Pathogenicity of Avian Polyomaviruses and Prospect of Vaccine Development
Abstract
1. Introduction
2. Major Pathogenic Difference between Avian and Mammalian Polyomaviruses
3. Clinical Signs and Disease Progression of Avian Polyomavirus Infection
4. Some Molecular Characteristics Involved in the Pathogenicity of Avian Polyomaviruses
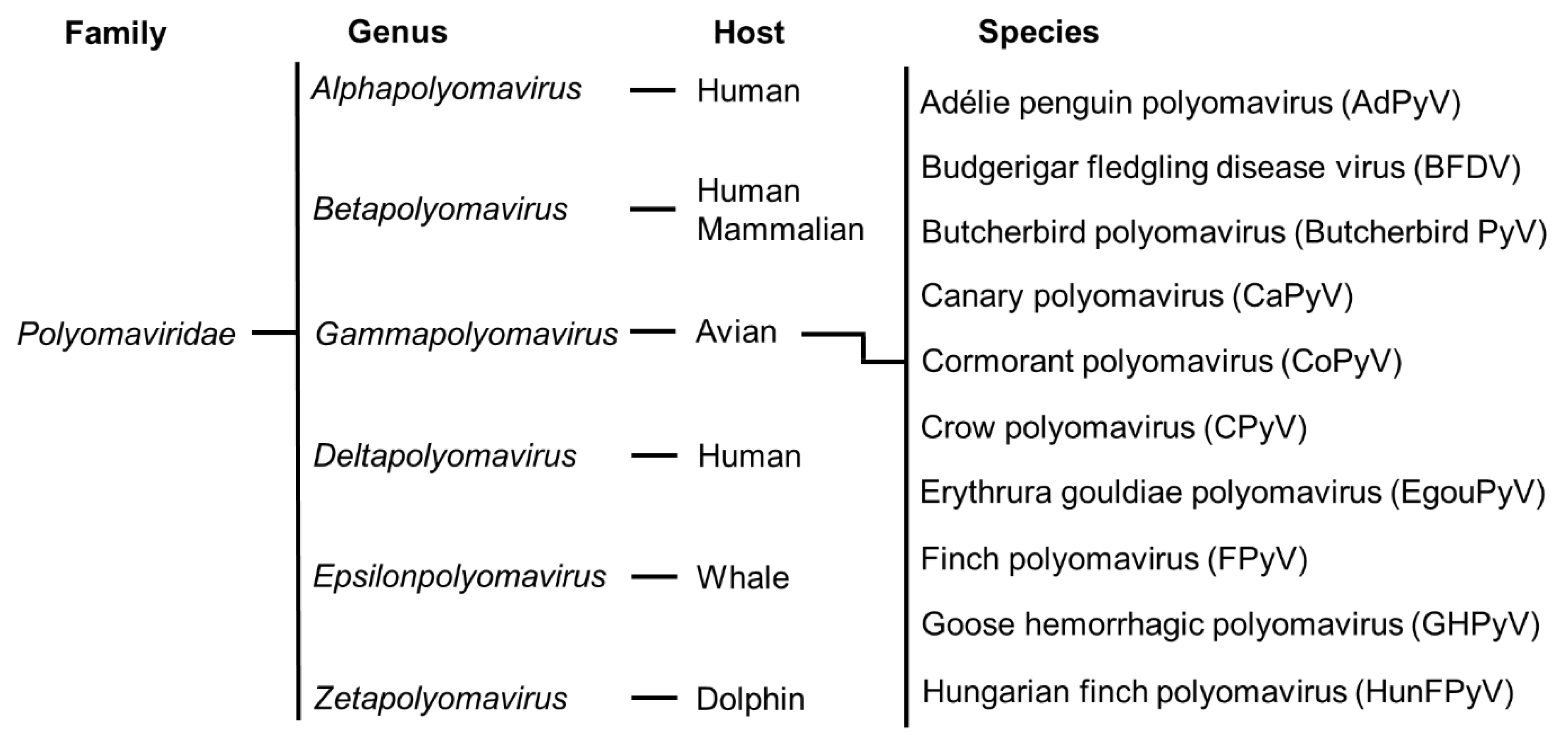
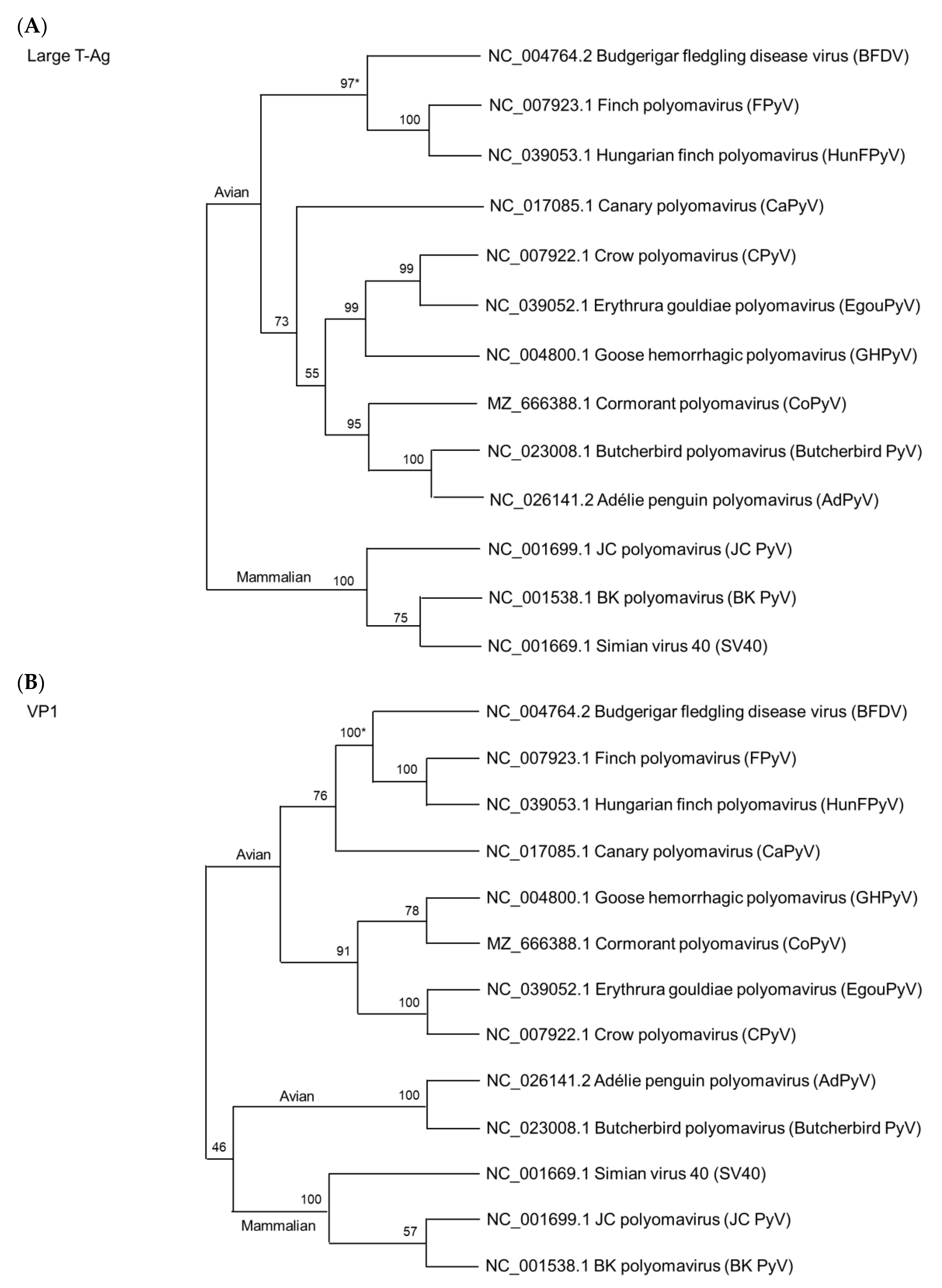
5. Phylogenetic Relationship among Avian Polyomaviruses
6. Historical Development of Vaccines for Avian Polyomaviruses
7. Some Prospects with Respect to the Development of Vaccines Utilizing the Epitope of Receptor-Binding Domain
- One monoclonal antibody can enhance the binding of another monoclonal antibody via cooperative binding, as proposed by Mao et al. (1983) [102,103]. This should not be limited to two antigenic domains because each epitope is relatively small, containing only approximately seven amino acid residues [106].
- The selection of a specific region of a viral protein for vaccine preparation should not only be made against a functional domain, but other structural proteins with “constant” regions or regions with minimal mutations should also be considered as a target to stabilize the antigen–antibody interaction. Other region(s) that will cause a drastic conformational change in the virus structure could also be considered as targets, such as precipitating antibodies found against LDL [104] that could directly immobilize viral activity.
- Finally, because the genomes of the 10 major avian polyomaviruses are relatively small, containing only six genes, it is possible to prepare a “cocktail” multivalent vaccine to produce “APV-capturing antibodies” within a single preparation.
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johne, R.; Paul, G.; Enderlein, D.; Stahl, T.; Grund, C.; Muller, H. Avian polyomavirus mutants with deletions in the VP4-encoding region show deficiencies in capsid assembly and virus release, and have reduced infectivity in chicken. J. Gen. Virol. 2007, 88 Pt 3, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Wittig, W.; Fernandez-de-Luco, D.; Hofle, U.; Muller, H. Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes. J. Virol. 2006, 80, 3523–3531. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Muller, H.; Tang, X.B.; Hobom, G. Expression and DNA binding of budgerigar fledgling disease virus large T antigen. J. Gen. Virol. 1994, 75 Pt 6, 1267–1280. [Google Scholar] [CrossRef] [PubMed]
- Rott, O.; Kroger, M.; Muller, H.; Hobom, G. The genome of budgerigar fledgling disease virus, an avian polyomavirus. Virology 1988, 165, 74–86. Available online: https://www.ncbi.nlm.nih.gov/pubmed/2838972 (accessed on 15 January 2022). [CrossRef]
- Pipas, J.M. Common and unique features of T antigens encoded by the polyomavirus group. J. Virol. 1992, 66, 3979–3985. [Google Scholar] [CrossRef] [PubMed]
- Butel, J.S.; Lednicky, J.A. Cell and molecular biology of simian virus 40: Implications for human infections and disease. J. Natl. Cancer Inst. 1999, 91, 119–134. [Google Scholar] [CrossRef]
- Gordon, J.; Del Valle, L.; Otte, J.; Khalili, K. Pituitary neoplasia induced by expression of human neurotropic polyomavirus, JCV, early genome in transgenic mice. Oncogene 2000, 19, 4840–4846. [Google Scholar] [CrossRef]
- Chen, X.S.; Stehle, T.; Harrison, S.C. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 1998, 17, 3233–3240. [Google Scholar] [CrossRef]
- Li, P.P.; Nakanishi, A.; Tran, M.A.; Salazar, A.M.; Liddington, R.C.; Kasamatsu, H. Role of simian virus 40 Vp1 cysteines in virion infectivity. J. Virol. 2000, 74, 11388–11393. [Google Scholar] [CrossRef]
- Johne, R.; Muller, H. Avian polyomavirus agnoprotein 1a is incorporated into the virus particle as a fourth structural protein, VP4. J. Gen. Virol. 2001, 82 Pt 4, 909–918. [Google Scholar] [CrossRef][Green Version]
- Johne, R.; Jungmann, A.; Muller, H. Agnoprotein 1a and agnoprotein 1b of avian polyomavirus are apoptotic inducers. J. Gen. Virol. 2000, 81 Pt 5, 1183–1190. [Google Scholar] [CrossRef]
- Calvignac-Spencer, S.; Feltkamp, M.C.; Daugherty, M.D.; Moens, U.; Ramqvist, T.; Johne, R.; Ehlers, B. A taxonomy update for the family Polyomaviridae. Arch. Virol. 2016, 161, 1739–1750. [Google Scholar] [CrossRef]
- Alley, M.R.; Rasiah, I.; Lee, E.A.; Howe, L.; Gartrell, B.D. Avian polyomavirus identified in a nestling Gouldian finch (Erythrura gouldiae) in New Zealand. N. Z. Vet. J. 2013, 61, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Ceccherelli, R.; Piersigilli, A.; Tarantino, C. Sertoli cell tumor associated with polyomavirus infection in a Gouldian finch (Erythrura gouldiae). Avian Dis. 2003, 47, 240–243. [Google Scholar] [CrossRef]
- Guerin, J.L.; Gelfi, J.; Dubois, L.; Vuillaume, A.; Boucraut-Baralon, C.; Pingret, J.L. A novel polyomavirus (goose hemorrhagic polyomavirus) is the agent of hemorrhagic nephritis enteritis of geese. J. Virol. 2000, 74, 4523–4529. [Google Scholar] [CrossRef]
- Pingret, J.L.; Boucraut-Baralon, C.; Guerin, J.L. Goose haemorrhagic polyomavirus infection in ducks. Vet. Rec. 2008, 162, 164. [Google Scholar] [CrossRef]
- Moens, U.; Calvignac-Spencer, S.; Lauber, C.; Ramqvist, T.; Feltkamp, M.C.W.; Daugherty, M.D.; Verschoor, E.J.; Ehlers, B.; Ictv Report, C. ICTV Virus Taxonomy Profile: Polyomaviridae. J. Gen. Virol. 2017, 98, 1159–1160. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; et al. Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch. Virol. 2020, 165, 2737–2748. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Van Doorslaer, K.; Peretti, A.; Geoghegan, E.M.; Tisza, M.J.; An, P.; Katz, J.P.; Pipas, J.M.; McBride, A.A.; Camus, A.C.; et al. The Ancient Evolutionary History of Polyomaviruses. PLoS Pathog. 2016, 12, e1005574. [Google Scholar] [CrossRef]
- Heenemann, K.; Sieg, M.; Rueckner, A.; Vahlenkamp, T.W. Complete Genome Sequence of a Novel Avian Polyomavirus Isolated from Gouldian Finch. Genome Announc. 2015, 3, e01001-15. [Google Scholar] [CrossRef]
- Feher, E.; Kaszab, E.; Bali, K.; Hoitsy, M.; Sos, E.; Banyai, K. A novel gammapolyomavirus in a great cormorant (Phalacrocorax carbo). Arch. Virol. 2022, 167, 1721–1724. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Erdelyi, K.; Dan, A.; Banyai, K.; Feher, E. Complete Genome Sequence of a Variant Pyrrhula pyrrhula polyomavirus 1 Strain Isolated from White-Headed Munia (Lonchura maja). Genome Announc. 2016, 4, e01172-16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Abend, J.R.; Johnson, S.F.; Imperiale, M.J. The role of polyomaviruses in human disease. Virology 2009, 384, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Buck, C.B.; Allander, T.; Atwood, W.J.; Garcea, R.L.; Imperiale, M.J.; Major, E.O.; Ramqvist, T.; Norkin, L.C. Taxonomical developments in the family Polyomaviridae. Arch. Virol. 2011, 156, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Assetta, B.; De Cecco, M.; O’Hara, B.; Atwood, W.J. JC Polyomavirus Infection of Primary Human Renal Epithelial Cells Is Controlled by a Type I IFN-Induced Response. mBio 2016, 7, e00903-16. [Google Scholar] [CrossRef] [PubMed]
- Co, J.K.; Verma, S.; Gurjav, U.; Sumibcay, L.; Nerurkar, V.R. Interferon-alpha and-Beta restrict polyomavirus JC replication in primary human fetal glial cells: Implications for progressive multifocal leukoencephalopathy therapy. J. Infect. Dis. 2007, 196, 712–718. [Google Scholar] [CrossRef]
- Ma, J.; Wu, R.; Tian, Y.; Zhang, M.; Wang, W.; Li, Y.; Tian, F.; Cheng, Y.; Yan, Y.; Sun, J. Isolation and characterization of an Aves polyomavirus 1 from diseased budgerigars in China. Vet. Microbiol. 2019, 237, 108397. [Google Scholar] [CrossRef]
- Varsani, A.; Kraberger, S.; Jennings, S.; Porzig, E.L.; Julian, L.; Massaro, M.; Pollard, A.; Ballard, G.; Ainley, D.G. A novel papillomavirus in Adelie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J. Gen. Virol. 2014, 95 Pt 6, 1352–1365. [Google Scholar] [CrossRef]
- Bennett, M.D.; Gillett, A. Butcherbird polyomavirus isolated from a grey butcherbird (Cracticus torquatus) in Queensland, Australia. Vet. Microbiol. 2014, 168, 302–311. [Google Scholar] [CrossRef]
- Johne, R.; Muller, H. Avian polymavirus in wild birds: Genome analysis of isolates from Falconiformes and Psittaciformes. Arch. Virol. 1998, 143, 1501–1512. [Google Scholar] [CrossRef]
- Krautwald, M.E.; Muller, H.; Kaleta, E.F. Polyomavirus infection in budgerigars (Melopsittacus undulatus): Clinical and aetiological studies. J. Vet. Med. Ser. B 1989, 36, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Halami, M.Y.; Dorrestein, G.M.; Couteel, P.; Heckel, G.; Muller, H.; Johne, R. Whole-genome characterization of a novel polyomavirus detected in fatally diseased canary birds. J. Gen. Virol. 2010, 91 Pt 12, 3016–3022. [Google Scholar] [CrossRef] [PubMed]
- Gawel, A.; Wozniakowski, G.; Samorek-Salamonowicz, E.; Kozdrun, W.; Bobrek, K.; Bobusia, K.; Nowak, M. Hemorrhagic nephritis and enteritis in a goose flock in Poland--disease course analysis and characterization of etiologic agent. Avian Dis. 2014, 58, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Garmyn, A.; Verlinden, M.; Bosseler, L.; Adriaensen, C.; Martel, A. Persistent Goose Hemorrhagic Polyomavirus Infection on a Belgian Goose Farm. Avian Dis. 2017, 61, 536–538. [Google Scholar] [CrossRef]
- Lacroux, C.; Andreoletti, O.; Payre, B.; Pingret, J.L.; Dissais, A.; Guerin, J.L. Pathology of spontaneous and experimental infections by Goose haemorrhagic polyomavirus. Avian Pathol. 2004, 33, 351–358. [Google Scholar] [CrossRef]
- Palya, V.; Ivanics, E.; Glavits, R.; Dan, A.; Mato, T.; Zarka, P. Epizootic occurrence of haemorrhagic nephritis enteritis virus infection of geese. Avian Pathol. 2004, 33, 244–250. [Google Scholar] [CrossRef]
- Tu, Y.C.; Li, W.T.; Lee, F.; Huang, C.W.; Chang, J.C.; Hsu, W.C.; Hu, S.C.; Chiou, C.J.; Chen, Y.P. Localization of goose haemorrhagic polyomavirus in naturally infected geese using in situ hybridization. Avian Pathol. 2021, 50, 41–51. [Google Scholar] [CrossRef]
- Corrand, L.; Gelfi, J.; Albaric, O.; Etievant, M.; Pingret, J.L.; Guerin, J.L. Pathological and epidemiological significance of goose haemorrhagic polyomavirus infection in ducks. Avian Pathol. 2011, 40, 355–360. [Google Scholar] [CrossRef]
- Davis, R.B.; Bozeman, L.H.; Gaudry, D.; Fletcher, O.J.; Lukert, P.D.; Dykstra, M.J. A viral disease of fledgling budgerigars. Avian Dis. 1981, 25, 179–183. Available online: https://www.ncbi.nlm.nih.gov/pubmed/7271654 (accessed on 15 January 2022). [CrossRef]
- Phalen, D.N.; Wilson, B.G.; Graham, D.L. Production of avian polyomavirus seronegative budgerigars (Melopsittacus undulatus) from seropositive adults. Avian Dis. 1995, 39, 897–899. Available online: https://www.ncbi.nlm.nih.gov/pubmed/8719226 (accessed on 15 January 2022). [CrossRef]
- Stoll, R.; Luo, D.; Kouwenhoven, B.; Hobom, G.; Muller, H. Molecular and biological characteristics of avian polyomaviruses: Isolates from different species of birds indicate that avian polyomaviruses form a distinct subgenus within the polyomavirus genus. J. Gen. Virol. 1993, 74 Pt 2, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.C.; Niu, K.; Sun, H.J.; Xia, Y.J.; Sun, S.J.; Li, J.; Wang, F.; Feng, Y.; Peng, X.W.; Zhu, L.Q.; et al. Complete Genome Sequence of an Avian Polyomavirus Strain First Isolated from a Pigeon in China. Microbiol. Resour. Ann. 2019, 8, e01490-18. [Google Scholar] [CrossRef] [PubMed]
- Bert, E.; Tomassone, L.; Peccati, C.; Navarrete, M.G.; Sola, S.C. Detection of beak and feather disease virus (BFDV) and avian polyomavirus (APV) DNA in psittacine birds in Italy. J. Vet. Med. Ser. B 2005, 52, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.M.; Ko, C.Y.; Tsaia, H.J. Detection and sequence analysis of avian polyomavirus and psittacine beak and feather disease virus from psittacine birds in Taiwan. Avian Dis. 2006, 50, 348–353. [Google Scholar] [CrossRef]
- Pass, D.A. A papova-like virus infection of lovebirds (Agapornis sp.). Aust. Vet. J. 1985, 62, 318–319. [Google Scholar] [CrossRef]
- Latimer, K.S.; Niagro, F.D.; Steffens, W.L., 3rd; Ritchie, B.W.; Campagnoli, R.P. Polyomavirus encephalopathy in a Ducorps’ cockatoo (Cacatua ducorpsii) with psittacine beak and feather disease. J. Vet. Diagn. Investig. 1996, 8, 291–295. [Google Scholar] [CrossRef]
- Sandmeier, P.; Gerlach, H.; Johne, R.; Muller, H. Polyomavirus infections in exotic birds in Switzerland. Schweiz. Arch. Tierheilkd. 1999, 141, 223–229. Available online: https://www.ncbi.nlm.nih.gov/pubmed/10354740 (accessed on 15 January 2022).
- Literak, I.; Smid, B.; Dubska, L.; Bryndza, L.; Valicek, L. An outbreak of the polyomavirus infection in budgerigars and cockatiels in Slovakia, including a genome analysis of an avian polyomavirus isolate. Avian Dis. 2006, 50, 120–123. [Google Scholar] [CrossRef]
- Kou, Z.; Zhang, Z.; Chen, S.; Fan, Z.; Tang, S.; Zhao, L.; Li, T. Molecular characterizations of avian polyomavirus isolated from budgerigar in China. Avian Dis. 2008, 52, 451–454. [Google Scholar] [CrossRef]
- Gilardi, K.V.; Lowenstine, L.J.; Gilardi, J.D.; Munn, C.A. A survey for selected viral, chlamydial, and parasitic diseases in wild dusky-headed parakeets (Aratinga weddellii) and tui parakeets (Brotogeris sanctithomae) in Peru. J. Wildl. Dis. 1995, 31, 523–528. [Google Scholar] [CrossRef]
- Riaz, A.; Yousaf, A.; Moaeen-Ud-Din, M.; Shah, M.A.A.; Zainab, T.; Masood, S.; Akhter, N.; Ali, A. First detection and molecular characterization of avian polyomavirus in young parrots in Pakistan. Vet. Res. Commun. 2019, 43, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Chahota, R.; Hagino, T.; Ohya, K.; Yamaguchi, T.; Fukushi, H. A survey of avian polyomavirus (APV) infection in imported and domestic bred psittacine birds in Japan. J. Vet. Med. Sci. 2006, 68, 743–745. [Google Scholar] [CrossRef] [PubMed]
- Feher, E.; Lengyel, G.; Dan, A.; Farkas, S.L.; Banyai, K. Whole genome sequence of a goose haemorrhagic polyomavirus detected in Hungary. Acta Microbiol. Immunol. Hung. 2014, 61, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Dobos-Kovacs, M.; Horvath, E.; Farsang, A.; Nagy, E.; Kovacs, A.; Szalai, F.; Bernath, S. Haemorrhagic nephritis and enteritis of geese: Pathomorphological investigations and proposed pathogenesis. Acta Vet. Hung. 2005, 53, 213–223. [Google Scholar] [CrossRef]
- Kaszab, E.; Marton, S.; Dan, A.; Farsang, A.; Balint, A.; Banyai, K.; Feher, E. Molecular epidemiology and phylodynamics of goose haemorrhagic polyomavirus. Transbound. Emerg. Dis. 2020, 67, 2602–2608. [Google Scholar] [CrossRef]
- Wan, C.; Chen, C.; Cheng, L.; Liu, R.; Fu, G.; Shi, S.; Chen, H.; Fu, Q.; Huang, Y. Genomic analysis of Sheldrake origin goose hemorrhagic polyomavirus, China. J. Vet. Sci. 2018, 19, 782–787. [Google Scholar] [CrossRef]
- Manarolla, G.; Liandris, E.; Pisoni, G.; Moroni, P.; Piccinini, R.; Rampin, T. Mycobacterium genavense and avian polyomavirus co-infection in a European goldfinch (Carduelis carduelis). Avian Pathol. 2007, 36, 423–426. [Google Scholar] [CrossRef][Green Version]
- Wittig, W.; Hoffmann, K.; Muller, H.; Johne, R. Detection of DNA of the finch polyomavirus in diseases of various types of birds in the order Passeriformes. Berl. Münch. Tierärztl. Wochenschr. 2007, 120, 113–119. Available online: https://www.ncbi.nlm.nih.gov/pubmed/17416133 (accessed on 15 January 2022).
- Sironi, G. Concurrent papovavirus-like and atoxoplasma infections in a goldfinch (Carduelis carduelis). Avian Pathol. 1991, 20, 725–729. [Google Scholar] [CrossRef]
- Fitzgerald, S.D.; Williams, S.M.; Reed, W.M. Development of a chicken model for studying avian polyomavirus infection. Avian Dis. 1996, 40, 377–381. Available online: https://www.ncbi.nlm.nih.gov/pubmed/8790889 (accessed on 15 January 2022). [CrossRef]
- Shivaprasad, H.L.; Kim, T.; Tripathy, D.; Woolcock, P.R.; Uzal, F. Unusual pathology of canary poxvirus infection associated with high mortality in young and adult breeder canaries (Serinus canaria). Avian Pathol. 2009, 38, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.; Latimer, K.S.; Niagro, F.D.; Norton, T.M.; Campagnoli, R.P.; Harmon, B.G.; Howerth, E.W.; Ritchie, B.W. Diagnosis of polyomavirus infection in seedcrackers (Pyrenestes sp.) and blue bills (Spermophaga haematina) using DNA in situ hybridization. Avian Pathol. 1994, 23, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Rinder, M.; Schmitz, A.; Peschel, A.; Moser, K.; Korbel, R. Identification and genetic characterization of polyomaviruses in estrildid and fringillid finches. Arch. Virol. 2018, 163, 895–909. [Google Scholar] [CrossRef]
- Varsani, A.; Porzig, E.L.; Jennings, S.; Kraberger, S.; Farkas, K.; Julian, L.; Massaro, M.; Ballard, G.; Ainley, D.G. Identification of an avian polyomavirus associated with Adelie penguins (Pygoscelis adeliae). J. Gen. Virol. 2015, 96 Pt 4, 851–857. [Google Scholar] [CrossRef]
- DeCaprio, J.A.; Garcea, R.L. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013, 11, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.L.; Chang, S.P.; Liu, H.J.; Liu, P.C.; Wang, C.Y. Genomic and phylogenetic analysis of avian polyomaviruses isolated from parrots in Taiwan. Virus Res. 2022, 308, 198634. [Google Scholar] [CrossRef]
- Carr, M.; Gonzalez, G.; Sasaki, M.; Ito, K.; Ishii, A.; Hang’ombe, B.M.; Mweene, A.S.; Orba, Y.; Sawa, H. Discovery of African bat polyomaviruses and infrequent recombination in the large T antigen in the Polyomaviridae. J. Gen. Virol. 2017, 98, 726–738. [Google Scholar] [CrossRef]
- Hu, X.; Cai, D.; Liu, S.; Li, Y.; Chen, L.; Luo, G.; Pu, H.; He, Y.; Liu, X.; Zhao, L.; et al. Molecular Characterization of a Novel Budgerigar Fledgling Disease Virus Strain From Budgerigars in China. Front. Vet. Sci. 2021, 8, 813397. [Google Scholar] [CrossRef]
- Freund, R.; Garcea, R.L.; Sahli, R.; Benjamin, T.L. A single-amino-acid substitution in polyomavirus VP1 correlates with plaque size and hemagglutination behavior. J. Virol. 1991, 65, 350–355. [Google Scholar] [CrossRef]
- Ahuja, D.; Saenz-Robles, M.T.; Pipas, J.M. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005, 24, 7729–7745. [Google Scholar] [CrossRef]
- Borchert, S.; Czech-Sioli, M.; Neumann, F.; Schmidt, C.; Wimmer, P.; Dobner, T.; Grundhoff, A.; Fischer, N. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J. Virol. 2014, 88, 3144–3160. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Kim, H.; Son, H.S. Codon usage patterns of LT-Ag genes in polyomaviruses from different host species. Virol. J. 2019, 16, 137. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Ohya, K.; Une, Y.; Yamaguchi, T.; Fukushi, H. Molecular characterization of avian polyomavirus isolated from psittacine birds based on the whole genome sequence analysis. Vet. Microbiol. 2009, 138, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Ben-nun-Shaul, O.; Bronfeld, H.; Reshef, D.; Schueler-Furman, O.; Oppenheim, A. The SV40 capsid is stabilized by a conserved pentapeptide hinge of the major capsid protein VP1. J. Mol. Biol. 2009, 386, 1382–1391. [Google Scholar] [CrossRef]
- Liddington, R.C.; Yan, Y.; Moulai, J.; Sahli, R.; Benjamin, T.L.; Harrison, S.C. Structure of simian virus 40 at 3.8-A resolution. Nature 1991, 354, 278–284. [Google Scholar] [CrossRef]
- Barouch, D.H.; Harrison, S.C. Interactions among the major and minor coat proteins of polyomavirus. J. Virol. 1994, 68, 3982–3989. [Google Scholar] [CrossRef]
- Griffith, J.P.; Griffith, D.L.; Rayment, I.; Murakami, W.T.; Caspar, D.L. Inside polyomavirus at 25-A resolution. Nature 1992, 355, 652–654. [Google Scholar] [CrossRef]
- Hurdiss, D.L.; Morgan, E.L.; Thompson, R.F.; Prescott, E.L.; Panou, M.M.; Macdonald, A.; Ranson, N.A. New Structural Insights into the Genome and Minor Capsid Proteins of BK Polyomavirus using Cryo-Electron Microscopy. Structure 2016, 24, 528–536. [Google Scholar] [CrossRef]
- Sapp, M.; Day, P.M. Structure, attachment and entry of polyoma- and papillomaviruses. Virology 2009, 384, 400–409. [Google Scholar] [CrossRef]
- Rodgers, R.E.; Consigli, R.A. Characterization of a calcium binding domain in the VP1 protein of the avian polyomavirus, budgerigar fledgling disease virus. Virus Res. 1996, 44, 123–135. [Google Scholar] [CrossRef]
- Haynes, J.I., 2nd; Chang, D.; Consigli, R.A. Mutations in the putative calcium-binding domain of polyomavirus VP1 affect capsid assembly. J. Virol. 1993, 67, 2486–2495. [Google Scholar] [CrossRef] [PubMed]
- Kaszab, E.; Marton, S.; Erdelyi, K.; Banyai, K.; Feher, E. Genomic evolution of avian polyomaviruses with a focus on budgerigar fledgling disease virus. Infect. Genet. Evol. 2021, 90, 104762. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ritchie, B.W.; Niagro, F.D.; Latimer, K.S.; Pritchard, N.; Campagnoli, R.P.; Lukert, P.D. An inactivated avian polyomavirus vaccine is safe and immunogenic in various Psittaciformes. Vaccine 1996, 14, 1103–1107. [Google Scholar] [CrossRef]
- Mato, T.; Penzes, Z.; Rueda, P.; Vela, C.; Kardi, V.; Zolnai, A.; Misak, F.; Palya, V. Recombinant subunit vaccine elicits protection against goose haemorrhagic nephritis and enteritis. Avian Pathol. 2009, 38, 233–237. [Google Scholar] [CrossRef]
- Gelfi, J.; Pappalardo, M.; Claverys, C.; Peralta, B.; Guerin, J.L. Safety and efficacy of an inactivated Carbopol-adjuvanted goose haemorrhagic polyomavirus vaccine for domestic geese. Avian Pathol. 2010, 39, 111–116. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaszab, E.; Szabadi, L.; Kepner, A.; Bajnoczi, P.; Lengyel, G.; Banyai, K.; Feher, E. Viral gene expression profile of goose haemorrhagic polyomavirus in susceptible primary cells. Avian Pathol. 2021, 50, 447–452. [Google Scholar] [CrossRef]
- Ritchie, B.W.; Vaughn, S.B.; Leger, J.S.; Rich, G.A.; Rupiper, D.J.; Forgey, G.; Greenacre, C.B.; Latimer, K.S.; Pesti, D.; Campagnoli, R.; et al. Use of an inactivated virus vaccine to control polyomavirus outbreaks in nine flocks of psittacine birds. J. Am. Vet. Med. Assoc. 1998, 212, 685–690. Available online: https://www.ncbi.nlm.nih.gov/pubmed/9524641 (accessed on 15 January 2022).
- Xie, Q.; Wang, W.; Kan, Q.; Mu, Y.; Zhang, W.; Chen, J.; Li, L.; Fu, H.; Li, T.; Wan, Z.; et al. FAdV-4 without Fiber-2 Is a Highly Attenuated and Protective Vaccine Candidate. Microbiol. Spectr. 2022, 10, e0143621. [Google Scholar] [CrossRef]
- Olbert, M.; Romer-Oberdorfer, A.; Herden, C.; Malberg, S.; Runge, S.; Staeheli, P.; Rubbenstroth, D. Viral vector vaccines expressing nucleoprotein and phosphoprotein genes of avian bornaviruses ameliorate homologous challenge infections in cockatiels and common canaries. Sci. Rep. 2016, 6, 36840. [Google Scholar] [CrossRef]
- Runge, S.; Olbert, M.; Herden, C.; Malberg, S.; Romer-Oberdorfer, A.; Staeheli, P.; Rubbenstroth, D. Viral vector vaccines protect cockatiels from inflammatory lesions after heterologous parrot bornavirus 2 challenge infection. Vaccine 2017, 35, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Rall, I.; Amann, R.; Malberg, S.; Herden, C.; Rubbenstroth, D. Recombinant Modified Vaccinia Virus Ankara (MVA) Vaccines Efficiently Protect Cockatiels Against Parrot Bornavirus Infection and Proventricular Dilatation Disease. Viruses 2019, 11, 1130. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, A.; Gedvilaite, A.; Ulrich, R.; Luschow, D.; Sasnauskas, K.; Muller, H.; Johne, R. Generation of virus-like particles consisting of the major capsid protein VP1 of goose hemorrhagic polyomavirus and their application in serological tests. Virus Res. 2006, 120, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, A.; Gedvilaite, A.; Reetz, J.; Rosler, U.; Muller, H.; Johne, R. Serological cross-reactions between four polyomaviruses of birds using virus-like particles expressed in yeast. J. Gen. Virol. 2012, 93 Pt 12, 2658–2667. [Google Scholar] [CrossRef] [PubMed]
- Grgacic, E.V.; Anderson, D.A. Virus-like particles: Passport to immune recognition. Methods 2006, 40, 60–65. [Google Scholar] [CrossRef]
- Ludwig, C.; Wagner, R. Virus-like particles-universal molecular toolboxes. Curr. Opin. Biotechnol. 2007, 18, 537–545. [Google Scholar] [CrossRef]
- Roldao, A.; Mellado, M.C.; Castilho, L.R.; Carrondo, M.J.; Alves, P.M. Virus-like particles in vaccine development. Expert Rev. Vaccines 2010, 9, 1149–1176. [Google Scholar] [CrossRef]
- Sasnauskas, K.; Bulavaite, A.; Hale, A.; Jin, L.; Knowles, W.A.; Gedvilaite, A.; Dargeviciute, A.; Bartkeviciute, D.; Zvirbliene, A.; Staniulis, J.; et al. Generation of recombinant virus-like particles of human and non-human polyomaviruses in yeast Saccharomyces cerevisiae. Intervirology 2002, 45, 308–317. [Google Scholar] [CrossRef]
- Muller, H.; Nitschke, R. A polyoma-like virus associated with an acute disease of fledgling budgerigars (Melopsittacus undulatus). Med. Microbiol. Immunol. 1986, 175, 1–13. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, D.; Wang, G.; Huang, L.; Zheng, Q.; Li, C.; Cheng, Z. A novel multi-variant epitope ensemble vaccine against avian leukosis virus subgroup J. Vaccine 2017, 35 Pt B, 6685–6690. [Google Scholar] [CrossRef]
- Qin, Y.; Tu, K.; Teng, Q.; Feng, D.; Zhao, Y.; Zhang, G. Identification of Novel T-Cell Epitopes on Infectious Bronchitis Virus N Protein and Development of a Multi-epitope Vaccine. J. Virol. 2021, 95, e0066721. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.J.; Patton, J.G.; Badimon, J.J.; Kottke, B.A.; Alley, M.C.; Cardin, A.D. Monoclonal antibodies to human plasma low-density lipoproteins. I. Enhanced binding of 125I-labeled low-density lipoproteins by combined use of two monoclonal antibodies. Clin. Chem. 1983, 29, 1890–1897. Available online: https://www.ncbi.nlm.nih.gov/pubmed/6627627 (accessed on 15 January 2022). [CrossRef]
- Patton, J.G.; Badimon, J.J.; Mao, S.J. Monoclonal antibodies to human plasma low-density lipoproteins. II. Evaluation for use in radioimmunoassay for apolipoprotein B in patients with coronary artery disease. Clin. Chem. 1983, 29, 1898–1903. Available online: https://www.ncbi.nlm.nih.gov/pubmed/6627628 (accessed on 15 January 2022). [CrossRef] [PubMed]
- Marcovina, S.; Kottke, B.A.; Mao, S.J. Monoclonal antibodies can precipitate low-density lipoprotein. II. Radioimmunoassays with single and combined monoclonal antibodies for determining apolipoprotein B in serum of patients with coronary artery disease. Clin. Chem. 1985, 31, 1659–1663. Available online: https://www.ncbi.nlm.nih.gov/pubmed/3930092 (accessed on 25 April 2022). [CrossRef] [PubMed]
- Forchette, L.; Sebastian, W.; Liu, T. A Comprehensive Review of COVID-19 Virology, Vaccines, Variants, and Therapeutics. Curr. Med. Sci. 2021, 41, 1037–1051. [Google Scholar] [CrossRef]
- Song, C.Y.; Chen, W.L.; Yang, M.C.; Huang, J.P.; Mao, S.J. Epitope mapping of a monoclonal antibody specific to bovine dry milk: Involvement of residues 66-76 of strand D in thermal denatured beta-lactoglobulin. J. Biol. Chem. 2005, 280, 3574–3582. [Google Scholar] [CrossRef]
- Winter, S.; Lechapt, E.; Gricourt, G.; N’Debi, M.; Boddaert, N.; Moshous, D.; Blauwblomme, T.; Kossorotoff, M.; Fouyssac, F.; Chareyre, J.; et al. Fatal encephalitis caused by Newcastle disease virus in a child. Acta Neuropathol. 2021, 142, 605–608. [Google Scholar] [CrossRef]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Adiguzel, M.C.; Timurkan, M.O.; Cengiz, S. Investigation and Sequence Analysis of Avian Polyomavirus and Psittacine Beak and Feather Disease Virus from Companion Birds in Eastern Turkey. J. Vet. Res. 2020, 64, 495–501. [Google Scholar] [CrossRef]
- Vitiello, A.; Ferrara, F. Brief review of the mRNA vaccines COVID-19. Inflammopharmacology 2021, 29, 645–649. [Google Scholar] [CrossRef]
- Szabo, G.T.; Mahiny, A.J.; Vlatkovic, I. COVID-19 mRNA vaccines: Platforms and current developments. Mol. Ther. 2022, 30, 1850–1868. [Google Scholar] [CrossRef] [PubMed]

| Nucleotide Size (bp) | Amino Acid Residues | Functions [Reference] | |
|---|---|---|---|
| Gene: gp5 Protein: Large T-Ag | 1995 (1995–2166) * | 599 (599–660) * | Interacts with the tandem repeat sequences in the noncoding control region to regulate DNA replication and RNA transcription [3,4] |
| Gene: gp6 Protein: Small T-Ag | 483 (483–537) | 145 (145–178) | The large T-Ag from the same ATG codon shares an N-terminal domain [5] and is involved in cell transformation and tumorigenicity [6,7] |
| Gene: gp4 Protein: VP1 | 1032 (1032–1083) | 343 (343–360) | VP1, a capsid protein, binds to the host cell receptor for infection and forms a pentamer for its stability [8,9], which is further reinforced with a single copy of VP2 and VP3. |
| Gene: gp2 Protein: VP2 | 1026 (981–1110) | 341 (331–369) | |
| Gene: gp3 Protein: VP3 | 708 (654–738) | 235 (217–245) | |
| Gene: gp1 Protein: VP4 | 675 (485–755) except canary polyomavirus and Adélie penguin polyomavirus | 176 (112–205) except canary polyomavirus and Adélie penguin polyomavirus | Suppresses immune responses, induces apoptosis, and increases in pathogenicity [1,10,11] and scaffolding function during virion assembly [1]. |
| Gene: gp1 Protein: VP4d | 675 | 112 | VP4 delta (deleted a.a. 69–132 in VP4) contains a leucine zipper-like motif [10]. Induces cell apoptosis and affects releases of viral progeny [1]. |
| Virus | Host | Clinical Symptoms [Reference] |
|---|---|---|
| Adélie penguin polyomavirus (AdPyV) | Adélie penguins (Pygoscelis adeliae) | Feather loss [28] |
| Butcherbird polyomavirus (Butcherbird PyV) | Grey butcherbird (Cracticus torquatus) | No apparent signs were observed except for periocular nodule growth. It is unclear whether these symptoms are directly associated with Butcherbird PyV [29] |
| Budgerigar fledgling disease virus (BFDV) | Parrots, chickens, vultures, falcons, canaries, ostriches, pigeons, ducks, geese, finches, gulls, common ravens, pheasants, Eurasian jays, and starlings | Development of hepatitis, ascites, pericardial effusion, and abdominal distension [30,31]; abnormal feather growth often occurs in adult and chronically infected parrots |
| Canary polyomavirus (CaPyV) | Canaries (Serinus canaria) | Subcutaneous bleeding; patosplenomegaly; extensive centrilobular degeneration with hemorrhage in the liver; splenic depletion; or polyomavirus-like intranuclear inclusion bodies in the liver and spleen, occasionally found in the epithelium of some renal glomeruli [32] |
| Cormorant polyomavirus (CoPyV) | Great cormorant (Phalacrocorax carbo) | Detection in the liver, but detailed pathological findings are not available [21] |
| Crow polyomavirus (CPyV) | Western jackdaws (Corvus monedula) | Enteritis and death with Salmonella co-infection [2] |
| Erythrura gouldiae polyomavirus (EgouPyV) | Gouldian finch (Erythrura gouldiae) | Detection in the liver, but detailed pathological findings are not available [20] |
| Finch polyomavirus (FPyV) | European goldfinches, grey-headed bullfinches, and Gouldian finches | Liver necrosis, membranous nephropathy, liver and renal cell nuclei hypertrophy, large amounts of transparent-to-basophilic intranuclear inclusion bodies, with no inclusion bodies in periarteriolar lymphoid sheaths [13,14] |
| Goose hemorrhagic polyomavirus (GHPyV) | Geese and ducks | Subcutaneous edema, ascites, renal pallor, and swelling; gastrointestinal hemorrhage in some cases [15,33,34,35]; in acute cases, hemorrhagic necrosis in the small intestine, with endothelial and vascular wall necrosis in the proventriculus, gizzard, and intestines; inflammatory cell infiltration, with mucosal bleeding in gizzard; in subacute and chronic disease cases, gout [35,36], renal and intestinal vascular necrosis, bleeding, and edema [15,33,35] are seen; distal renal tubule necrosis is more severe than proximal tubule necrosis, resulting in urate deposition, calcification, and fibrosis [36,37]; ducks are considered asymptomatic carriers [16,38] |
| Hungarian finch polyomavirus (HunFPyV) | White-headed munia (Lonchura maja) | Liver failure, nephritis, and myocarditis [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.-W.; Chen, Y.-L.; Mao, S.J.T.; Lin, T.-C.; Wu, C.-W.; Thongchan, D.; Wang, C.-Y.; Wu, H.-Y. Pathogenicity of Avian Polyomaviruses and Prospect of Vaccine Development. Viruses 2022, 14, 2079. https://doi.org/10.3390/v14092079
Wang C-W, Chen Y-L, Mao SJT, Lin T-C, Wu C-W, Thongchan D, Wang C-Y, Wu H-Y. Pathogenicity of Avian Polyomaviruses and Prospect of Vaccine Development. Viruses. 2022; 14(9):2079. https://doi.org/10.3390/v14092079
Chicago/Turabian StyleWang, Chen-Wei, Yung-Liang Chen, Simon J. T. Mao, Tzu-Chieh Lin, Ching-Wen Wu, Duangsuda Thongchan, Chi-Young Wang, and Hung-Yi Wu. 2022. "Pathogenicity of Avian Polyomaviruses and Prospect of Vaccine Development" Viruses 14, no. 9: 2079. https://doi.org/10.3390/v14092079
APA StyleWang, C.-W., Chen, Y.-L., Mao, S. J. T., Lin, T.-C., Wu, C.-W., Thongchan, D., Wang, C.-Y., & Wu, H.-Y. (2022). Pathogenicity of Avian Polyomaviruses and Prospect of Vaccine Development. Viruses, 14(9), 2079. https://doi.org/10.3390/v14092079







