Cap Is the Protease of the Porcine Circovirus 2
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plasmids Construction
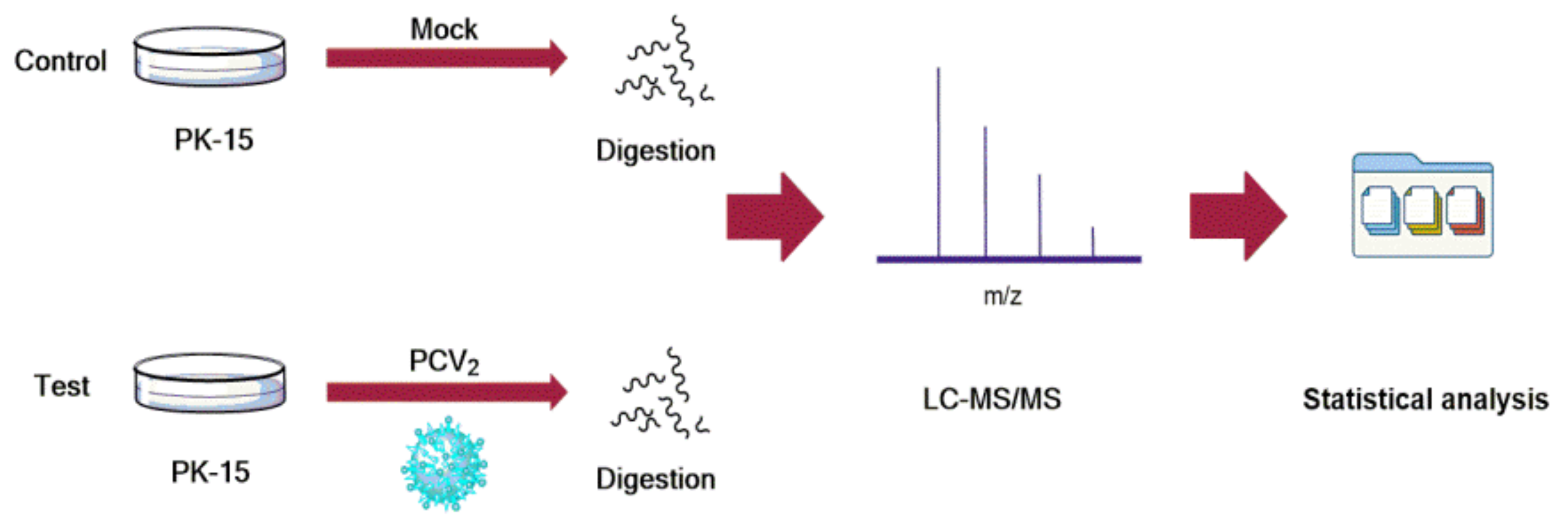
2.3. The Analysis of Host Protein Proteome
2.4. Proteomics Analysis
2.5. Cell Transfection
2.6. SDS-PAGE and Western Blotting
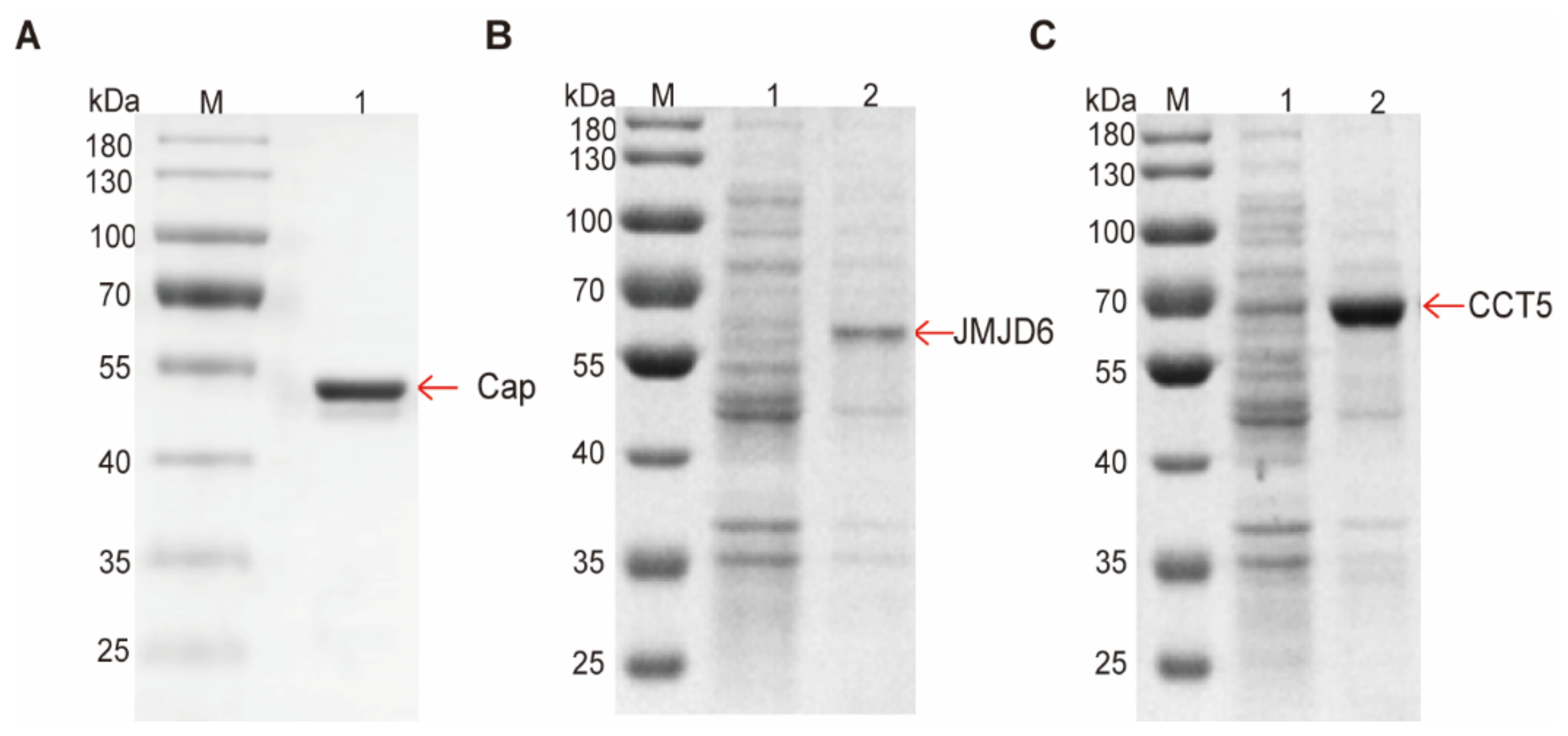
2.7. Protein Expression and Purification
2.8. Enzymatic Activity Assay of the Cap with the Host Proteins
2.9. Statistical Analysis
3. Results and Discussion
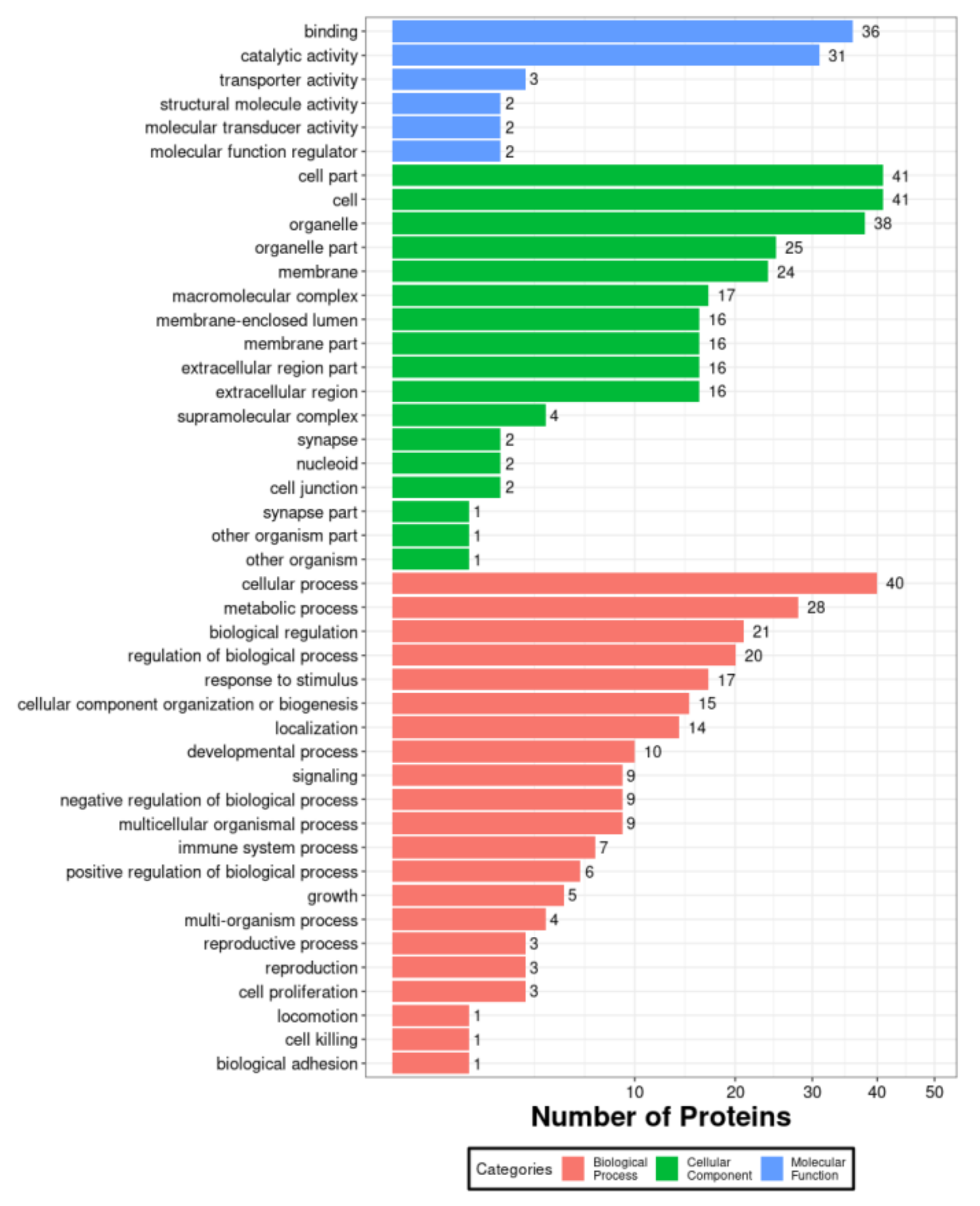
3.1. Proteomic Analysis of Host Proteins in PK-15 Cells Infected by PCV2
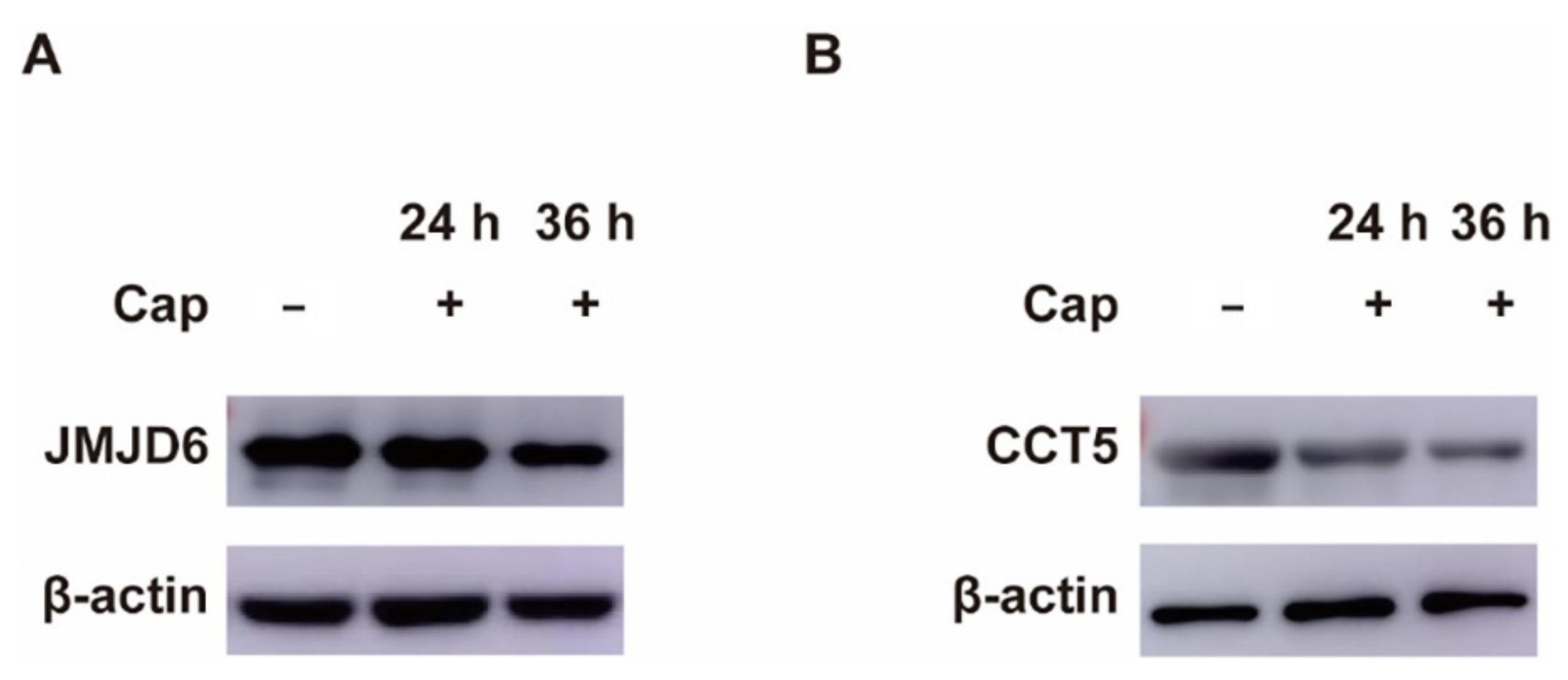
3.2. Validation That Down-Regulation of Host Proteins Is Due to Cap
3.3. Validation That Cap down Regulates Host Proteins by Directly Hydrolyzing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.J. Porcine circovirus type 2 (PCV2): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 2013, 1, 43–64. [Google Scholar] [CrossRef] [PubMed]
- Finsterbusch, T.; Mankertz, A. Porcine circoviruses–small but powerful. Virus Res. 2009, 143, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Gelderblom, H.; Vettermann, W.; Koch, A.M. A very small porcine virus with circular single-stranded-DNA. Nature 1982, 295, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.H.J.; Clark, E.; Harding, J.; Allan, G.; Willson, P.; Strokappe, J.; Martin, K.; McNeilly, F.; Meehan, B.; Todd, D.; et al. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 1998, 39, 44–51. [Google Scholar] [PubMed]
- Nawagitgul, I.M.P.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81 Pt 9, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, I.; Kwang, J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 2005, 79, 8262–8274. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cao, J.; Zhou, N.; Jin, Y.; Wu, J.; Zhou, J. Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J. Virol. 2013, 87, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Guo, K.; Xu, H.; Wang, T.; Zhang, Y. Identification of Putative ORF5 Protein of Porcine Circovirus Type 2 and Functional Analysis of GFP-Fused ORF5 Protein. PLoS ONE 2015, 10, e0127859. [Google Scholar]
- Agbowuro, A.A.; Huston, W.M.; Gamble, A.B.; Tyndall, J.D.A. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2018, 38, 1295–1331. [Google Scholar] [CrossRef] [PubMed]
- Ivanoff, T.T.L.A.; Ray, J.; Korant, B.D.; Petteway, S.R., Jr. Expression and site-specific mutagenesis of the poliovirus 3C protease in Escherichia coli. Proc. Natl. Acad. Sci. USA 1986, 83, 5392–5396. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.R.; Roque-Rosell, N.; Birtley, J.R.; Leatherbarrow, R.J.; Curry, S. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J. Virol. 2007, 81, 115–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaletsky, R.L.; Simmons, G.; Bates, P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007, 81, 13378–13384. [Google Scholar] [CrossRef] [PubMed]
- Misasi, J.; Chandran, K.; Yang, J.Y.; Considine, B.; Filone, C.M.; Cote, M.; Sullivan, N.; Fabozzi, G.; Hensley, L.; Cunningham, J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J. Virol. 2012, 86, 3284–3292. [Google Scholar] [CrossRef] [PubMed]
- Elahi, S.M.; Oualikene, W.; Naghdi, L.; O’Connor-McCourt, M.; Massie, B. Adenovirus-based libraries: Efficient generation of recombinant adenoviruses by positive selection with the adenovirus protease. Gene Ther. 2002, 9, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Tandon, R.; Mocarski, E.S.; Conway, J.F. The A, B, Cs of herpesvirus capsids. Viruses 2015, 7, 899–914. [Google Scholar] [CrossRef] [PubMed]
- Tong, L. Viruse protease. Chem. Rev. 2002, 102, 4609–4626. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Kelleher, L.N. Precision proteomics: The case for high resolution and high mass accuracy.pdf. Proc. Natl. Acad. Sci. USA 2008, 105, 18132–18138. [Google Scholar] [CrossRef] [PubMed]
- Magrane, M.; UniProt Consortium. UniProt Knowledgebase: A hub of integrated protein data. Database 2011, 2011, bar009. [Google Scholar] [CrossRef] [PubMed]


| Accession No. | Protein | Functions |
|---|---|---|
| tr|A0A287BND4 | TOMM70 | Innate immune pathway |
| tr|A0A480ZTV3 | ENDOD1 | endonuclease activity |
| tr|A0A4X1W398 | LOC100525876 | mitochondrion organization |
| tr|A0A480SUM8 | SARS2 | serine-tRNA ligase activity |
| tr|A0A480PJ32 | EIF2B4 | translation initiation factor activity |
| tr|F1SHH7 | API5 | fibroblast growth factor binding |
| tr|A0A5G2R9Q7 | LSM14A | DNA/RNA binding |
| tr|A0A4X1TKG0 | LMAN1 | endoplasmic reticulum organization |
| tr|Q95N04 | DLAT | pyruvate metabolic process |
| tr|A0A287B466 | PSEN2 | intracellular signal transduction |
| Accession No. | Protein | Functions |
|---|---|---|
| tr|A5A758 | KRT1 | associated with bullous congenital ichthyosiform erythroderma |
| tr|A0A4X1TWN6 | TMX3 | protein-disulfide reductase activity |
| tr|A0A4X1SP89 | N/A | actin binding |
| tr|A0A5G2R9A9 | PFN1 | actin binding |
| tr|I3LD72 | EHD2 | ATP binding |
| tr|A0A4X1WBN6 | N/A | N/A |
| tr|A0A287AAG0 | UAP1L1 | UDP-N-acetylglucosamine diphosphorylase activity |
| tr|A0A4X1V6Q9 | DDX39A | ATP binding |
| tr|A0A480MAF3 | MRPL16 | rRNA binding |
| tr|A0A480Z0T5 | KLC1 | regulate organelle transport |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Yang, W.; Zhang, W.; Li, J.; Yang, G.; Zhao, S.; Zheng, Y. Cap Is the Protease of the Porcine Circovirus 2. Viruses 2022, 14, 1550. https://doi.org/10.3390/v14071550
Yang X, Yang W, Zhang W, Li J, Yang G, Zhao S, Zheng Y. Cap Is the Protease of the Porcine Circovirus 2. Viruses. 2022; 14(7):1550. https://doi.org/10.3390/v14071550
Chicago/Turabian StyleYang, Xuechen, Wei Yang, Wei Zhang, Jiamei Li, Guoyu Yang, Shuhong Zhao, and Yueting Zheng. 2022. "Cap Is the Protease of the Porcine Circovirus 2" Viruses 14, no. 7: 1550. https://doi.org/10.3390/v14071550
APA StyleYang, X., Yang, W., Zhang, W., Li, J., Yang, G., Zhao, S., & Zheng, Y. (2022). Cap Is the Protease of the Porcine Circovirus 2. Viruses, 14(7), 1550. https://doi.org/10.3390/v14071550






