.; Cameron, C.E.; Evans, D.J. Biochemical and Genetic Analysis of the Role of the Viral Polymerase in Enterovirus Recombination. Nucleic Acids Res. 2016, 44, 6883–6895. [Google Scholar] [CrossRef] [PubMed]
- Rowe, C.L.; Fleming, J.O.; Nathan, M.J.; Sgro, J.Y.; Palmenberg, A.C.; Baker, S.C. Generation of Coronavirus Spike Deletion Variants by High-Frequency Recombination at Regions of Predicted RNA Secondary Structure. J. Virol. 1997, 71, 6183–6190. [Google Scholar] [CrossRef] [PubMed]
- Runckel, C.; Westesson, O.; Andino, R.; DeRisi, J.L. Identification and Manipulation of the Molecular Determinants Influencing Poliovirus Recombination. PLoS Pathog. 2013, 9, e1003164. [Google Scholar] [CrossRef] [PubMed]
- Lowry, K.; Woodman, A.; Cook, J.; Evans, D.J. Recombination in Enteroviruses Is a Biphasic Replicative Process Involving the Generation of Greater-than Genome Length “Imprecise” Intermediates. PLoS Pathog. 2014, 10, e1004191. [Google Scholar] [CrossRef] [PubMed]
- Taucher, C.; Berger, A.; Mandl, C.W. A Trans -Complementing Recombination Trap Demonstrates a Low Propensity of Flaviviruses for Intermolecular Recombination. J. Virol. 2010, 84, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Marshall, N.; Priyamvada, L.; Ende, Z.; Steel, J.; Lowen, A.C. Influenza Virus Reassortment Occurs with High Frequency in the Absence of Segment Mismatch. PLoS Pathog. 2013, 9, e1003421. [Google Scholar] [CrossRef]
- Bancroft, C.T.; Parslow, T.G. Evidence for Segment-Nonspecific Packaging of the Influenza a Virus Genome. J. Virol. 2002, 76, 7133–7139. [Google Scholar] [CrossRef]
- Essere, B.; Yver, M.; Gavazzi, C.; Terrier, O.; Isel, C.; Fournier, E.; Giroux, F.; Textoris, J.; Julien, T.; Socratous, C.; et al. Critical Role of Segment-Specific Packaging Signals in Genetic Reassortment of Influenza a Viruses. Proc. Natl. Acad. Sci. USA 2013, 110, E3840–E3848. [Google Scholar] [CrossRef]
- Baker, S.F.; Nogales, A.; Finch, C.; Tuffy, K.M.; Domm, W.; Perez, D.R.; Topham, D.J.; Martínez-Sobrido, L. Influenza a and B Virus Intertypic Reassortment through Compatible Viral Packaging Signals. J. Virol. 2014, 88, 10778–10791. [Google Scholar] [CrossRef]
- Klempa, B. Reassortment Events in the Evolution of Hantaviruses. Virus Genes 2018, 54, 638–646. [Google Scholar] [CrossRef]
- Boni, M.F.; Zhou, Y.; Taubenberger, J.K.; Holmes, E.C. Homologous Recombination Is Very Rare or Absent in Human Influenza a Virus. J. Virol. 2008, 82, 4807–4811. [Google Scholar] [CrossRef][Green Version]
- Varsani, A.; Lefeuvre, P.; Roumagnac, P.; Martin, D. Notes on Recombination and Reassortment in Multipartite/Segmented Viruses. Curr. Opin. Virol. 2018, 33, 156–166. [Google Scholar] [CrossRef]
- Johnson, T.; Barton, N.H. The Effect of Deleterious Alleles on Adaptation in Asexual Populations. Genetics 2002, 162, 395–411. [Google Scholar] [CrossRef]
- Peck, J.R. A Ruby in the Rubbish: Beneficial Mutations, Deleterious Mutations and the Evolution of Sex. Genetics 1994, 137, 597–606. [Google Scholar] [CrossRef]
- Elena, S.F.; Sanjuan, R. Adaptive Value of High Mutation Rates of RNA Viruses: Separating Causes from Consequences. J. Virol. 2005, 79, 11555–11558. [Google Scholar] [CrossRef]
- Bonhoeffer, S.; Chappey, C.; Parkin, N.T.; Whitcomb, J.M.; Petropoulos, C.J. Evidence for Positive Epistasis in HIV-1. Science 2004, 306, 1547–1550. [Google Scholar] [CrossRef]
- Chare, E.R.; Gould, E.A.; Holmes, E.C. Phylogenetic Analysis Reveals a Low Rate of Homologous Recombination in Negative-Sense RNA Viruses. J. Gen. Virol. 2003, 84, 2691–2703. [Google Scholar] [CrossRef]
- Han, G.-Z.; Worobey, M. Homologous Recombination in Negative Sense RNA Viruses. Viruses 2011, 3, 1358–1373. [Google Scholar] [CrossRef]
- Twiddy, S.S. The Extent of Homologous Recombination in Members of the Genus Flavivirus. J. Gen. Virol. 2003, 84, 429–440. [Google Scholar] [CrossRef]
- Pickett, B.E.; Lefkowitz, E.J. Recombination in West Nile Virus: Minimal Contribution to Genomic Diversity. Virol. J. 2009, 6, 165. [Google Scholar] [CrossRef] [PubMed]
- González-Candelas, F.; López-Labrador, F.X.; Bracho, M.A. Recombination in Hepatitis c Virus. Viruses 2011, 3, 2006–2024. [Google Scholar] [CrossRef] [PubMed]
- Master Species Lists|ICTV. Available online: https://ictv.global/msl (accessed on 5 November 2022).
- Bonilauri, P.; Rugna, G. Animal Coronaviruses and SARS-COV-2 in Animals, What Do We Actually Know? Life 2021, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Hamre, D.; Procknow, J.J. A New Virus Isolated from the Human Respiratory Tract. Exp. Biol. Med. 1966, 121, 190–193. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, K.; Dees, J.H.; Becker, W.B.; Kapikian, A.Z.; Chanock, R.M. Recovery in Tracheal Organ Cultures of Novel Viruses from Patients with Respiratory Disease. Proc. Natl. Acad. Sci. USA 1967, 57, 933–940. [Google Scholar] [CrossRef]
- Peiris, J.; Lai, S.; Poon, L.; Guan, Y.; Yam, L.; Lim, W.; Nicholls, J.; Yee, W.; Yan, W.; Cheung, M.; et al. Coronavirus as a Possible Cause of Severe Acute Respiratory Syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; Yuen, K.Y.; Osterhaus, A.D.M.E.; Stöhr, K. The Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 349, 2431–2441. [Google Scholar] [CrossRef]
- Snijder, E.J.; Bredenbeek, P.J.; Dobbe, J.C.; Thiel, V.; Ziebuhr, J.; Poon, L.L.M.; Guan, Y.; Rozanov, M.; Spaan, W.J.M.; Gorbalenya, A.E. Unique and Conserved Features of Genome and Proteome of SARS-Coronavirus, an Early Split-off from the Coronavirus Group 2 Lineage. J. Mol. Biol. 2003, 331, 991–1004. [Google Scholar] [CrossRef]
- van der Hoek, L.; Pyrc, K.; Berkhout, B. Human Coronavirus NL63, a New Respiratory Virus. FEMS Microbiol. Rev. 2006, 30, 760–773. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.; Chan, K.; Tsoi, H.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2004, 79, 884–895. [Google Scholar] [CrossRef]
- Monto, A.S. Medical Reviews. Coronaviruses. Yale J. Biol. Med. 1974, 47, 234–251. [Google Scholar]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- MERS-CoV Worldwide Overview. Available online: https://www.ecdc.europa.eu/en/middle-east-respiratory-syndrome-coronavirus-mers-cov-situation-update (accessed on 10 November 2022).
- World Health Organization (WHO). Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Available online: https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov) (accessed on 10 November 2022).
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 1–9. [Google Scholar]
- World Health Organization. WHO COVID-19 Dashboard. Available online: https://covid19.who.int/ (accessed on 2 November 2022).
- Nayak, B.; Kumar, S.; Collins, P.L.; Samal, S.K. Molecular Characterization and Complete Genome Sequence of Avian Paramyxovirus Type 4 Prototype Strain Duck/Hong Kong/D3/75. Virol. J. 2008, 5, 124. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Tahir, M. Coronavirus Genomic Nsp14-ExoN, Structure, Role, Mechanism, and Potential Application as a Drug Target. J. Med. Virol. 2021, 93, 4258–4264. [Google Scholar] [CrossRef]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA Virus 3′->5′ Exoribonuclease That Is Critically Involved in Coronavirus RNA Synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef]
- Eckerle, L.D.; Becker, M.M.; Halpin, R.A.; Li, K.; Venter, E.; Lu, X.; Scherbakova, S.; Graham, R.L.; Baric, R.S.; Stockwell, T.B.; et al. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. PLoS Pathog. 2010, 6, e1000896. [Google Scholar] [CrossRef]
- Zuo, Y. Exoribonuclease Superfamilies: Structural Analysis and Phylogenetic Distribution. Nucleic Acids Res. 2001, 29, 1017–1026. [Google Scholar] [CrossRef]
- Berrio, A.; Gartner, V.; Wray, G.A. Positive Selection within the Genomes of SARS-CoV-2 and Other Coronaviruses Independent of Impact on Protein Function. PeerJ 2020, 8, e10234. [Google Scholar] [CrossRef] [PubMed]
- Jaroszewski, L.; Iyer, M.; Alisoltani, A.; Sedova, M.; Godzik, A. The Interplay of SARS-CoV-2 Evolution and Constraints Imposed by the Structure and Functionality of Its Proteins. PLoS Comput. Biol. 2021, 17, e1009147. [Google Scholar] [CrossRef] [PubMed]
- Magazine, N.; Zhang, T.; Wu, Y.; McGee, M.C.; Veggiani, G.; Huang, W. Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses 2022, 14, 640. [Google Scholar] [CrossRef] [PubMed]
- Amicone, M.; Borges, V.; Alves, M.J.; Isidro, J.; Zé-Zé, L.; Duarte, S.; Vieira, L.; Guiomar, R.; Gomes, J.P.; Gordo, I. Mutation Rate of SARS-CoV-2 and Emergence of Mutators during Experimental Evolution. Evol. Med. Public Health 2022, 10, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.M.; Baric, R.S.; Makino, S.; Keck, J.G.; Egbert, J.; Leibowitz, J.L.; Stohlman, S.A. Recombination between Nonsegmented RNA Genomes of Murine Coronaviruses. J. Virol. 1985, 56, 449–456. [Google Scholar] [CrossRef]
- Makino, S.; Keck, J.G.; Stohlman, S.A.; Lai, M.M. High-Frequency RNA Recombination of Murine Coronaviruses. J. Virol. 1986, 57, 729–737. [Google Scholar] [CrossRef]
- Koetzner, C.A.; Parker, M.M.; Ricard, C.S.; Sturman, L.S.; Masters, P.S. Repair and Mutagenesis of the Genome of a Deletion Mutant of the Coronavirus Mouse Hepatitis Virus by Targeted RNA Recombination. J. Virol. 1992, 66, 1841–1848. [Google Scholar] [CrossRef]
- Baric, R.S.; Fu, K.; Schaad, M.C.; Stohlman, S.A. Establishing a Genetic Recombination Map for Murine Coronavirus Strain A59 Complementation Groups. Virology 1990, 177, 646–656. [Google Scholar] [CrossRef]
- Goldstein, S.A.; Brown, J.; Pedersen, B.S.; Quinlan, A.R.; Elde, N.C. Extensive Recombination-Driven Coronavirus Diversification Expands the Pool of Potential Pandemic Pathogens. Genome Biol. Evol. 2021, 14, evac161. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.-Y.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary Origins of the SARS-CoV-2 Sarbecovirus Lineage Responsible for the COVID-19 Pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Huang, Y.; Yuen, K.-Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 428–440. [Google Scholar]
- Pfefferle, S.; Oppong, S.; Drexler, J.F.; Gloza-Rausch, F.; Ipsen, A.; Seebens, A.; Müller, M.A.; Annan, A.; Vallo, P.; Adu-Sarkodie, Y.; et al. Distant Relatives of Severe Acute Respiratory Syndrome Coronavirus and Close Relatives of Human Coronavirus 229E in Bats, Ghana. Emerg. Infect. Dis. 2009, 15, 1377–1384. [Google Scholar] [CrossRef]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef]
- Stout, A.E.; Millet, J.K.; Stanhope, M.J.; Whittaker, G.R. Furin Cleavage Sites in the Spike Proteins of Bat and Rodent Coronaviruses: Implications for Virus Evolution and Zoonotic Transfer from Rodent Species. One Health 2021, 13, 100282. [Google Scholar] [CrossRef]
- Zhu, Z.; Meng, K.; Meng, G. Genomic Recombination Events May Reveal the Evolution of Coronavirus and the Origin of SARS-CoV-2. Sci. Rep. 2020, 10, 21617. [Google Scholar] [CrossRef]
- Khalafalla, A.I.; Lu, X.; Al-Mubarak, A.I.A.; Dalab, A.H.S.; Al-Busadah, K.A.S.; Erdman, D.D. MERS-CoV in Upper Respiratory Tract and Lungs of Dromedary Camels, Saudi Arabia, 2013–2014. Emerg. Infect. Dis. 2015, 21, 1153–1158. [Google Scholar] [CrossRef]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; AlHakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East Respiratory Syndrome Coronavirus in Bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef]
- Song, H.-D.; Tu, C.-C.; Zhang, G.-W.; Wang, S.-Y.; Zheng, K.; Lei, L.-C.; Chen, Q.-X.; Gao, Y.-W.; Zhou, H.-Q.; Xiang, H.; et al. Cross-Host Evolution of Severe Acute Respiratory Syndrome Coronavirus in Palm Civet and Human. Proc. Natl. Acad. Sci. USA 2005, 102, 2430–2435. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351. [Google Scholar] [CrossRef]
- Poon, L.L.M.; Chu, D.K.W.; Chan, K.H.; Wong, O.K.; Ellis, T.M.; Leung, Y.H.C.; Lau, S.K.P.; Woo, P.C.Y.; Suen, K.Y.; Yuen, K.Y.; et al. Identification of a Novel Coronavirus in Bats. J. Virol. 2005, 79, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.A.; Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.; Bosch, B.-J.; Lattwein, E.; Hilali, M.; Musa, B.E.; et al. MERS Coronavirus Neutralizing Antibodies in Camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014, 20, 2093–2095. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zeng, L.-P.; Yang, X.-L.; Ge, X.-Y.; Zhang, W.; Li, B.; Xie, J.-Z.; Shen, X.-R.; Zhang, Y.-Z.; Wang, N.; et al. Discovery of a Rich Gene Pool of Bat SARS-Related Coronaviruses Provides New Insights into the Origin of SARS Coronavirus. PLOS Pathog. 2017, 13, e1006698. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.-C.; Lam, T.-Y.; Shi, Z.-L.; Drummond, A.J.; Yip, C.-W.; Zeng, F.; Lam, P.-Y.; Leung, F.C.-C. Evidence of the Recombinant Origin of a Bat Severe Acute Respiratory Syndrome (SARS)-like Coronavirus and Its Implications on the Direct Ancestor of SARS Coronavirus. J. Virol. 2007, 82, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the Sars-Cov-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Ge, X.-Y.; Li, J.-L.; Yang, X.-L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and Characterization of a Bat SARS-like Coronavirus That Uses the ACE2 Receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Yang, X.-L.; Hu, B.; Wang, B.; Wang, M.-N.; Zhang, Q.; Zhang, W.; Wu, L.-J.; Ge, X.-Y.; Zhang, Y.-Z.; Daszak, P.; et al. Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2015, 90, 3253–3256. [Google Scholar] [CrossRef]
- Starr, T.N.; Zepeda, S.K.; Walls, A.C.; Greaney, A.J.; Alkhovsky, S.; Veesler, D.; Bloom, J.D. ACE2 Binding Is an Ancestral and Evolvable Trait of Sarbecoviruses. Nature 2022, 603, 913–918. [Google Scholar] [CrossRef]
- Makarenkov, V.; Mazoure, B.; Rabusseau, G.; Legendre, P. Horizontal Gene Transfer and Recombination Analysis of SARS-CoV-2 Genes Helps Discover Its Close Relatives and Shed Light on Its Origin. BMC Ecol. Evol. 2021, 21, 5. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The Spike Glycoprotein of the New Coronavirus 2019-NCoV Contains a Furin-like Cleavage Site Absent in CoV of the Same Clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Kadam, S.B.; Sukhramani, G.S.; Bishnoi, P.; Pable, A.A.; Barvkar, V.T. SARS-CoV-2, the Pandemic Coronavirus: Molecular and Structural Insights. J. Basic Microbiol. 2021, 61, 180–202. [Google Scholar] [CrossRef]
- The Sarbecovirus Origin of SARS-CoV-2′s Furin Cleavage Site. Available online: https://virological.org/t/the-sarbecovirus-origin-of-sars-cov-2-s-furin-cleavage-site/536 (accessed on 15 November 2022).
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef]
- Garvin, M.R.; Prates, E.T.; Pavicic, M.; Jones, P.; Amos, B.K.; Geiger, A.; Shah, M.B.; Streich, J.; Felipe Machado Gazolla, J.G.; Kainer, D.; et al. Potentially Adaptive SARS-CoV-2 Mutations Discovered with Novel Spatiotemporal and Explainable AI Models. Genome Biol. 2020, 21, 304. [Google Scholar] [CrossRef]
- MacLean, O.A.; Lytras, S.; Weaver, S.; Singer, J.B.; Boni, M.F.; Lemey, P.; Kosakovsky Pond, S.L.; Robertson, D.L. Natural Selection in the Evolution of SARS-CoV-2 in Bats Created a Generalist Virus and Highly Capable Human Pathogen. PLOS Biol. 2021, 19, e3001115. [Google Scholar] [CrossRef]
- Hill, V.; Du Plessis, L.; Peacock, T.P.; Aggarwal, D.; Colquhoun, R.; Carabelli, A.M.; Ellaby, N.; Gallagher, E.; Groves, N.; Jackson, B.; et al. The Origins and Molecular Evolution of SARS-CoV-2 Lineage B.1.1.7 in the UK. Virus Evol. 2022, 8, e73896. [Google Scholar] [CrossRef]
- WHO. Tracking SARS-CoV-2 variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 20 November 2022).
- Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel set of Spike Mutations. Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 20 November 2022).
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; da, S. Candido, D.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Singh, J.; Rahman, S.A.; Ehtesham, N.Z.; Hira, S.; Hasnain, S.E. SARS-CoV-2 Variants of Concern Are Emerging in India. Nat. Med. 2021, 27, 1131–1133. [Google Scholar] [CrossRef]
- Tian, D.; Sun, Y.; Zhou, J.; Ye, Q. The Global Epidemic of the SARS-CoV-2 Delta Variant, Key Spike Mutations and Immune Escape. Front. Immunol. 2021, 12, 751778. [Google Scholar] [CrossRef]
- Novelli, G.; Colona, V.; Pandolfi, P. A Focus on the Spread of the Delta Variant of SARS-CoV-2 in India. Indian J. Med. Res. 2021, 153, 537–541. [Google Scholar] [CrossRef]
- Cameroni, E.; Bowen, J.E.; Rosen, L.E.; Saliba, C.; Zepeda, S.K.; Culap, K.; Pinto, D.; VanBlargan, L.A.; De Marco, A.; di Iulio, J.; et al. Broadly Neutralizing Antibodies Overcome SARS-CoV-2 Omicron Antigenic Shift. Nature 2021, 602, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Lytras, S.; Lucaci, A.G.; Maier, W.; Grüning, B.; Shank, S.D.; Weaver, S.; MacLean, O.A.; Orton, R.J.; Lemey, P.; et al. Selection Analysis Identifies Clusters of Unusual Mutational Changes in Omicron Lineage BA.1 That Likely Impact Spike Function. Mol. Biol. Evol. 2022, 39, msac061. [Google Scholar]
- Chen, J.; Wei, G.W. Omicron BA.2 (B.1.1.529.2): High potential to becoming the next dominating variant. arXiv 2022, arXiv:2202.05031v1. [Google Scholar]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody Evasion by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef]
- Telenti, A.; Hodcroft, E.B.; Robertson, D.L. The Evolution and Biology of SARS-CoV-2 Variants. Cold Spring Harb. Perspect. Med. 2022, 12, a041390. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-Likelihood Phylodynamic Analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef]
- Dearlove, B.; Lewitus, E.; Bai, H.; Li, Y.; Reeves, D.B.; Joyce, M.G.; Scott, P.T.; Amare, M.F.; Vasan, S.; Michael, N.L.; et al. A SARS-CoV-2 Vaccine Candidate Would Likely Match All Currently Circulating Variants. Proc. Natl. Acad. Sci. USA 2020, 117, 23652–23662. [Google Scholar] [CrossRef]
- Nealon, J.; Cowling, B.J. Omicron Severity: Milder but Not Mild. Lancet 2022, 399, 412–413. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šimičić, P.; Židovec-Lepej, S. A Glimpse on the Evolution of RNA Viruses: Implications and Lessons from SARS-CoV-2. Viruses 2023, 15, 1. https://doi.org/10.3390/v15010001
Šimičić P, Židovec-Lepej S. A Glimpse on the Evolution of RNA Viruses: Implications and Lessons from SARS-CoV-2. Viruses. 2023; 15(1):1. https://doi.org/10.3390/v15010001
Chicago/Turabian StyleŠimičić, Petra, and Snježana Židovec-Lepej. 2023. "A Glimpse on the Evolution of RNA Viruses: Implications and Lessons from SARS-CoV-2" Viruses 15, no. 1: 1. https://doi.org/10.3390/v15010001
APA StyleŠimičić, P., & Židovec-Lepej, S. (2023). A Glimpse on the Evolution of RNA Viruses: Implications and Lessons from SARS-CoV-2. Viruses, 15(1), 1. https://doi.org/10.3390/v15010001




