Isolation of a Human Betaretrovirus from Patients with Primary Biliary Cholangitis
Abstract
1. Introduction
1.1. Human Betaretrovirus
1.2. Primary Biliary Cholangitis
1.3. Linking HBRV with PBC
2. Materials and Methods
2.1. Co-Cultivation Studies
2.2. Detection of HBRV
2.3. Informatic Analysis of Integration Sites
3. Results
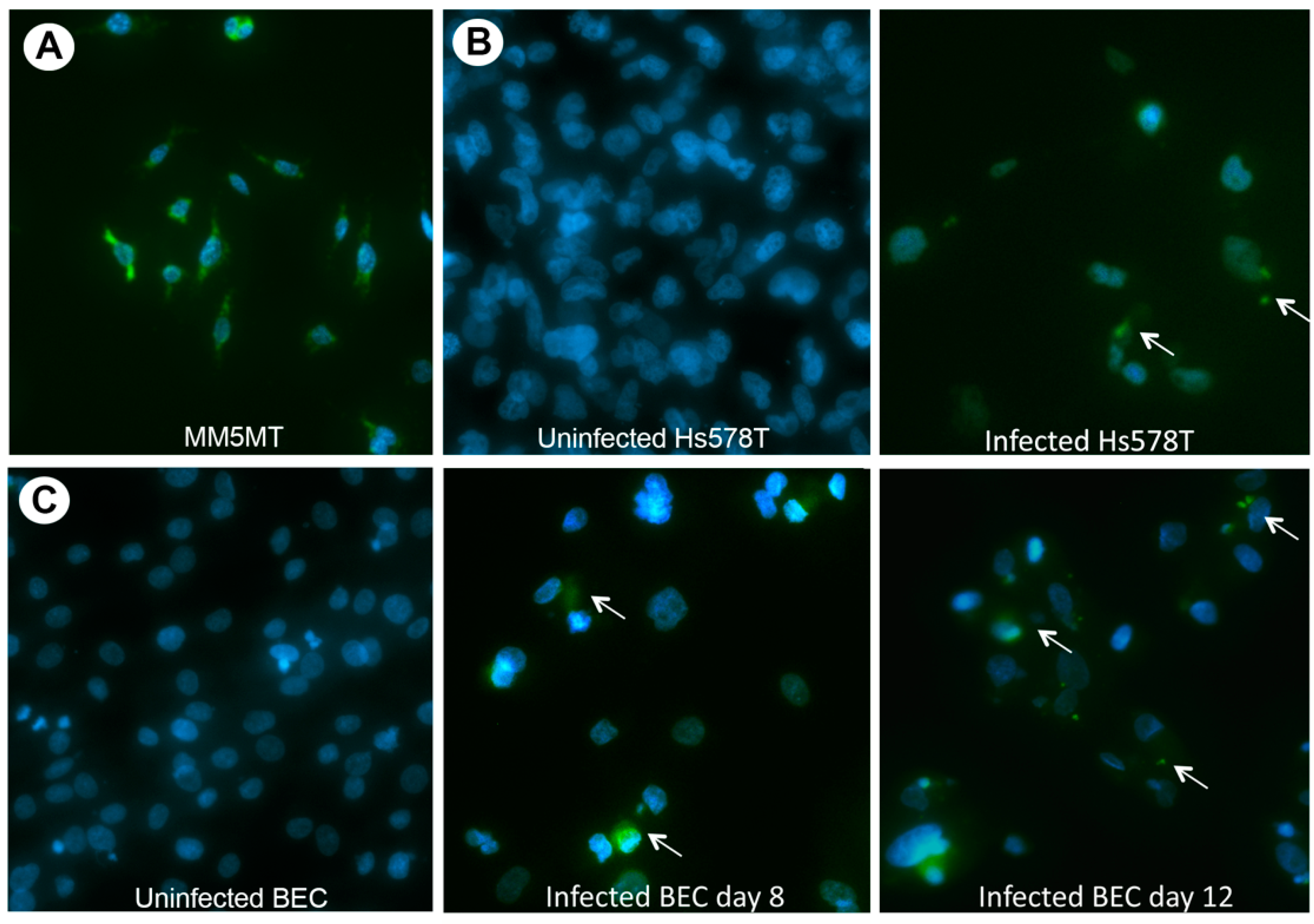
3.1. HBRV Co-Culture Assay

3.2. Characterization of HBRV
3.3. Cloning of HBRV Proviral Genome
3.4. Evaluation of HBRV Proviral Integration Sites
3.4.1. HBRV Integration: Transcription Start Sites (TSS) and CpG Islands
3.4.2. HBRV Common Insertion Sites (CIS) Genes
3.4.3. HBRV In Vitro Genome Clusters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Melana, S.M.; Nepomnaschy, I.; Hasa, J.; Djougarian, A.; Djougarian, A.; Holland, J.F.; Pogo, B.G. Detection of human mammary tumor virus proteins in human breast cancer cells. J. Virol. Methods 2010, 163, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Nartey, T.; Mazzanti, C.M.; Melana, S.; Glenn, W.K.; Bevilacqua, G.; Holland, J.F.; Whitaker, N.J.; Lawson, J.S.; Pogo, B.G. Mouse mammary tumor-like virus (MMTV) is present in human breast tissue before development of virally associated breast cancer. Infect. Agent. Cancer 2017, 12, 1. [Google Scholar] [CrossRef]
- Nartey, T.; Moran, H.; Marin, T.; Arcaro, K.F.; Anderton, D.L.; Etkind, P.; Holland, J.F.; Melana, S.M.; Pogo, B.G. Human mammary tumor virus (HMTV) sequences in human milk. Infect. Agent Cancer 2014, 9, 20. [Google Scholar] [CrossRef][Green Version]
- Johal, H.; Faedo, M.; Faltas, J.; Lau, A.; Mousina, R.; Cozzi, P.; deFazio, A.; Rawlinson, W.D. DNA of mouse mammary tumor virus-like virus is present in human tumors influenced by hormones. J. Med. Virol. 2010, 82, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Johal, H.; Ford, C.; Glenn, W.; Heads, J.; Lawson, J.; Rawlinson, W. Mouse mammary tumor like virus sequences in breast milk from healthy lactating women. Breast Cancer Res. Treat. 2011, 129, 149–155. [Google Scholar] [CrossRef]
- Lawson, J.S.; Glenn, W.K.; Salmons, B.; Ye, Y.; Heng, B.; Moody, P.; Johal, H.; Rawlinson, W.D.; Delprado, W.; Lutze-Mann, L.; et al. Mouse mammary tumor virus-like sequences in human breast cancer. Cancer Res. 2010, 70, 3576–3585. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.E.; Faedo, M.; Rawlinson, W.D. Mouse mammary tumor virus-like RNA transcripts and DNA are found in affected cells of human breast cancer. Clin. Cancer Res. 2004, 10, 7284–7289. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.E.; Tran, D.; Deng, Y.; Ta, V.T.; Rawlinson, W.D.; Lawson, J.S. Mouse mammary tumor virus-like gene sequences in breast tumors of Australian and Vietnamese women. Clin. Cancer Res. 2003, 9, 1118–1120. [Google Scholar] [PubMed]
- Lawson, J.S.; Mazzanti, C.; Civita, P.; Menicagli, M.; Ngan, C.C.; Whitaker, N.J.; Hochman, J.; Braitbard, O.; Yosufi, B.; Glenn, W.K. Association of mouse mammary tumor virus with human breast cancer: Histology, immunohistochemistry and polymerase chain reaction analyses. Front. Oncol. 2018, 8, 141. [Google Scholar] [CrossRef]
- Lawson, J.S.; Tran, D.D.; Carpenter, E.; Ford, C.E.; Rawlinson, W.D.; Whitaker, N.J.; Delprado, W. Presence of mouse mammary tumour-like virus gene sequences may be associated with morphology of specific human breast cancer. J. Clin. Pathol. 2006, 59, 1287–1292. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Y.; Melana, S.M.; Pelisson, I.; Najfeld, V.; Holland, J.F.; Pogo, B.G. Identification of a proviral structure in human breast cancer. Cancer Res. 2001, 61, 1754–1759. [Google Scholar] [PubMed]
- Wang, Y.; Jiang, J.D.; Xu, D.; Li, Y.; Qu, C.; Holland, J.F.; Pogo, B.G. A mouse mammary tumor virus-like long terminal repeat superantigen in human breast cancer. Cancer Res. 2004, 64, 4105–4111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pelisson, I.; Melana, S.M.; Go, V.; Holland, J.F.; Pogo, B.G. MMTV-like env gene sequences in human breast cancer. Arch. Virol. 2001, 146, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pelisson, I.; Melana, S.M.; Holland, J.F.; Pogo, B.G. Detection of MMTV-like LTR and LTR-env gene sequences in human breast cancer. Int. J. Oncol. 2001, 18, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Zammarchi, F.; Pistello, M.; Piersigilli, A.; Murr, R.; Cristofano, C.D.; Naccarato, A.G.; Bevilacqua, G. MMTV-like sequences in human breast cancer: A fluorescent PCR/laser microdissection approach. J. Pathol. 2006, 209, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.L.; Gilady, S.Y.; Mackey, J.R. Mouse mammary tumor virus in human breast cancer red herring or smoking gun? Am. J. Pathol. 2011, 179, 1588–1590. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.H.; Charney, J.; Kramarsky, B.; Lasfargues, E.Y.; Sarkar, N.H.; Brennan, M.J.; Burrows, J.H.; Sirsat, S.M.; Paymaster, J.C.; Vaidya, A.B. Search for a human breast cancer virus. Nature 1971, 229, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Holland, J.F.; Bleiweiss, I.J.; Melana, S.; Liu, X.; Pelisson, I.; Cantarella, A.; Stellrecht, K.; Mani, S.; Pogo, B.G. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995, 55, 5173–5179. [Google Scholar] [PubMed]
- Wang, W.; Indik, S.; Wasilenko, S.T.; Faschinger, A.; Carpenter, E.J.; Tian, Z.; Zhang, Y.; Wong, G.K.; Mason, A.L. Frequent proviral integration of the human betaretrovirus in biliary epithelium of patients with autoimmune and idiopathic liver disease. Aliment. Pharmacol. Ther. 2015, 41, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Sakalian, M.; Shen, Z.; Loss, G.; Neuberger, J.; Mason, A. Cloning the human betaretrovirus proviral genome from patients with primary biliary cirrhosis. Hepatology 2004, 39, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Shen, Z.; Guo, L.; Fodera, B.; Keogh, A.; Joplin, R.; O’Donnell, B.; Aitken, J.; Carman, W.; Neuberger, J.; et al. Does a betaretrovirus infection trigger primary biliary cirrhosis? Proc. Natl. Acad. Sci. USA 2003, 100, 8454–8459. [Google Scholar] [CrossRef]
- Johal, H.; Scott, G.M.; Jones, R.; Camaris, C.; Riordan, S.; Rawlinson, W.D. Mouse mammary tumour virus-like virus (MMTV-LV) is present within the liver in a wide range of hepatic disorders and unrelated to nuclear p53 expression or hepatocarcinogenesis. J. Hepatol. 2009, 50, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Mason, A. Is PBC a viral infectious disease? Best Pract. Res. Clin. Gastroenterol. 2018, 34–35, 27–39. [Google Scholar] [CrossRef]
- Garcia, M.; Wellinger, R.; Vessaz, A.; Diggelmann, H. A new site of integration for mouse mammary tumor virus proviral DNA common to BALB/cf(C3H) mammary and kidney adenocarcinomas. EMBO J. 1986, 5, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Sharon, D.; Chen, M.; Zhang, G.; Girgis, S.; Sis, B.; Graham, D.; McDougall, C.; Wasilenko, S.T.; Montano-Loza, A.; Mason, A.L. Impact of combination antiretroviral therapy in the NOD.c3c4 mouse model of autoimmune biliary disease. Liver Int. 2015, 35, 1442–1450. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Graham, D.; Subsin, B.; McDougall, C.; Gilady, S.; Kneteman, M.; Law, L.; Swain, M.; Trauner, M.; et al. Mouse mammary tumor virus in anti-mitochondrial antibody producing mouse models. J. Hepatol. 2011, 55, 876–884. [Google Scholar] [CrossRef]
- Grandi, N.; Cadeddu, M.; Pisano, M.P.; Esposito, F.; Blomberg, J.; Tramontano, E. Identification of a novel HERV-K(HML10): Comprehensive characterization and comparative analysis in non-human primates provide insights about HML10 proviruses structure and diffusion. Mob. DNA 2017, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.L. The evidence supports a viral aetiology for primary biliary cirrhosis. J. Hepatol. 2011, 54, 1312–1314. [Google Scholar] [CrossRef]
- Selmi, C. The evidence does not support a viral etiology for primary biliary cirrhosis. J. Hepatol. 2011, 54, 1315–1316. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazzanti, C.M.; Lessi, F.; Armogida, I.; Zavaglia, K.; Franceschi, S.; Al Hamad, M.; Roncella, M.; Ghilli, M.; Boldrini, A.; Aretini, P.; et al. Human saliva as route of inter-human infection for mouse mammary tumor virus. Oncotarget 2015, 6, 18355–18363. [Google Scholar] [CrossRef]
- Lessi, F.; Grandi, N.; Mazzanti, C.M.; Civita, P.; Scatena, C.; Aretini, P.; Bandiera, P.; Fornaciari, A.; Giuffra, V.; Fornaciari, G.; et al. A human MMTV-like betaretrovirus linked to breast cancer has been present in humans at least since the copper age. Aging 2020, 12, 15978–15994. [Google Scholar] [CrossRef]
- Ross, S.R.; Schofield, J.J.; Farr, C.J.; Bucan, M. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 2002, 99, 12386–12390. [Google Scholar] [CrossRef]
- Mant, C.; Cason, J. A human murine mammary tumour virus-like agent is an unconvincing aetiological agent for human breast cancer. Rev. Med. Virol. 2004, 14, 169–177. [Google Scholar] [CrossRef]
- Indik, S.; Günzburg, W.H.; Kulich, P.; Salmons, B.; Rouault, F. Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology 2007, 4, 73. [Google Scholar] [CrossRef]
- Indik, S.; Gunzburg, W.H.; Salmons, B.; Rouault, F. Mouse mammary tumor virus infects human cells. Cancer Res. 2005, 65, 6651–6659. [Google Scholar] [CrossRef] [PubMed]
- Faschinger, A.; Rouault, F.; Sollner, J.; Lukas, A.; Salmons, B.; Gunzburg, W.H.; Indik, S. Mouse mammary tumor virus integration site selection in human and mouse genomes. J. Virol. 2008, 82, 1360–1367. [Google Scholar] [CrossRef] [PubMed]
- Carey, E.J.; Ali, A.H.; Lindor, K.D. Primary biliary cirrhosis. Lancet 2015, 386, 1565–1575. [Google Scholar] [CrossRef]
- Mason, A.L.; Xu, L.; Guo, L.; Munoz, S.; Jaspan, J.B.; Bryer-Ash, M.; Cao, Y.; Sander, D.M.; Shoenfeld, Y.; Ahmed, A.; et al. Detection of retroviral antibodies in primary biliary cirrhosis and other idiopathic biliary disorders. Lancet 1998, 351, 1620–1624. [Google Scholar] [CrossRef]
- Xu, L.; Guo, L.; Shen, Z.; Loss, G.; Gish, R.; Wasilenko, S.; Mason, A.L. Duplication of MER115 on chromosome 4 in patients with primary biliary cirrhosis. Liver Int. 2009, 29, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Selmi, C.; Ross, S.R.; Ansari, A.A.; Invernizzi, P.; Podda, M.; Coppel, R.L.; Gershwin, M.E. Lack of immunological or molecular evidence for a role of mouse mammary tumor retrovirus in primary biliary cirrhosis. Gastroenterology 2004, 127, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Voisset, C.; Weiss, R.A.; Griffiths, D.J. Human RNA “Rumor” Viruses: The Search for Novel Human Retroviruses in Chronic Disease. Microbiol. Mol. Biol. Rev. 2008, 72, 157–196. [Google Scholar] [CrossRef]
- Abofayed, H.; Syed, H.; Rahbari, M.; Mason, A. Cellular immune responses to human betaretrovirus in patients with primary biliary cholangitis [ILC abstract]. J. Hepatol. 2021, 75, PO-1822. [Google Scholar]
- Zhang, G.; Bashiri, K.; Kneteman, M.; Cave, K.; Hong, Y.; Mackay, J.; Alter, H.; Mason, A. Seroprevalence of human betaretrovirus surface protein antibodies in patients with breast cancer and liver disease. J. Oncol. 2020, 2020, 8958192. [Google Scholar]
- Lytvyak, E.; Hosamani, I.; Montano-Loza, A.; Saxinger, L.; Mason, A.L. Randomized controlled trial: Combination antiretroviral therapy with Tenofovir-Emtricitabine and Lopinavir-Ritonavir in patients with primary biliary cholangitis. Can. Liver J. 2019, 2, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Lytvyak, E.; Montano-Loza, A.J.; Mason, A.L. Combination antiretroviral studies for patients with primary biliary cirrhosis. World J. Gastroenterol. 2016, 22, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.L.; Lindor, K.D.; Bacon, B.R.; Vincent, C.; Neuberger, J.M.; Wasilenko, S.T. Clinical trial: Randomized controlled study of zidovudine and lamivudine for patients with primary biliary cirrhosis stabilized on ursodiol. Aliment. Pharmacol. Ther. 2008, 28, 886–894. [Google Scholar] [CrossRef]
- Joplin, R.; Gershwin, M.E. Ductular expression of autoantigens in primary biliary cirrhosis. Semin. Liver Dis. 1997, 17, 97–103. [Google Scholar] [CrossRef]
- Sadamoto, T.; Joplin, R.; Keogh, A.; Mason, A.; Carman, W.; Neuberger, J. Expression of pyruvate-dehydrogenase complex PDC-E2 on biliary epithelial cells induced by lymph nodes from primary biliary cirrhosis. Lancet 1998, 352, 1595–1596. [Google Scholar] [CrossRef]
- Konstantoulas, C.J.; Indik, S. C3H strain of mouse mammary tumour virus, like GR strain, infects human mammary epithelial cells, albeit less efficiently than murine mammary epithelial cells. J. Gen. Virol. 2015, 96 Pt 3, 650–662. [Google Scholar] [CrossRef]
- Konstantoulas, C.J.; Lamp, B.; Rumenapf, T.H.; Indik, S. Single amino acid substitution (G42E) in the receptor binding domain of mouse mammary tumour virus envelope protein facilitates infection of non-murine cells in a transferrin receptor 1-independent manner. Retrovirology 2015, 12, 43. [Google Scholar] [CrossRef][Green Version]
- Hackett, A.J.; Smith, H.S.; Springer, E.L.; Owens, R.B.; Nelson-Rees, W.A.; Riggs, J.L.; Gardner, M.B. Two syngeneic cell lines from human breast tissue: The aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J. Natl. Cancer Inst. 1977, 58, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Callahan, R.; Mudunur, U.; Bargo, S.; Raafat, A.; McCurdy, D.; Boulanger, C.; Lowther, W.; Stephens, R.; Luke, B.T.; Stewart, C.; et al. Genes affected by mouse mammary tumor virus (MMTV) proviral insertions in mouse mammary tumors are deregulated or mutated in primary human mammary tumors. Oncotarget 2012, 3, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Joplin, R. Isolation and culture of biliary epithelial cells. Gut 1994, 35, 875–878. [Google Scholar] [CrossRef] [PubMed]
- McDermid, J.; Chen, M.; Li, Y.; Wasilenko, S.; Bintner, J.; McDougall, C.; Pang, X.; Bain, V.G.; Mason, A.L. Reverse transcriptase activity in patients with primary biliary cirrhosis and other autoimmune liver disorders. Aliment. Pharmacol. Ther. 2007, 26, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Kearney, M.F.; Spindler, J.; Wiegand, A.; Shao, W.; Anderson, E.M.; Maldarelli, F.; Ruscetti, F.W.; Mellors, J.W.; Hughes, S.H.; Le Grice, S.F.; et al. Multiple sources of contamination in samples from patients reported to have XMRV infection. PLoS ONE 2012, 7, e30889. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Shan, J.; Kearney, M.F.; Wu, X.; Maldarelli, F.; Mellors, J.W.; Luke, B.; Coffin, J.M.; Hughes, S.H. Retrovirus Integration Database (RID): A public database for retroviral insertion sites into host genomes. Retrovirology 2016, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.J.; Gazin, C.; Lawson, N.D.; Pagès, H.; Lin, S.M.; Lapointe, D.S.; Green, M.R. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 2010, 11, 237. [Google Scholar] [CrossRef] [PubMed]
- Mikkers, H.; Allen, J.; Knipscheer, P.; Romeijn, L.; Hart, A.; Vink, E.; Berns, A. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 2002, 32, 153–159. [Google Scholar] [CrossRef]
- Abbott, K.L.; Nyre, E.T.; Abrahante, J.; Ho, Y.Y.; Isaksson Vogel, R.; Starr, T.K. The Candidate Cancer Gene Database: A database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res. 2015, 43, D844–D848. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Hung, W.S.; Chan, H.H.; Tsai, S.J.; Sun, H.S. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics 2013, 29, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jiménez, F.; Muiños, F.; Sentís, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 2020, 20, 555–572. [Google Scholar] [CrossRef]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Stewart, T.H.; Sage, R.D.; Stewart, A.F.; Cameron, D.W. Breast cancer incidence highest in the range of one species of house mouse, Mus domesticus. Br. J. Cancer 2000, 82, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Hagen, B.; Kraase, M.; Indikova, I.; Indik, S. A high rate of polymerization during synthesis of mouse mammary tumor virus DNA alleviates hypermutation by APOBEC3 proteins. PLoS Pathog. 2019, 15, e1007533. [Google Scholar] [CrossRef] [PubMed]
- Wally, N.; Schneider, M.; Thannesberger, J.; Kastner, M.T.; Bakonyi, T.; Indik, S.; Rattei, T.; Bedarf, J.; Hildebrand, F.; Law, J.; et al. Plasmid DNA contaminant in molecular reagents. Sci. Rep. 2019, 9, 1652. [Google Scholar] [CrossRef] [PubMed]
- De Rijck, J.; de Kogel, C.; Demeulemeester, J.; Vets, S.; El Ashkar, S.; Malani, N.; Bushman, F.D.; Landuyt, B.; Husson, S.J.; Busschots, K.; et al. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013, 5, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.S.; Beitzel, B.F.; Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004, 2, E234. [Google Scholar] [CrossRef]
- Gilroy, K.L.; Terry, A.; Naseer, A.; de Ridder, J.; Allahyar, A.; Wang, W.; Carpenter, E.; Mason, A.; Wong, G.K.; Cameron, E.R.; et al. Gamma-Retrovirus Integration Marks Cell Type-Specific Cancer Genes: A Novel Profiling Tool in Cancer Genomics. PLoS ONE 2016, 11, e0154070. [Google Scholar] [CrossRef] [PubMed]
- Shun, M.C.; Raghavendra, N.K.; Vandegraaff, N.; Daigle, J.E.; Hughes, S.; Kellam, P.; Cherepanov, P.; Engelman, A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007, 21, 1767–1778. [Google Scholar] [CrossRef]
- Hill, V.K.; Ricketts, C.; Bieche, I.; Vacher, S.; Gentle, D.; Lewis, C.; Maher, E.R.; Latif, F. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res. 2011, 71, 2988–2999. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Hussain, T.; Manzoor, S.; Saalim, M.; Khaliq, S. PTEN: A potential prognostic marker in virus-induced hepatocellular carcinoma. Tumour Biol. 2017, 39, 1010428317705754. [Google Scholar] [CrossRef] [PubMed]
- Wendt, C.; Margolin, S. Identifying breast cancer susceptibility genes—A review of the genetic background in familial breast cancer. Acta Oncol. 2019, 58, 135–146. [Google Scholar] [CrossRef]
- Wang, X.; Huang, H.; Young, K.H. The PTEN tumor suppressor gene and its role in lymphoma pathogenesis. Aging 2015, 7, 1032–1049. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Xie, W.; Wang, S.; Li, Q.; Wei, X.; Chen, B.; Hua, Y.; Li, S.; Peng, B.; Shen, S. High SPINK1 Expression Predicts Poor Prognosis and Promotes Cell Proliferation and Metastasis of Hepatocellular Carcinoma. J. Investig. Surg. 2021, 34, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Jin, X.; Liu, Y.; Wang, S.; Bian, F.; Zhao, Q.; Shi, H.; Gao, Z. Long noncoding RNA ZFPM2-AS1 promotes the proliferation, migration, and invasion of hepatocellular carcinoma cells by regulating the miR-576-3p/HIF-1α axis. Anticancer Drugs 2021, 32, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.F.; Li, L.; Li, H.J.; Yang, F.R.; Liu, Z.K.; Hu, X.W.; Wang, Q. LncRNA ZFPM2-AS1 aggravates the malignant development of breast cancer via upregulating JMJD6. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11139–11147. [Google Scholar] [PubMed]
- Sun, X.; Zhao, S.; Li, H.; Chang, H.; Huang, Z.; Ding, Z.; Dong, L.; Chen, J.; Zang, Y.; Zhang, J. MicroRNA-30b Suppresses Epithelial-Mesenchymal Transition and Metastasis of Hepatoma Cells. J. Cell Physiol. 2017, 232, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.H.; Bachelier, R.; Allioli, N.; Hong, S.S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis-Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef]
- Uren, A.G.; Kool, J.; Berns, A.; van Lohuizen, M. Retroviral insertional mutagenesis: Past, present and future. Oncogene 2005, 24, 7656–7672. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y.; Wang, B.; Lan, H.; Liu, Y.; Chen, F.; Zhang, J.; Luo, J. Sumoylation in p27kip1 via RanBP2 promotes cancer cell growth in cholangiocarcinoma cell line QBC939. BMC Mol. Biol. 2017, 18, 23. [Google Scholar] [CrossRef]
- Blondel-Tepaz, E.; Leverve, M.; Sokrat, B.; Paradis, J.S.; Kosic, M.; Saha, K.; Auffray, C.; Lima-Fernandes, E.; Zamborlini, A.; Poupon, A.; et al. The RanBP2/RanGAP1-SUMO complex gates β-arrestin2 nuclear entry to regulate the Mdm2-p53 signaling axis. Oncogene 2021, 40, 2243–2257. [Google Scholar] [CrossRef]
- Xu, C.; Qi, X.; Du, X.; Zou, H.; Gao, F.; Feng, T.; Lu, H.; Li, S.; An, X.; Zhang, L.; et al. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 722–727. [Google Scholar] [CrossRef]
- Suo, H.D.; Tao, Z.; Zhang, L.; Jin, Z.N.; Li, X.Y.; Ma, W.; Wang, Z.; Qiu, Y.; Jin, F.; Chen, B.; et al. Coexpression Network Analysis of Genes Related to the Characteristics of Tumor Stemness in Triple-Negative Breast Cancer. Biomed Res. Int. 2020, 2020, 7575862. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.; Bonuccelli, G.; Ozsvari, B.; Peiris-Pages, M.; Fiorillo, M.; Smith, D.L.; Bevilacqua, G.; Mazzanti, C.M.; McDonnell, L.A.; Naccarato, A.G.; et al. Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: Understanding WNT/FGF-driven anabolic signaling. Oncotarget 2015, 6, 30453–30471. [Google Scholar] [CrossRef] [PubMed]
- Callahan, R.; Smith, G.H. MMTV-induced mammary tumorigenesis: Gene discovery, progression to malignancy and cellular pathways. Oncogene 2000, 19, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Shackel, N.A.; McGuinness, P.H.; Abbott, C.A.; Gorrell, M.D.; McCaughan, G.W. Identification of novel molecules and pathogenic pathways in primary biliary cirrhosis: cDNA array analysis of intrahepatic differential gene expression. Gut 2001, 49, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Yang, Z.; Zhong, R. Primary biliary cirrhosis and cancer risk: A systematic review and meta-analysis. Hepatology 2012, 56, 1409–1417. [Google Scholar] [CrossRef]
- Trivedi, P.J.; Lammers, W.J.; van Buuren, H.R.; Pares, A.; Floreani, A.; Janssen, H.L.; Invernizzi, P.; Battezzati, P.M.; Ponsioen, C.Y.; Corpechot, C.; et al. Stratification of hepatocellular carcinoma risk in primary biliary cirrhosis: A multicentre international study. Gut 2016, 65, 321–329. [Google Scholar] [CrossRef]
- Wang, S.; Dong, V.; Montano-Loza, A.J.; Mason, A.L. Autoimmune liver diseases, hepatocellular carcinoma, and recurrence of autoimmunity post-liver transplantation. Hepatoma Res. 2021, 7, 42. [Google Scholar] [CrossRef]
- Mowat, M.; Cheng, A.; Kimura, N.; Bernstein, A.; Benchimol, S. Rearrangements of the cellular p53 gene in erythroleukaemic cells transformed by Friend virus. Nature 1985, 314, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Ranzani, M.; Annunziato, S.; Adams, D.J.; Montini, E. Cancer gene discovery: Exploiting insertional mutagenesis. Mol. Cancer Res. 2013, 11, 1141–1158. [Google Scholar] [CrossRef]
- Caparica, R.; Richard, F.; Brandão, M.; Awada, A.; Sotiriou, C.; de Azambuja, E. Prognostic and Predictive Impact of Beta-2 Adrenergic Receptor Expression in HER2-Positive Breast Cancer. Clin. Breast Cancer 2020, 20, 262–273.e7. [Google Scholar] [CrossRef] [PubMed]
- Sigin, V.O.; Kalinkin, A.I.; Kuznetsova, E.B.; Simonova, O.A.; Chesnokova, G.G.; Litviakov, N.V.; Slonimskaya, E.M.; Tsyganov, M.M.; Ibragimova, M.K.; Volodin, I.V.; et al. DNA methylation markers panel can improve prediction of response to neoadjuvant chemotherapy in luminal B breast cancer. Sci. Rep. 2020, 10, 9239. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, A.; Rahman, M.T.; Upadhyay, K.; Loke, J.; Ostrer, H. Ectopic Otoconin 90 expression in triple negative breast cancer cell lines is associated with metastasis functions. PLoS ONE 2019, 14, e0211737. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.K.; Zhang, C.; Sanda, T. Oncogenic transcriptional program driven by TAL1 in T-cell acute lymphoblastic leukemia. Int J. Hematol. 2019, 109, 5–17. [Google Scholar] [CrossRef]
- Yu, Q.; Cao, S.; Tang, H.; Li, J.; Guo, W.; Zhang, S. Clinical significance of aberrant DEUP1 promoter methylation in hepatocellular carcinoma. Oncol. Lett. 2019, 18, 1356–1364. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, S.; Li, Y.; Yao, J.; Ruan, J.; Zheng, Y.; Deng, Y.; Li, N.; Wei, B.; Wu, Y.; et al. Prediction of Overall Survival Among female patients with breast cancer using a prognostic signature based on 8 DNA repair-related genes. JAMA Netw. Open 2020, 3, e2014622. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | Integration Orientation | Candidate Cancer Gene Database | Tumor Associated Gene | Integrative Oncogenomics | COSMIC Cancer Gene Census | Relevant Cancer Type |
|---|---|---|---|---|---|---|
| PTEN * | (+) | Y ** | Y | Y | Y-Tier 1/Hallmark | Liver [71], Breast [72], Lymphoma [73] |
| RANBP2 * | (+) | NL | Y | Y-Tier 1/Hallmark | Liver [80], Breast [81] | |
| PRAMEF8 | (+) | Y | ||||
| MB21D2 * | (+) | Y | Y | Y-Tier 2 | Liver [82] | |
| CLEC2L | (+) | Y | ||||
| KLRG2 | (+) | Y | ||||
| ZFAT | (+) | Y | ||||
| MAGEB5 | (+) | Y | ||||
| HEATR1 | (+)/(-) | Y | ||||
| ORC1 * | (+)/(-) | Y | Breast [83] | |||
| FARP1 | (+)/(-) | Y | ||||
| STK24 | (+)/(-) | Y | ||||
| BRMS1L | (+)/(-) | Y | ||||
| DCAKD * | (+)/(-) | Y | ||||
| SSB * | (+)/(-) | Y | ||||
| SPOCK3 * | (+)/(-) | Y | ||||
| FAM73B | (+)/(-) | Y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goubran, M.; Wang, W.; Indik, S.; Faschinger, A.; Wasilenko, S.T.; Bintner, J.; Carpenter, E.J.; Zhang, G.; Nuin, P.; Macintyre, G.; et al. Isolation of a Human Betaretrovirus from Patients with Primary Biliary Cholangitis. Viruses 2022, 14, 886. https://doi.org/10.3390/v14050886
Goubran M, Wang W, Indik S, Faschinger A, Wasilenko ST, Bintner J, Carpenter EJ, Zhang G, Nuin P, Macintyre G, et al. Isolation of a Human Betaretrovirus from Patients with Primary Biliary Cholangitis. Viruses. 2022; 14(5):886. https://doi.org/10.3390/v14050886
Chicago/Turabian StyleGoubran, Mariam, Weiwei Wang, Stanislav Indik, Alexander Faschinger, Shawn T. Wasilenko, Jasper Bintner, Eric J. Carpenter, Guangzhi Zhang, Paulo Nuin, Georgina Macintyre, and et al. 2022. "Isolation of a Human Betaretrovirus from Patients with Primary Biliary Cholangitis" Viruses 14, no. 5: 886. https://doi.org/10.3390/v14050886
APA StyleGoubran, M., Wang, W., Indik, S., Faschinger, A., Wasilenko, S. T., Bintner, J., Carpenter, E. J., Zhang, G., Nuin, P., Macintyre, G., Wong, G. K.-S., & Mason, A. L. (2022). Isolation of a Human Betaretrovirus from Patients with Primary Biliary Cholangitis. Viruses, 14(5), 886. https://doi.org/10.3390/v14050886





