CAPG Is Required for Ebola Virus Infection by Controlling Virus Egress from Infected Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Culture
2.2. Reagents and Antibodies
2.3. Plasmids
2.4. Virus Infections
2.5. Calculation of Infection Efficiency
2.6. Small Interfering RNA (siRNA)
2.7. Generation of Knockout (KO) Clonal Cell Lines
2.8. Cell Viability Assay
2.9. Virus-Like Particle Release Assay
2.10. Immunoblot Assay
2.11. Immunofluorescence Based Detection of Virus Infection and Uptake of Virus into Cells
2.12. Proximity Ligation Assay
2.13. Tripartite Split-GFP Assay
2.14. Real-Time Quantitative PCR (RT-qPCR)
2.15. In Vitro Pull Down
2.16. Statistical Analysis
3. Results
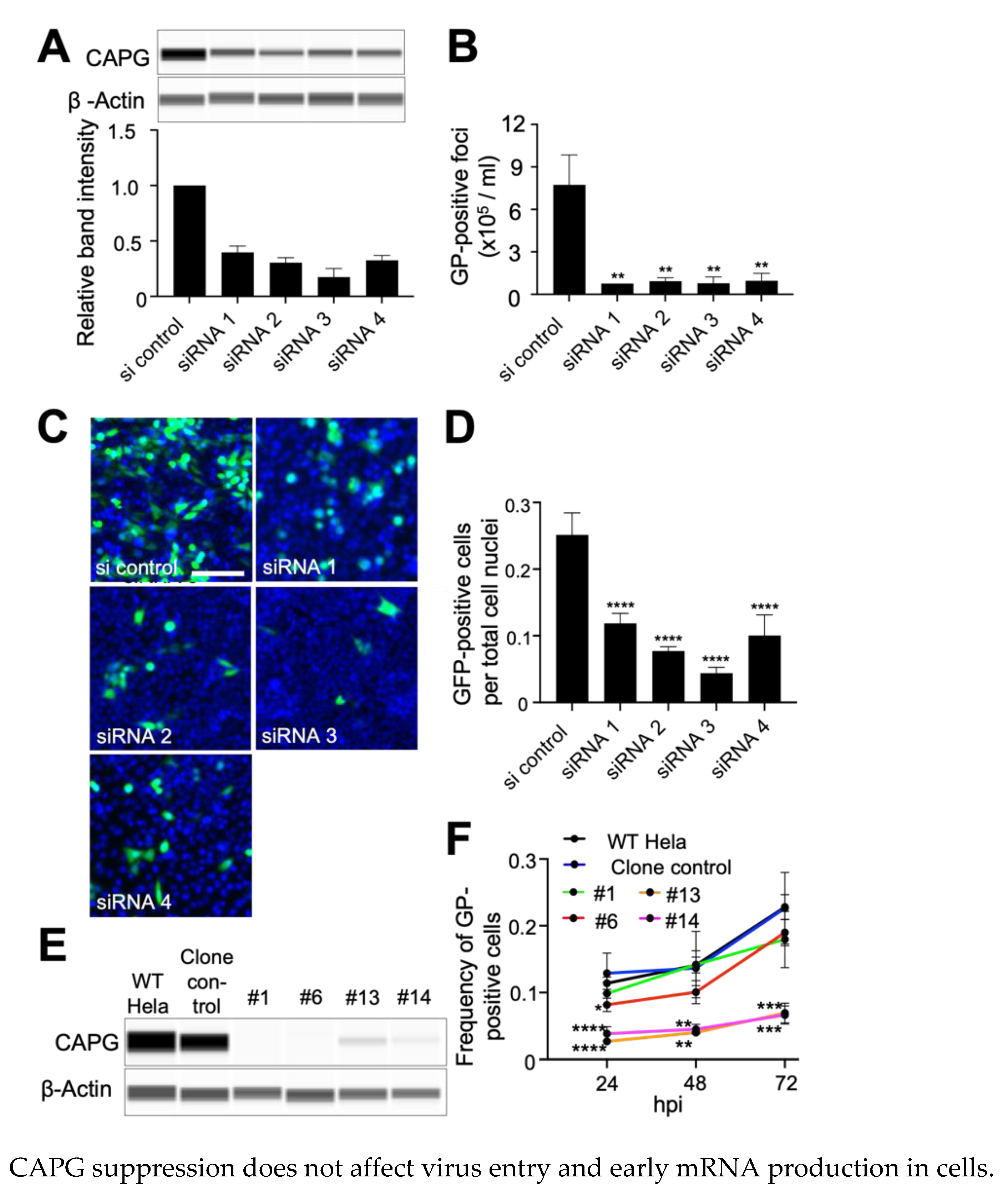
3.1. Suppression of CAPG Expression Impairs EBOV Infection
3.2. CAPG Is Required for Efficient Production of Virus from Infected Cells
3.3. VP40 and GP Are Found in Close Proximity to CAPG in the Cytoplasm
3.4. Identification of CAPG Subdomains Involved in Actin Interaction Are Important for Association with VP40
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baseler, L.; Chertow, D.S.; Johnson, K.M.; Feldmann, H.; Morens, D.M. The Pathogenesis of Ebola Virus Disease. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 387–418. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, H.; Sprecher, A.; Geisbert, T.W. Ebola. N. Engl. J. Med. 2020, 382, 1832–1842. [Google Scholar] [CrossRef] [PubMed]
- Geisbert, T.W.; Jahrling, P.B. Differentiation of Filoviruses by Electron Microscopy. Virus Res. 1995, 39, 129–150. [Google Scholar] [CrossRef]
- Elliott, L.H.; Kiley, M.P.; McCormick, J.B. Descriptive Analysis of Ebola Virus Proteins. Virology 1985, 147, 169–176. [Google Scholar] [CrossRef]
- Noda, T.; Hagiwara, K.; Sagara, H.; Kawaoka, Y. Characterization of the Ebola Virus Nucleoprotein-RNA Complex. J. Gen. Virol. 2010, 91, 1478–1483. [Google Scholar] [CrossRef] [PubMed]
- Harty, R.N.; Brown, M.E.; Wang, G.; Huibregtse, J.; Hayes, F.P. A PPxY Motif within the VP40 Protein of Ebola Virus Interacts Physically and Functionally with a Ubiquitin Ligase: Implications for Filovirus Budding. Proc. Natl. Acad. Sci. USA 2000, 97, 13871–13876. [Google Scholar] [CrossRef]
- Wool-Lewis, R.J.; Bates, P. Characterization of Ebola Virus Entry by Using Pseudotyped Viruses: Identification of Receptor-Deficient Cell Lines. J. Virol. 1998, 72, 3155–3160. [Google Scholar] [CrossRef]
- Nanbo, A.; Imai, M.; Watanabe, S.; Noda, T.; Takahashi, K.; Neumann, G.; Halfmann, P.; Kawaoka, Y. Ebolavirus Is Internalized into Host Cells via Macropinocytosis in a Viral Glycoprotein-Dependent Manner. PLoS Pathog. 2010, 6, e1001121. [Google Scholar] [CrossRef]
- Saeed, M.F.; Kolokoltsov, A.A.; Albrecht, T.; Davey, R.A. Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes. PLoS Pathog. 2010, 6, e1001110. [Google Scholar] [CrossRef]
- Mylvaganam, S.; Freeman, S.A.; Grinstein, S. The Cytoskeleton in Phagocytosis and Macropinocytosis. Curr. Biol. 2021, 31, R619–R632. [Google Scholar] [CrossRef]
- Saeed, M.F.; Kolokoltsov, A.A.; Freiberg, A.N.; Holbrook, M.R.; Davey, R.A. Phosphoinositide-3 Kinase-Akt Pathway Controls Cellular Entry of Ebola Virus. PLoS Pathog. 2008, 4, e1000141. [Google Scholar] [CrossRef] [PubMed]
- Quinn, K.; Brindley, M.A.; Weller, M.L.; Kaludov, N.; Kondratowicz, A.; Hunt, C.L.; Sinn, P.L.; McCray, P.B.; Stein, C.S.; Davidson, B.L.; et al. Rho GTPases Modulate Entry of Ebola Virus and Vesicular Stomatitis Virus Pseudotyped Vectors. J. Virol. 2009, 83, 10176–10186. [Google Scholar] [CrossRef] [PubMed]
- Schudt, G.; Dolnik, O.; Kolesnikova, L.; Biedenkopf, N.; Herwig, A.; Becker, S. Transport of Ebolavirus Nucleocapsids Is Dependent on Actin Polymerization: Live-Cell Imaging Analysis of Ebolavirus-Infected Cells. J. Infect. Dis. 2015, 212, S160–S166. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xu, L.; Sun, Y.; Nabel, G.J. The Assembly of Ebola Virus Nucleocapsid Requires Virion-Associated Proteins 35 and 24 and Posttranslational Modification of Nucleoprotein. Mol. Cell 2002, 10, 307–316. [Google Scholar] [CrossRef]
- Mateo, M.; Carbonnelle, C.; Martinez, M.J.; Reynard, O.; Page, A.; Volchkova, V.A.; Volchkov, V.E. Knockdown of Ebola Virus VP24 Impairs Viral Nucleocapsid Assembly and Prevents Virus Replication. J. Infect. Dis. 2011, 204, S892–S896. [Google Scholar] [CrossRef]
- Watanabe, S.; Noda, T.; Halfmann, P.; Jasenosky, L.; Kawaoka, Y. Ebola Virus (EBOV) VP24 Inhibits Transcription and Replication of the EBOV Genome. J. Infect. Dis. 2007, 196, S284–S290. [Google Scholar] [CrossRef]
- Grikscheit, K.; Dolnik, O.; Takamatsu, Y.; Pereira, A.R.; Becker, S. Ebola Virus Nucleocapsid-Like Structures Utilize Arp2/3 Signaling for Intracellular Long-Distance Transport. Cells 2020, 9, 1728. [Google Scholar] [CrossRef]
- Noda, T.; Sagara, H.; Suzuki, E.; Takada, A.; Kida, H.; Kawaoka, Y. Ebola Virus VP40 Drives the Formation of Virus-Like Filamentous Particles Along with GP. J. Virol. 2002, 76, 4855–4865. [Google Scholar] [CrossRef]
- Licata, J.M.; Simpson-Holley, M.; Wright, N.T.; Han, Z.; Paragas, J.; Harty, R.N. Overlapping Motifs (PTAP and PPEY) within the Ebola Virus VP40 Protein Function Independently as Late Budding Domains: Involvement of Host Proteins TSG101 and VPS-4. J. Virol. 2003, 77, 1812–1819. [Google Scholar] [CrossRef]
- Adu-Gyamfi, E.; Digman, M.A.; Gratton, E.; Stahelin, R.V. Single-Particle Tracking Demonstrates That Actin Coordinates the Movement of the Ebola Virus Matrix Protein. Biophys. J. 2012, 103, L41–L43. [Google Scholar] [CrossRef]
- Han, Z.; Harty, R. Packaging of Actin into Ebola Virus VLPs. Virol. J. 2005, 2, 92. [Google Scholar] [CrossRef]
- Izadi, M.; Hou, W.; Qualmann, B.; Kessels, M.M. Direct Effects of Ca2+/Calmodulin on Actin Filament Formation. Biochem. Biophys. Res. Commun. 2018, 506, 355–360. [Google Scholar] [CrossRef]
- Cooper, J.A.; Schafer, D.A. Control of Actin Assembly and Disassembly at Filament Ends. Curr. Opin. Cell Biol. 2000, 12, 97–103. [Google Scholar] [CrossRef]
- Yu, F.-X.; Johnston, P.A.; Südhof, T.C.; Yin, H.L. GCap39, a Calcium Ion- and Polyphosphoinositide-Regulated Actin Capping Protein. Science 1990, 250, 1413–1415. [Google Scholar] [CrossRef]
- Weed, S.A.; Parsons, J.T. Cortactin: Coupling Membrane Dynamics to Cortical Actin Assembly. Oncogene 2001, 20, 6418–6434. [Google Scholar] [CrossRef] [PubMed]
- Shtanko, O.; Sakurai, Y.; Reyes, A.N.; Noël, R.; Cintrat, J.-C.; Gillet, D.; Barbier, J.; Davey, R.A. Retro-2 and Its Dihydroquinazolinone Derivatives Inhibit Filovirus Infection. Antiviral Res. 2018, 149, 154–163. [Google Scholar] [CrossRef]
- Ciancanelli, M.J.; Basler, C.F. Mutation of YMYL in the Nipah Virus Matrix Protein Abrogates Budding and Alters Subcellular Localization. J. Virol. 2006, 80, 12070–12078. [Google Scholar] [CrossRef]
- Mühlberger, E.; Weik, M.; Volchkov, V.E.; Klenk, H.-D.; Becker, S. Comparison of the Transcription and Replication Strategies of Marburg Virus and Ebola Virus by Using Artificial Replication Systems. J. Virol. 1999, 73, 2333–2342. [Google Scholar] [CrossRef]
- Cabantous, S.; Nguyen, H.B.; Pedelacq, J.-D.; Koraïchi, F.; Chaudhary, A.; Ganguly, K.; Lockard, M.A.; Favre, G.; Terwilliger, T.C.; Waldo, G.S. A New Protein-Protein Interaction Sensor Based on Tripartite Split-GFP Association. Sci. Rep. 2013, 3, 2854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Schepis, A.; Huang, H.; Yang, J.; Ma, W.; Torra, J.; Zhang, S.-Q.; Yang, L.; Wu, H.; Nonell, S.; et al. Designing a Green Fluorogenic Protease Reporter by Flipping a Beta Strand of GFP for Imaging Apoptosis in Animals. J. Am. Chem. Soc. 2019, 141, 4526–4530. [Google Scholar] [CrossRef]
- Mori, H.; Davey, R.A. CellProfiler Pipelines Used for CAPG-Ebola Analysis. Zenodo 2022. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Anantpadma, M.; Kouznetsova, J.; Wang, H.; Huang, R.; Kolokoltsov, A.; Guha, R.; Lindstrom, A.R.; Shtanko, O.; Simeonov, A.; Maloney, D.J.; et al. Large-Scale Screening and Identification of Novel Ebola Virus and Marburg Virus Entry Inhibitors. Antimicrob. Agents Chemother. 2016, 60, 4471–4481. [Google Scholar] [CrossRef] [PubMed]
- Batra, J.; Hultquist, J.F.; Liu, D.; Shtanko, O.; Von Dollen, J.; Satkamp, L.; Jang, G.M.; Luthra, P.; Schwarz, T.M.; Small, G.I.; et al. Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication. Cell 2018, 175, 1917–1930.e13. [Google Scholar] [CrossRef]
- Huang, Y.; Wei, H.; Wang, Y.; Shi, Z.; Raoul, H.; Yuan, Z. Rapid Detection of Filoviruses by Real-Time TaqMan Polymerase Chain Reaction Assays. Virol. Sin. 2012, 27, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Gibb, T.R.; Norwood, D.A.; Woollen, N.; Henchal, E.A. Development and Evaluation of a Fluorogenic 5′ Nuclease Assay To Detect and Differentiate between Ebola Virus Subtypes Zaire and Sudan. J. Clin. Microbiol. 2001, 39, 4125–4130. [Google Scholar] [CrossRef] [PubMed]
- Ro, Y.-T.; Ticer, A.; Carrion, R.; Patterson, J.L. Rapid Detection and Quantification of Ebola Zaire Virus by One-Step Real-Time Quantitative Reverse Transcription-Polymerase Chain Reaction: Ebola Virus Detection by QRT-PCR Assay. Microbiol. Immunol. 2017, 61, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Watanabe, S.; Halfmann, P.; Kawaoka, Y. The Spatio-Temporal Distribution Dynamics of Ebola Virus Proteins and RNA in Infected Cells. Sci. Rep. 2013, 3, 1206. [Google Scholar] [CrossRef]
- Mishra, V.S.; Henske, E.P.; Kwiatkowski, D.J.; Southwick, F.S. The Human Actin-Regulatory Protein Cap G: Gene Structure and Chromosome Location. Genomics 1994, 23, 560–565. [Google Scholar] [CrossRef]
- Salanga, C.M.; Salanga, M.C. Genotype to Phenotype: CRISPR Gene Editing Reveals Genetic Compensation as a Mechanism for Phenotypic Disjunction of Morphants and Mutants. Int. J. Mol. Sci. 2021, 22, 3472. [Google Scholar] [CrossRef]
- Söderberg, O.; Gullberg, M.; Jarvius, M.; Ridderstråle, K.; Leuchowius, K.-J.; Jarvius, J.; Wester, K.; Hydbring, P.; Bahram, F.; Larsson, L.-G.; et al. Direct Observation of Individual Endogenous Protein Complexes in Situ by Proximity Ligation. Nat. Methods 2006, 3, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Kirchdoerfer, R.N.; Moyer, C.L.; Abelson, D.M.; Saphire, E.O. The Ebola Virus VP30-NP Interaction Is a Regulator of Viral RNA Synthesis. PLOS Pathog. 2016, 12, e1005937. [Google Scholar] [CrossRef] [PubMed]
- Van Impe, K.; Bethuyne, J.; Cool, S.; Impens, F.; Ruano-Gallego, D.; De Wever, O.; Vanloo, B.; Van Troys, M.; Lambein, K.; Boucherie, C.; et al. A Nanobody Targeting the F-Actin Capping Protein CapG Restrains Breast Cancer Metastasis. Breast Cancer Res. 2013, 15, R116. [Google Scholar] [CrossRef]
- Kesari, A.S.; Aryal, U.K.; LaCount, D.J. A Novel Proximity Biotinylation Assay Based on the Self-Associating Split GFP1-10/11. Proteomes 2020, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. HIV-1 and Ebola Virus Encode Small Peptide Motifs That Recruit Tsg101 to Sites of Particle Assembly to Facilitate Egress. Nat. Med. 2001, 7, 1313–1319. [Google Scholar] [CrossRef]
- Burkel, B.M.; von Dassow, G.; Bement, W.M. Versatile Fluorescent Probes for Actin Filaments Based on the Actin-Binding Domain of Utrophin. Cell Motil. Cytoskeleton 2007, 64, 822–832. [Google Scholar] [CrossRef]
- Zhang, Y.; Vorobiev, S.M.; Gibson, B.G.; Hao, B.; Sidhu, G.S.; Mishra, V.S.; Yarmola, E.G.; Bubb, M.R.; Almo, S.C.; Southwick, F.S. A CapG Gain-of-Function Mutant Reveals Critical Structural and Functional Determinants for Actin Filament Severing. EMBO J. 2006, 25, 4458–4467. [Google Scholar] [CrossRef][Green Version]
- Kumari, A.; Kesarwani, S.; Javoor, M.G.; Vinothkumar, K.R.; Sirajuddin, M. Structural Insights into Actin Filament Recognition by Commonly Used Cellular Actin Markers. EMBO J. 2020, 39, e104006. [Google Scholar] [CrossRef]
- Smith, B.A.; Daugherty-Clarke, K.; Goode, B.L.; Gelles, J. Pathway of Actin Filament Branch Formation by Arp2/3 Complex Revealed by Single-Molecule Imaging. Proc. Natl. Acad. Sci. USA 2013, 110, 1285–1290. [Google Scholar] [CrossRef]
- Winter, S.L.; Chlanda, P. Dual-Axis Volta Phase Plate Cryo-Electron Tomography of Ebola Virus-like Particles Reveals Actin-VP40 Interactions. J. Struct. Biol. 2021, 213, 107742. [Google Scholar] [CrossRef]
- Witke, W.; Li, W.; Kwiatkowski, D.J.; Southwick, F.S. Comparisons of CapG and Gelsolin-Null Macrophages. J. Cell Biol. 2001, 154, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Martinez, O.; Johnson, J.C.; Honko, A.; Yen, B.; Shabman, R.S.; Hensley, L.E.; Olinger, G.G.; Basler, C.F. Ebola Virus Exploits a Monocyte Differentiation Program To Promote Its Entry. J. Virol. 2013, 87, 3801–3814. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pierre, V.; Liu, C.; Senapati, S.; Park, P.S.-H.; Senyo, S.E. Exogenous Extracellular Matrix Proteins Decrease Cardiac Fibroblast Activation in Stiffening Microenvironment through CAPG. J. Mol. Cell. Cardiol. 2021, 159, 105–119. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mori, H.; Connell, J.P.; Donahue, C.J.; Boytz, R.; Nguyen, Y.T.K.; Leung, D.W.; LaCount, D.J.; Davey, R.A. CAPG Is Required for Ebola Virus Infection by Controlling Virus Egress from Infected Cells. Viruses 2022, 14, 1903. https://doi.org/10.3390/v14091903
Mori H, Connell JP, Donahue CJ, Boytz R, Nguyen YTK, Leung DW, LaCount DJ, Davey RA. CAPG Is Required for Ebola Virus Infection by Controlling Virus Egress from Infected Cells. Viruses. 2022; 14(9):1903. https://doi.org/10.3390/v14091903
Chicago/Turabian StyleMori, Hiroyuki, James P. Connell, Callie J. Donahue, RuthMabel Boytz, Yen Thi Kim Nguyen, Daisy W. Leung, Douglas J. LaCount, and Robert A. Davey. 2022. "CAPG Is Required for Ebola Virus Infection by Controlling Virus Egress from Infected Cells" Viruses 14, no. 9: 1903. https://doi.org/10.3390/v14091903
APA StyleMori, H., Connell, J. P., Donahue, C. J., Boytz, R., Nguyen, Y. T. K., Leung, D. W., LaCount, D. J., & Davey, R. A. (2022). CAPG Is Required for Ebola Virus Infection by Controlling Virus Egress from Infected Cells. Viruses, 14(9), 1903. https://doi.org/10.3390/v14091903








