Characterization and Immunogenicity of Influenza H7N9 Vaccine Antigens Produced Using a Serum-Free Suspension MDCK Cell-Based Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Preparation of Vaccine Antigens
2.2. Freeze-Drying
2.3. Electron Microscopy and Nanoparticle Size Assays
2.4. Single Radial Immunodiffusion Assay
2.5. Hemagglutination (HA) Assay
2.6. SDS-Page and Western Blot Assays of Deglycosylated Vaccine Antigens
2.7. Mouse Immunization Studies
2.8. Hemagglutinin Inhibition (HI) Assay
2.9. Neutralizing Antibody (NT) Assay
2.10. Statistical Analysis
2.11. Ethics Statement
3. Results
3.1. Biochemical Characteristics of H7N9 Vaccine Antigens
3.2. Quality Control of Vaccine Antigens
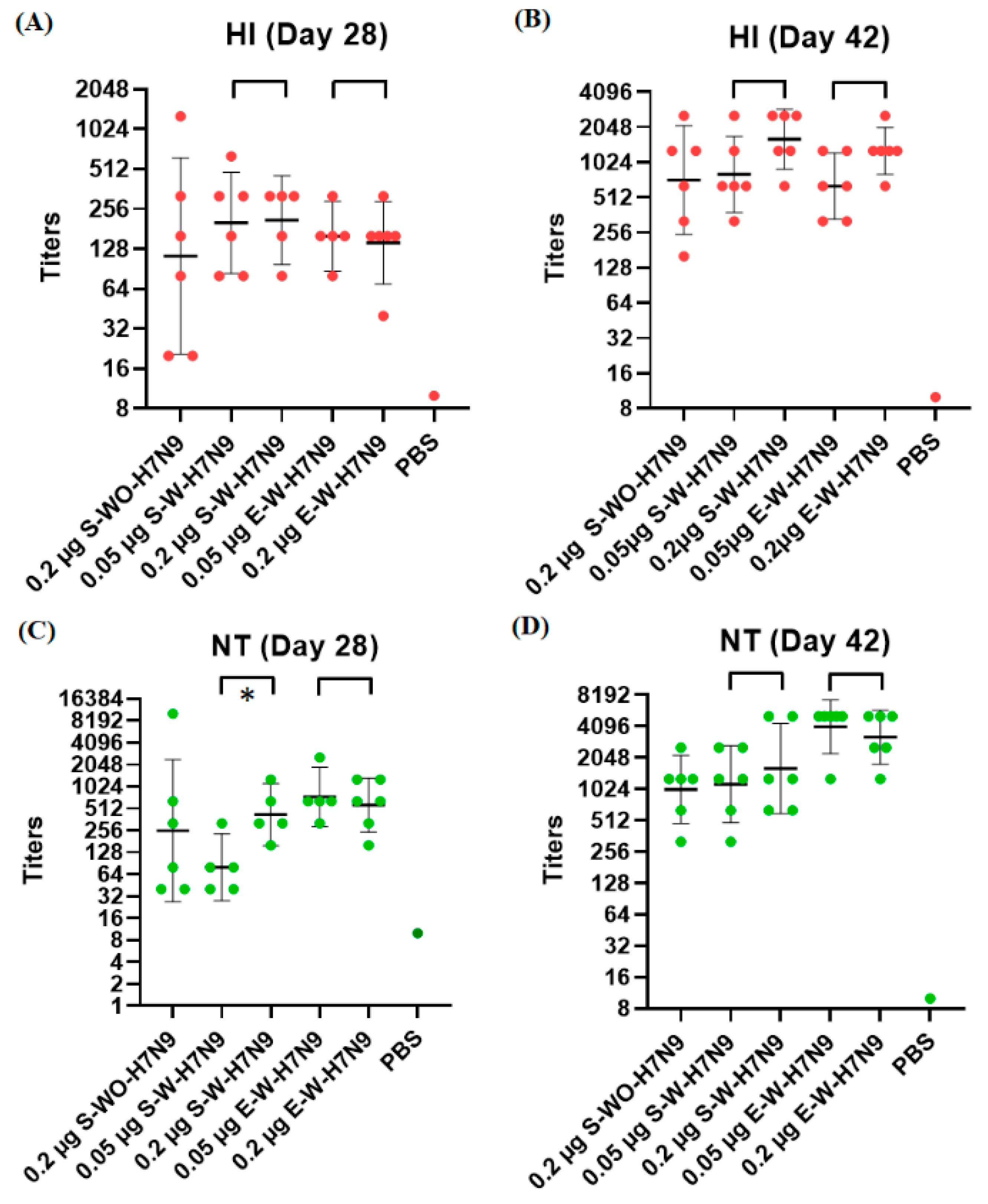
3.3. Immunogenicity in Mice
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Lam, T.T.; Wang, J.; Shen, Y.; Zhou, B.; Duan, L.; Cheung, C.L.; Ma, C.; Lycett, S.J.; Leung, C.Y.; Chen, X.; et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 2013, 502, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lam, T.T.; Smith, D.K.; Guan, Y. Emergence and development of H7N9 influenza viruses in China. Curr. Opin. Virol. 2016, 16, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Belser, J.A.; Pappas, C.; Pulit-Penaloza, J.A.; Brock, N.; Zeng, H.; Creager, H.M.; Le, S.; Wilson, M.; Lewis, A.; et al. Risk assessment of fifth-wave H7N9 influenza A viruses in mammalian models. J. Virol. 2018, 93, e01740-18. [Google Scholar] [CrossRef]
- Liu, W.J.; Xiao, H.; Dai, L.; Liu, D.; Chen, J.; Qi, X.; Bi, Y.; Shi, Y.; Gao, G.F.; Liu, Y. Avian influenza A (H7N9) virus: From low pathogenic to highly pathogenic. Front. Med. 2021, 15, 507–527. [Google Scholar] [CrossRef] [PubMed]
- Quan, C.; Shi, W.; Yang, Y.; Yang, Y.; Liu, X.; Xu, W.; Li, H.; Li, J.; Wang, Q.; Tong, Z.; et al. New Threats from H7N9 Influenza Virus: Spread and Evolution of High- and Low-Pathogenicity Variants with High Genomic Diversity in Wave Five. J. Virol. 2018, 92, e00301-18. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Hu, A.Y. A cell-based backup to speed up pandemic influenza vaccine production. Trends Microbiol. 2012, 20, 103–105. [Google Scholar] [CrossRef]
- Hegde, N.R. Cell culture-based influenza vaccines: A necessary and indispensable investment for the future. Hum. Vaccin. Immunother. 2015, 11, 1223–1234. [Google Scholar] [CrossRef]
- van Wielink, R.; Kant-Eenbergen, H.C.; Harmsen, M.M.; Martens, D.E.; Wijffels, R.H.; Coco-Martin, J.M. Adaptation of a Madin-Darby canine kidney cell line to suspension growth in serum-free media and comparison of its ability to produce avian influenza virus to Vero and BHK21 cell lines. J. Virol. Methods 2011, 171, 53–60. [Google Scholar] [CrossRef]
- Hu, A.Y.; Weng, T.C.; Tseng, Y.F.; Chen, Y.S.; Wu, C.H.; Hsiao, S.; Chou, A.H.; Chao, H.J.; Gu, A.; Wu, S.C.; et al. Microcarrier-based MDCK cell culture system for the production of influenza H5N1 vaccines. Vaccine 2008, 26, 5736–5740. [Google Scholar] [CrossRef]
- Peschel, B.; Frentzel, S.; Laske, T.; Genzel, Y.; Reichl, U. Comparison of influenza virus yields and apoptosis-induction in an adherent and a suspension MDCK cell line. Vaccine 2013, 31, 5693–5699. [Google Scholar] [CrossRef]
- Huang, D.; Peng, W.J.; Ye, Q.; Liu, X.P.; Zhao, L.; Fan, L.; Xia-Hou, K.; Jia, H.J.; Luo, J.; Zhou, L.T.; et al. Serum-Free Suspension Culture of MDCK Cells for Production of Influenza H1N1 Vaccines. PLoS ONE 2015, 10, e0141686. [Google Scholar] [CrossRef] [PubMed]
- Onions, D.; Egan, W.; Jarrett, R.; Novicki, D.; Gregersen, J.P. Validation of the safety of MDCK cells as a substrate for the production of a cell-derived influenza vaccine. Biologicals 2010, 38, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.S.; Noh, J.Y.; Song, J.Y.; Cheong, H.J.; Wie, S.H.; Lee, J.S.; Lee, J.; Kim, S.W.; Jeong, H.W.; Jung, S.I.; et al. Immunogenicity and safety of a cell culture-derived inactivated quadrivalent influenza vaccine (NBP607-QIV): A randomized, double-blind, multi-center, phase III clinical trial in adults and elderly subjects. Hum. Vaccin. Immunother. 2017, 13, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.E.; Choi, U.Y.; Eun, B.W.; Lee, T.J.; Kim, K.H.; Kim, D.H.; Kim, N.H.; Jo, D.S.; Shin, S.H.; Kim, K.H.; et al. A Randomized, Double-blind, Active-controlled Clinical Trial of a Cell Culture-derived Inactivated Trivalent Influenza Vaccine (NBP607) in Healthy Children 6 Months Through 18 Years of Age. Pediatr. Infect. Dis. J. 2018, 37, 605–611. [Google Scholar] [CrossRef]
- Barr, I.G.; Donis, R.O.; Katz, J.M.; McCauley, J.W.; Odagiri, T.; Trusheim, H.; Tsai, T.F.; Wentworth, D.E. Cell culture-derived influenza vaccines in the severe 2017-2018 epidemic season: A step towards improved influenza vaccine effectiveness. NPJ Vaccines 2018, 3, 44. [Google Scholar] [CrossRef]
- Tomar, J.; Born, P.A.; Frijlink, H.W.; Hinrichs, W.L. Dry influenza vaccines: Towards a stable, effective and convenient alternative to conventional parenteral influenza vaccination. Expert Rev. Vaccines 2016, 15, 1431–1447. [Google Scholar] [CrossRef]
- Flood, A.; Estrada, M.; McAdams, D.; Ji, Y.; Chen, D. Development of a Freeze-Dried, Heat-Stable Influenza Subunit Vaccine Formulation. PLoS ONE 2016, 11, e0164692. [Google Scholar] [CrossRef]
- Amorij, J.P.; Meulenaar, J.; Hinrichs, W.L.; Stegmann, T.; Huckriede, A.; Coenen, F.; Frijlink, H.W. Rational design of an influenza subunit vaccine powder with sugar glass technology: Preventing conformational changes of haemagglutinin during freezing and freeze-drying. Vaccine 2007, 25, 6447–6457. [Google Scholar] [CrossRef]
- Geeraedts, F.; Saluja, V.; ter Veer, W.; Amorij, J.P.; Frijlink, H.W.; Wilschut, J.; Hinrichs, W.L.; Huckriede, A. Preservation of the immunogenicity of dry-powder influenza H5N1 whole inactivated virus vaccine at elevated storage temperatures. AAPS J. 2010, 12, 215–222. [Google Scholar] [CrossRef]
- Oleszycka, E.; Lavelle, E.C. Immunomodulatory properties of the vaccine adjuvant alum. Curr. Opin. Immunol. 2014, 28, 1–5. [Google Scholar] [CrossRef]
- Tseng, Y.F.; Weng, T.C.; Lai, C.C.; Chen, P.L.; Lee, M.S.; Hu, A.Y. A fast and efficient purification platform for cell-based influenza viruses by flow-through chromatography. Vaccine 2018, 36, 3146–3152. [Google Scholar] [CrossRef]
- Chia, M.Y.; Hu, A.Y.; Tseng, Y.F.; Weng, T.C.; Lai, C.C.; Lin, J.Y.; Chen, P.L.; Wang, Y.F.; Chao, S.R.; Chang, J.Y.; et al. Evaluation of MDCK cell-derived influenza H7N9 vaccine candidates in ferrets. PLoS ONE 2015, 10, e0120793. [Google Scholar] [CrossRef]
- Wu, U.I.; Hsieh, S.M.; Lee, W.S.; Wang, N.C.; Kung, H.C.; Ou, T.Y.; Chen, F.L.; Lin, T.Y.; Chen, Y.C.; Chang, S.C. Safety and immunogenicity of an inactivated cell culture-derived H7N9 influenza vaccine in healthy adults: A phase I/II, prospective, randomized, open-label trial. Vaccine 2017, 35, 4099–4104. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, T.T.; Lai, C.C.; Weng, T.C.; Cyue, M.H.; Tsai, S.Y.; Tseng, Y.F.; Sung, W.C.; Lee, M.S.; Hu, A.Y. The stability and immunogenicity of inactivated MDCK cell-derived influenza H7N9 viruses. Vaccine 2019, 37, 7117–7122. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, T.T.; Chen, P.L.; Weng, T.C.; Tsai, S.Y.; Lai, C.C.; Chou, H.I.; Chen, P.W.; Lu, C.C.; Liu, M.T.; Sung, W.C.; et al. Development of high-growth influenza H7N9 prepandemic candidate vaccine viruses in suspension MDCK cells. J. Biomed. Sci. 2020, 27, 47. [Google Scholar] [CrossRef]
- Chia, M.Y.; Chung, W.Y.; Wang, C.H.; Chang, W.H.; Lee, M.S. Development of a high-growth enterovirus 71 vaccine candidate inducing cross-reactive neutralizing antibody responses. Vaccine 2018, 36, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.M.; Schild, G.C.; Newman, R.W.; Seagroatt, V. An improved single-radial-immunodiffusion technique for the assay of influenza haemagglutinin antigen: Application for potency determinations of inactivated whole virus and subunit vaccines. J. Biol. Stand. 1977, 5, 237–247. [Google Scholar] [CrossRef]
- Harvey, R.; Hamill, M.; Robertson, J.S.; Minor, P.D.; Vodeiko, G.M.; Weir, J.P.; Takahashi, H.; Harada, Y.; Itamura, S.; Bamford, P.; et al. Application of deglycosylation to SDS PAGE analysis improves calibration of influenza antigen standards. Biologicals 2012, 40, 96–99. [Google Scholar] [CrossRef]
- Lin, C.Y.; Chia, M.Y.; Chen, P.L.; Yeh, C.T.; Cheng, M.C.; Su, I.J.; Lee, M.S. Assessment of pathogenicity and antigenicity of American lineage influenza H5N2 viruses in Taiwan. Virology 2017, 508, 159–163. [Google Scholar] [CrossRef]
- Partridge, J.; Kieny, M.P.; World Health Organization, H.N.i.v.T.F. Global production of seasonal and pandemic (H1N1) influenza vaccines in 2009-2010 and comparison with previous estimates and global action plan targets. Vaccine 2010, 28, 4709–4712. [Google Scholar] [CrossRef]
- Manini, I.; Trombetta, C.M.; Lazzeri, G.; Pozzi, T.; Rossi, S.; Montomoli, E. Egg-Independent Influenza Vaccines and Vaccine Candidates. Vaccines 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Suzuki, Y.; Takada, A.; Kawamoto, A.; Otsuki, K.; Masuda, H.; Yamada, M.; Suzuki, T.; Kida, H.; Kawaoka, Y. Differences in sialic acid-galactose linkages in the chicken egg amnion and allantois influence human influenza virus receptor specificity and variant selection. J. Virol. 1997, 71, 3357–3362. [Google Scholar] [CrossRef] [PubMed]
- Emoto, Y. Cellular aggregation facilitates anoikis in MDCK cells. J. Physiol. Sci. 2008, 58, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Lohr, V.; Genzel, Y.; Behrendt, I.; Scharfenberg, K.; Reichl, U. A new MDCK suspension line cultivated in a fully defined medium in stirred-tank and wave bioreactor. Vaccine 2010, 28, 6256–6264. [Google Scholar] [CrossRef]
- Tsutsumi, R.; Fujisaki, S.; Shozushima, M.; Saito, K.; Sato, S. Anoikis-resistant MDCK cells carrying susceptibilities to TNF-alpha and verotoxin that are suitable for influenza virus cultivation. Cytotechnology 2006, 52, 71–85. [Google Scholar] [CrossRef]
- Chu, C.; Lugovtsev, V.; Golding, H.; Betenbaugh, M.; Shiloach, J. Conversion of MDCK cell line to suspension culture by transfecting with human siat7e gene and its application for influenza virus production. Proc. Natl. Acad. Sci. USA 2009, 106, 14802–14807. [Google Scholar] [CrossRef]
- Hansen, L.J.J.; Daoussi, R.; Vervaet, C.; Remon, J.P.; De Beer, T.R.M. Freeze-drying of live virus vaccines: A review. Vaccine 2015, 33, 5507–5519. [Google Scholar] [CrossRef]
- Assegehegn, G.; Brito-de la Fuente, E.; Franco, J.M.; Gallegos, C. The Importance of Understanding the Freezing Step and Its Impact on Freeze-Drying Process Performance. J. Pharm. Sci. 2018, 108, 1378–1395. [Google Scholar] [CrossRef]
- Minor, P.D. Assaying the Potency of Influenza Vaccines. Vaccines 2015, 3, 90–104. [Google Scholar] [CrossRef]
- Lai, C.C.; Weng, T.C.; Chen, P.L.; Tseng, Y.F.; Lin, C.Y.; Chia, M.Y.; Sung, W.C.; Lee, M.S.; Hu, A.Y. Development and characterization of standard reagents for cell-based prepandemic influenza vaccine products. Hum. Vaccin. Immunother. 2020, 16, 2245–2251. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Shao, M.; Cui, X.; Song, Y.; Li, J.; Yuan, L.; Fang, H.; Liang, Z.; Cyr, T.D.; Li, F.; et al. Application of deglycosylation and electrophoresis to the quantification of influenza viral hemagglutinins facilitating the production of 2009 pandemic influenza (H1N1) vaccines at multiple manufacturing sites in China. Biologicals 2010, 38, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Coudeville, L.; Bailleux, F.; Riche, B.; Megas, F.; Andre, P.; Ecochard, R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: Development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 2010, 10, 18. [Google Scholar] [CrossRef]
- Rudenko, L.; Kiseleva, I.; Naykhin, A.N.; Erofeeva, M.; Stukova, M.; Donina, S.; Petukhova, G.; Pisareva, M.; Krivitskaya, V.; Grudinin, M.; et al. Assessment of human immune responses to H7 avian influenza virus of pandemic potential: Results from a placebo-controlled, randomized double-blind phase I study of live attenuated H7N3 influenza vaccine. PLoS ONE 2014, 9, e87962. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Gao, F.; Zhao, C.; Ding, Y.; Cao, Y.; Yang, T.; Xu, X.; Chen, Z. Comparative effectiveness of H7N9 vaccines in healthy individuals. Hum. Vaccin. Immunother. 2019, 15, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Hovden, A.O.; Cox, R.J.; Haaheim, L.R. Whole influenza virus vaccine is more immunogenic than split influenza virus vaccine and induces primarily an IgG2a response in BALB/c mice. Scand. J. Immunol. 2005, 62, 36–44. [Google Scholar] [CrossRef]
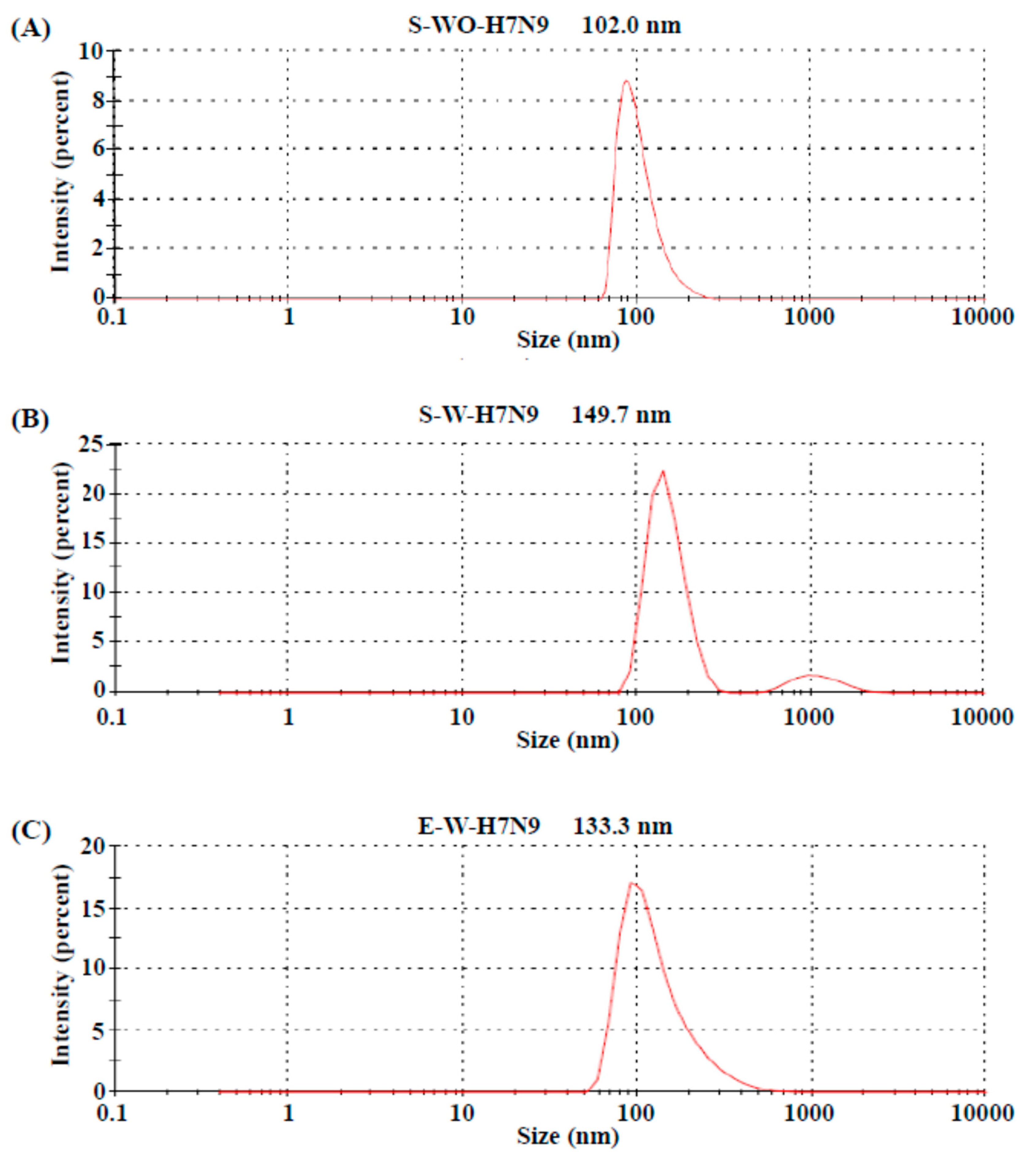
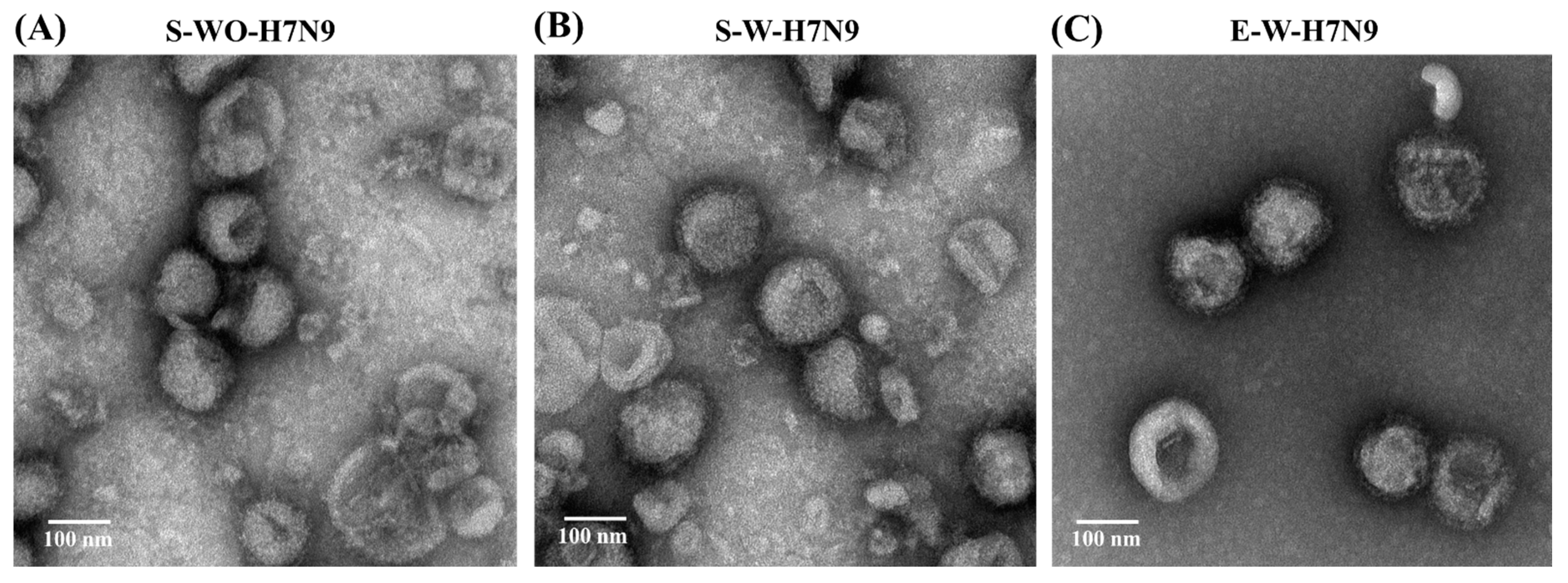
| S-WO-H7N9 | S-W-H7N9 | E-W-H7N9 | |
|---|---|---|---|
| HAU (50 μL) | 8192 | 8192 | 128 |
| SRD (μg/mL) | 108.9 | 106.1 | 37.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chia, M.-Y.; Lin, C.-Y.; Chen, P.-L.; Lai, C.-C.; Weng, T.-C.; Sung, W.-C.; Hu, A.Y.-C.; Lee, M.-S. Characterization and Immunogenicity of Influenza H7N9 Vaccine Antigens Produced Using a Serum-Free Suspension MDCK Cell-Based Platform. Viruses 2022, 14, 1937. https://doi.org/10.3390/v14091937
Chia M-Y, Lin C-Y, Chen P-L, Lai C-C, Weng T-C, Sung W-C, Hu AY-C, Lee M-S. Characterization and Immunogenicity of Influenza H7N9 Vaccine Antigens Produced Using a Serum-Free Suspension MDCK Cell-Based Platform. Viruses. 2022; 14(9):1937. https://doi.org/10.3390/v14091937
Chicago/Turabian StyleChia, Min-Yuan, Chun-Yang Lin, Po-Ling Chen, Chia-Chun Lai, Tsai-Chuan Weng, Wang-Chou Sung, Alan Yung-Chih Hu, and Min-Shi Lee. 2022. "Characterization and Immunogenicity of Influenza H7N9 Vaccine Antigens Produced Using a Serum-Free Suspension MDCK Cell-Based Platform" Viruses 14, no. 9: 1937. https://doi.org/10.3390/v14091937
APA StyleChia, M.-Y., Lin, C.-Y., Chen, P.-L., Lai, C.-C., Weng, T.-C., Sung, W.-C., Hu, A. Y.-C., & Lee, M.-S. (2022). Characterization and Immunogenicity of Influenza H7N9 Vaccine Antigens Produced Using a Serum-Free Suspension MDCK Cell-Based Platform. Viruses, 14(9), 1937. https://doi.org/10.3390/v14091937





