The N-Terminal α-Helix of Potato Virus X-Encoded RNA-Dependent RNA Polymerase Is Required for Membrane Association and Multimerization
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Vector Construction
2.3. Transient Expression
2.4. Microsomal Fractionation
2.5. Protein Purification from Plants
2.6. Western Blotting
2.7. Laser Scanning Confocal Microscopy
2.8. Glutaraldehyde Cross-Linking Assay
2.9. Bioinformatics Analyses
3. Results
3.1. N-Terminal Part of PVX RdRp Is Peripherally Associated with ER
3.2. Prediction of Membrane-Immersed Amino Acids
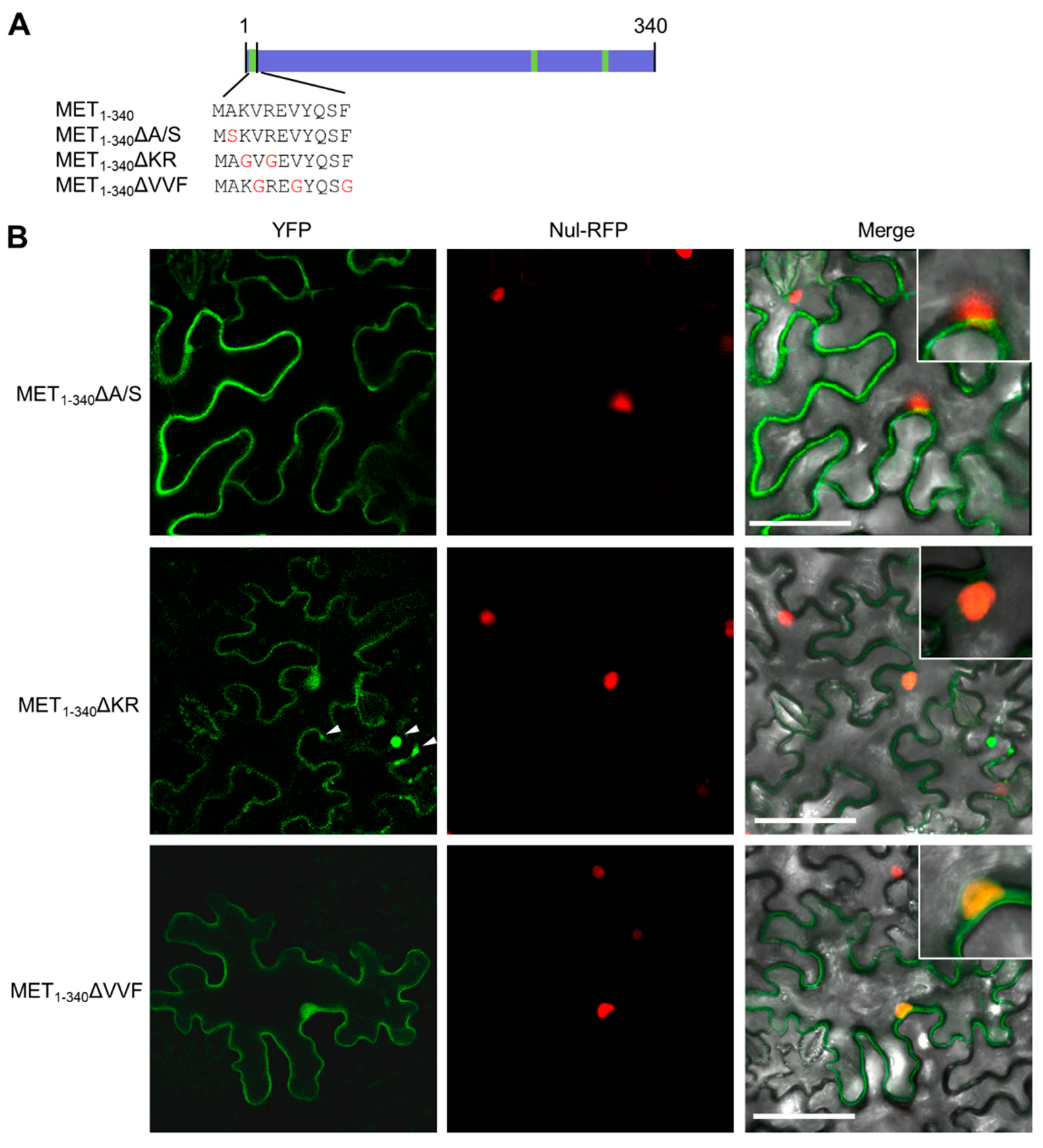
3.3. The First α-Helix Is Essential for Membrane Targeting
3.4. Hydrophobic Residues in the First α-Helix Are Required for Multimerization
3.5. MET Forms Multimers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nguyen-Dinh, V.; Herker, E. Ultrastructural features of membranous replication organelles induced by positive-stranded RNA viruses. Cells 2021, 10, 2407. [Google Scholar] [CrossRef]
- Den Boon, J.A.; Ahlquist, P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010, 64, 241–256. [Google Scholar] [CrossRef]
- Den Boon, J.A.; Diaz, A.; Ahlquist, P. Cytoplasmic viral replication complexes. Cell Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef]
- Wolff, G.; Melia, C.E.; Snijder, E.J.; Bárcena, M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020, 28, 1022–1033. [Google Scholar] [CrossRef]
- de Castro, I.F.; Volonté, L.; Risco, C. Virus factories: Biogenesis and structural design. Cell. Microbiol. 2013, 15, 24–34. [Google Scholar] [CrossRef]
- Salonen, A.; Ahola, T.; Kaariainen, L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005, 285, 139–173. [Google Scholar]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.-M.; Gale, M.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef]
- Blevins, T.; Rajeswaran, R.; Shivaprasad, P.V.; Beknazariants, D.; Si-Ammour, A.; Park, H.S.; Vazquez, F.; Robertson, D.; Meins, F., Jr.; Hohn, T.; et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006, 34, 6233–6246. [Google Scholar] [CrossRef]
- Peisley, A.; Hur, S. Multi-level regulation of cellular recognition of viral dsRNA. Cell. Mol. Life Sci. 2013, 70, 1949–1963. [Google Scholar] [CrossRef]
- Kumar, A.; Haque, J.; Lacoste, J.; Hiscott, J.; Williams, B.R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl. Acad. Sci. USA 1994, 91, 6288–6292. [Google Scholar] [CrossRef]
- Schmidt-Mende, J.; Bieck, E.; Hügle, T.; Penin, F.; Rice, C.M.; Blum, H.E.; Moradpour, D. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 2001, 276, 44052–44063. [Google Scholar] [CrossRef]
- Miller, D.J.; Ahlquist, P. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 2002, 76, 9856–9867. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; de Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef]
- Jones, R.; Bragagnolo, G.; Arranz, R.; Reguera, J. Capping pores of alphavirus nsP1 gate membranous viral replication factories. Nature 2021, 589, 615–619. [Google Scholar] [CrossRef]
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008, 6, 363–374. [Google Scholar] [CrossRef]
- Liu, L.; Westler, W.M.; den Boon, J.A.; Wang, X.; Diaz, A.; Steinberg, H.A.; Ahlquist, P. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 2009, 5, e1000351. [Google Scholar] [CrossRef]
- Ertel, K.J.; Benefield, D.; Castaño-Diez, D.; Pennington, J.G.; Horswill, M.; den Boon, J.A.; Otegui, M.S.; Ahlquist, P. Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife 2017, 6, e25940. [Google Scholar] [CrossRef]
- Unchwaniwala, N.; Zhan, H.; den Boon, J.A.; Ahlquist, P. Cryo-electron microscopy of nodavirus RNA replication organelles illuminates positive-strand RNA virus genome replication. Curr. Opin. Virol. 2021, 51, 74–79. [Google Scholar] [CrossRef]
- Zhang, K.; Law, Y.-S.; Law, M.C.Y.; Tan, Y.B.; Wirawan, M.; Luo, D. Structural insights into viral RNA capping and plasma membrane targeting by Chikungunya virus nonstructural protein 1. Cell Host Microbe 2021, 29, 757–764.e3. [Google Scholar] [CrossRef]
- Kumar, M.; Altan-Bonnet, N. Viral pores are everywhere. Mol. Cell 2021, 81, 2061–2063. [Google Scholar] [CrossRef]
- Unchwaniwala, N.; Zhan, H.; Pennington, J.; Horswill, M.; den Boon, J.A.; Ahlquist, P. Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication. Proc. Natl. Acad. Sci. USA 2020, 117, 18680–18691. [Google Scholar] [CrossRef] [PubMed]
- Davenport, G.F.; Baulcombe, D.C. Mutation of the GKS motif of the RNA-dependent RNA polymerase from potato virus X disables or eliminates virus replication. J. Gen. Virol. 1997, 78, 1247–1251. [Google Scholar] [CrossRef][Green Version]
- Verchot-Lubicz, J.; Torrance, L.; Solovyev, A.G.; Morozov, S.Y.; Jackson, A.O.; Gilmer, D. Varied movement strategies employed by triple gene block–encoding viruses. Mol. Plant. Microbe Interact. 2010, 23, 1231–1247. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Verchot-Lubicz, J.; Ye, C.-M.; Bamunusinghe, D. Molecular biology of potexviruses: Recent advances. J. Gen. Virol. 2007, 88, 1643–1655. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cheng, X. Intercellular movement of plant RNA viruses: Targeting replication complexes to the plasmodesma for both accuracy and efficiency. Traffic 2020, 21, 725–732. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Chai, M.; Wang, J.; Li, D.; Wang, A.; Cheng, X.F. The Potato virus X TGBp2 protein plays dual functional roles in viral replication and movement. J. Virol. 2019, 93, e01635-18. [Google Scholar] [CrossRef]
- Doronin, S.V.; Hemenway, C. Synthesis of potato virus X RNAs by membrane-containing extracts. J. Virol. 1996, 70, 4795–4799. [Google Scholar] [CrossRef]
- Bamunusinghe, D.; Hemenway, C.L.; Nelson, R.S.; Sanderfoot, A.A.; Ye, C.M.; Silva, M.A.T.; Payton, M.; Verchot-Lubicz, J. Analysis of Potato virus X replicase and TGBp3 subcellular locations. Virology 2009, 393, 272–285. [Google Scholar] [CrossRef]
- Komatsu, K.; Sasaki, N.; Yoshida, T.; Suzuki, K.; Masujima, Y.; Hashimoto, M.; Watanabe, S.; Tochio, N.; Kigawa, T.; Yamaji, Y.; et al. Identification of a proline-kinked amphipathic α-helix downstream from the methyltransferase domain of a potexvirus replicase and its role in virus replication and perinuclear complex formation. J. Virol. 2021, 95, e0190620. [Google Scholar] [CrossRef]
- Lee, H.-C.; Huang, Y.-P.; Huang, Y.-W.; Hu, C.-C.; Lee, C.-W.; Chang, C.-H.; Lin, N.-S.; Hsu, Y.-H. Voltage-dependent anion channel proteins associate with dynamic Bamboo mosaic virus-induced complexes. Plant Physiol. 2021, 188, 1061–1080. [Google Scholar] [CrossRef] [PubMed]
- Schaad, M.C.; Jensen, P.E.; Carrington, J.C. Formation of plant RNA virus replication complexes on membranes: Role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997, 16, 4049–4059. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Wu, X.; Liu, J.; Fang, Y.; Luan, Y.; Cui, X.; Zhou, X.; Wang, A.; Cheng, X. P3N-PIPO interacts with P3 via the shared N-terminal domain to recruit viral replication vesicles for cell-to-cell movement. J. Virol. 2020, 94, e01898-19. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, X.; Wu, J.; Briddon, R.W.; Zhou, X. βC1 encoded by tomato yellow leaf curl China betasatellite forms multimeric complexes in vitro and in vivo. Virology 2011, 409, 156–162. [Google Scholar] [CrossRef]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef]
- Chatzigoulas, A.; Cournia, Z. Predicting protein–membrane interfaces of peripheral membrane proteins using ensemble machine learning. Brief. Bioinform. 2022, 23, bbab518. [Google Scholar] [CrossRef]
- Romano, J.D.; Schmidt, W.K.; Michaelis, S. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol. Biol. Cell 1998, 9, 2231–2247. [Google Scholar] [CrossRef]
- Honma, H.; Tsushima, D.; Kawakami, H.; Fujihara, N.; Tsusaka, T.; Kawashimo, M.; Nishimura, T.; Fuji, S. Complete nucleotide sequence of a new potexvirus, ‘Cnidium virus X’, isolated from Cnidium officinale in Japan. Arch. Virol. 2019, 164, 1931–1935. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Luan, Y.; Chai, M.; Yang, Y.; Wang, Y.; Deng, W.; Li, Y.; Cheng, X.; Wu, X. The N-Terminal α-Helix of Potato Virus X-Encoded RNA-Dependent RNA Polymerase Is Required for Membrane Association and Multimerization. Viruses 2022, 14, 1907. https://doi.org/10.3390/v14091907
Jiang X, Luan Y, Chai M, Yang Y, Wang Y, Deng W, Li Y, Cheng X, Wu X. The N-Terminal α-Helix of Potato Virus X-Encoded RNA-Dependent RNA Polymerase Is Required for Membrane Association and Multimerization. Viruses. 2022; 14(9):1907. https://doi.org/10.3390/v14091907
Chicago/Turabian StyleJiang, Xue, Yameng Luan, Mengzhu Chai, Yingshuai Yang, Yuting Wang, Wenjia Deng, Yonggang Li, Xiaofei Cheng, and Xiaoyun Wu. 2022. "The N-Terminal α-Helix of Potato Virus X-Encoded RNA-Dependent RNA Polymerase Is Required for Membrane Association and Multimerization" Viruses 14, no. 9: 1907. https://doi.org/10.3390/v14091907
APA StyleJiang, X., Luan, Y., Chai, M., Yang, Y., Wang, Y., Deng, W., Li, Y., Cheng, X., & Wu, X. (2022). The N-Terminal α-Helix of Potato Virus X-Encoded RNA-Dependent RNA Polymerase Is Required for Membrane Association and Multimerization. Viruses, 14(9), 1907. https://doi.org/10.3390/v14091907





