Porcine Intestinal Organoids: Overview of the State of the Art
Abstract
:1. Introduction
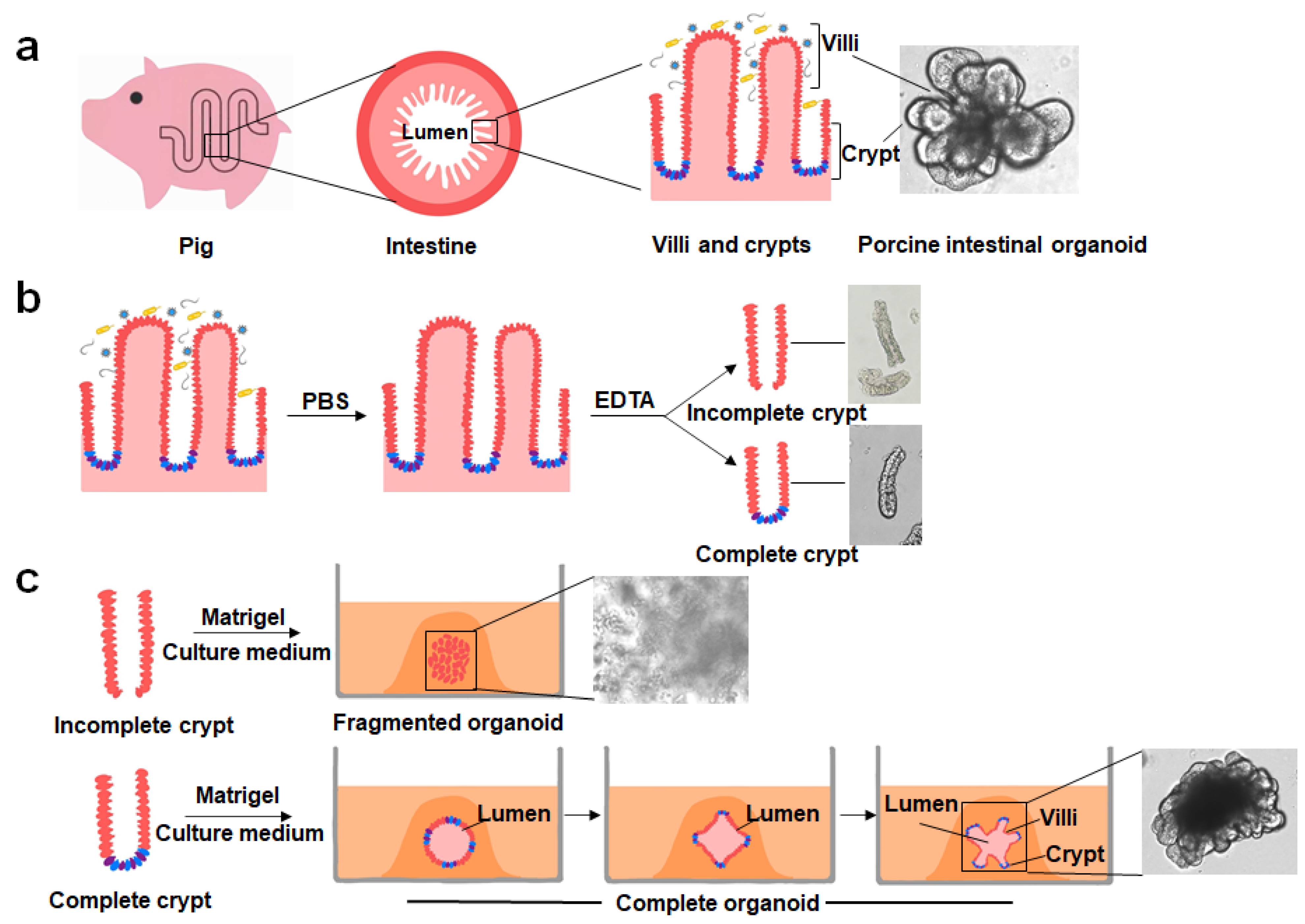
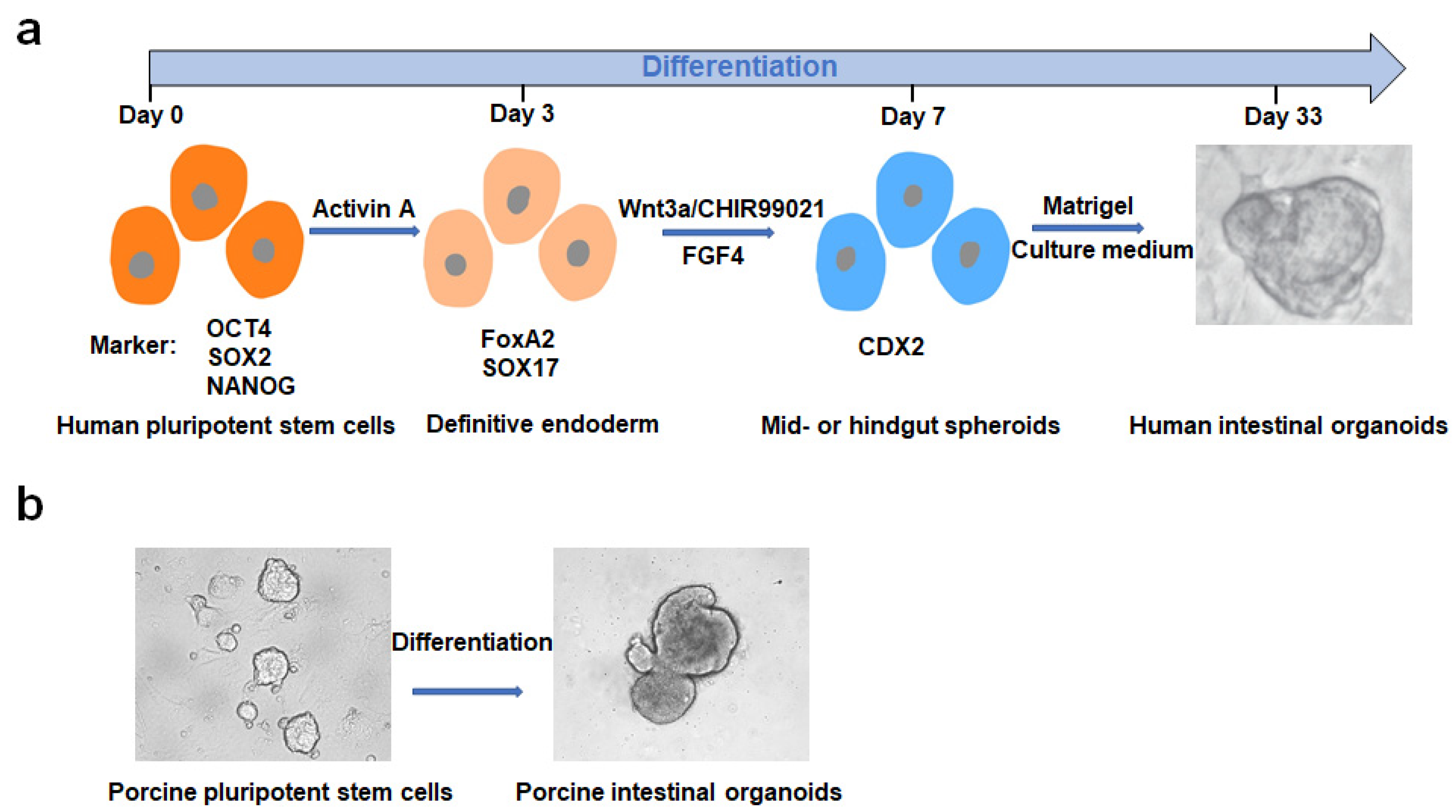
2. Culturing, Establishment and Development of PIOs
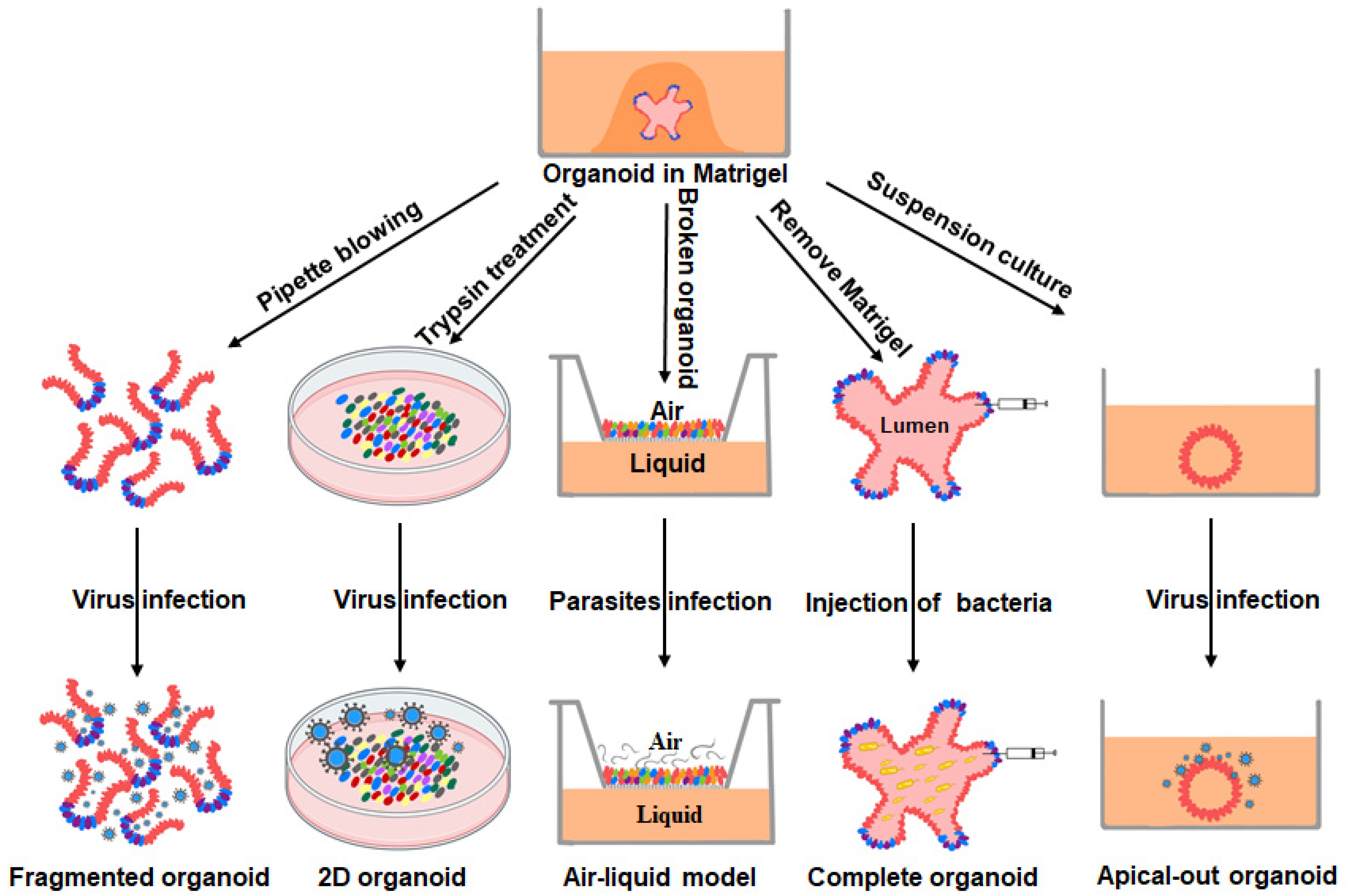
3. PIOs as Models for the Study of Intestinal Pathogen–Host Interactions
3.1. Host–Viral Interactions
3.2. Host–Bacterial Interactions
3.3. Host–Parasitic Interactions
4. Other Applications of PIOs
4.1. The Study of Intestinal Nutritional Development
4.2. Drug Discovery
4.3. Gene Editing
5. Concluding Remarks and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Foulke-Abel, J.; In, J.; Yin, J.Y.; Zachos, N.C.; Kovbasnjuk, O.; Estes, M.K.; de Jonge, H.; Donowitz, M. Human Enteroids as a Model of Upper Small Intestinal Ion Transport Physiology and Pathophysiology. Gastroenterology 2016, 150, 638–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.W.; Qiao, S.Y.; Li, D.F. Amino acids and gut function. Amino Acids 2009, 37, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Hussain, M.M. Intestinal lipid absorption. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1183–E1194. [Google Scholar] [CrossRef] [Green Version]
- Helander, H.F.; Fandriks, L. Surface area of the digestive tract-revisited. Scand. J. Gastroenterol. 2014, 49, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Rizk, P.; Barker, N. Gut stem cells in tissue renewal and disease: Methods, markers, and myths. WIREs Syst. Biol. Med. 2012, 4, 475–496. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Prabhu, P.N.; Benefiel, A.C.; Miller, M.J.; Chow, J.; Davis, S.R.; Gaskins, H.R. Galacto-oligosaccharides may directly enhance intestinal barrier function through the modulation of goblet cells. Mol. Nutr. Food Res. 2015, 59, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Holly, M.K.; Smith, J.G. Paneth Cells during Viral Infection and Pathogenesis. Viruses 2018, 10, 225. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Millar, S.E. Wnt/beta-catenin Signaling in Oral Tissue Development and Disease. J. Dent. Res. 2010, 89, 318–330. [Google Scholar] [CrossRef]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [Green Version]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The pig: A model for human infectious diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Miyazaki, A.; Hu, H.; Saif, L.J. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet. Microbiol. 2018, 221, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Fang, L.R.; Liu, S.D.; Ke, W.T.; Wang, D.; Peng, G.Q.; Xiao, I.B. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses. Vet. Microbiol. 2019, 233, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.L.; Zhang, H.; Ding, Z.; Luo, R.; An, K.; Liu, L.Z.; Bi, J.; Chen, H.C.; Xiao, S.B.; Fang, L.R. Proteome analysis of porcine epidemic diarrhea virus (PEDV)-infected Vero cells. Proteomics 2015, 15, 1819–1828. [Google Scholar] [CrossRef]
- Kaur, G.; Dufour, J.M. Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Duell, B.L.; Cripps, A.W.; Schembri, M.A.; Ulett, G.C. Epithelial cell coculture models for studying infectious diseases: Benefits and limitations. J. Biomed. Biotechnol. 2011, 2011, 852419. [Google Scholar] [CrossRef] [Green Version]
- Desmyter, J.; Melnick, J.L.; Rawls, W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 1968, 2, 955–961. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Ke, H.; Blikslager, A.; Fujita, T.; Yoo, D. Type III Interferon Restriction by Porcine Epidemic Diarrhea Virus and the Role of Viral Protein nsp1 in IRF1 Signaling. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- van der Hee, B.; Madsen, O.; Vervoort, J.; Smidt, H.; Wells, J.M. Congruence of Transcription Programs in Adult Stem Cell-Derived Jejunum Organoids and Original Tissue During Long-Term Culture. Front. Cell Dev. Biol. 2020, 8, 375. [Google Scholar] [CrossRef]
- Derricott, H.; Luu, L.; Fong, W.Y.; Hartley, C.S.; Johnston, L.J.; Armstrong, S.D.; Randle, N.; Duckworth, C.A.; Campbell, B.J.; Wastling, J.M.; et al. Developing a 3D intestinal epithelium model for livestock species. Cell Tissue Res. 2019, 375, 409–424. [Google Scholar] [CrossRef] [Green Version]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [Green Version]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; van Es, J.H.; van den Brink, S.; van Houdt, W.J.; Pronk, A.; van Gorp, J.; Siersema, P.D.; et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Chandra, L.; Borcherding, D.C.; Kingsbury, D.; Atherly, T.; Ambrosini, Y.M.; Bourgois-Mochel, A.; Yuan, W.; Kimber, M.; Qi, Y.; Wang, Q.; et al. Derivation of adult canine intestinal organoids for translational research in gastroenterology. BMC Biol. 2019, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.M.; Williamson, I.; Piedrahita, J.A.; Blikslager, A.T.; Magness, S.T. Cell Lineage Identification and Stem Cell Culture in a Porcine Model for the Study of Intestinal Epithelial Regeneration. PLoS ONE 2013, 8, e66465. [Google Scholar] [CrossRef] [Green Version]
- Kramer, N.; Pratscher, B.; Meneses, A.M.C.; Tschulenk, W.; Walter, I.; Swoboda, A.; Kruitwagen, H.S.; Schneeberger, K.; Penning, L.C.; Spee, B.; et al. Generation of Differentiating and Long-Living Intestinal Organoids Reflecting the Cellular Diversity of Canine Intestine. Cells 2020, 9, 822. [Google Scholar] [CrossRef] [Green Version]
- Powell, R.H.; Behnke, M.S. WRN conditioned media is sufficient for in vitro propagation of intestinal organoids from large farm and small companion animals. Biol. Open 2017, 6, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, C.; Liu, X.J.; Chiu, M.C.; Zhao, X.Y.; Wang, D.; Wei, Y.X.; Lee, A.R.; Zhang, A.J.; Chu, H.; et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef]
- Mori-Akiyama, Y.; Van den Born, M.; Van Es, J.H.; Hamilton, S.R.; Adams, H.P.; Zhang, J.X.; Clevers, H.; De Crombrugghe, B. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 2007, 133, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Milano, J.; McKay, J.; Dagenais, C.; Foster-Brown, L.; Pognan, F.; Gadient, R.; Jacobs, R.T.; Zacco, A.; Greenberg, B.; Ciaccio, P.J. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 2004, 82, 341–358. [Google Scholar] [CrossRef] [PubMed]
- He, X.C.; Zhang, J.W.; Tong, W.G.; Tawfik, O.; Ross, J.; Scoville, D.H.; Tian, Q.; Zeng, X.; He, X.; Wiedemann, L.M.; et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004, 36, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, H.; Irshad, S.; Bansal, M.; Rafferty, H.; Boitsova, T.; Bardella, C.; Jaeger, E.; Lewis, A.; Freeman-Mills, L.; Giner, F.C.; et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med. 2015, 21, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Bommer, G.T.; Zhao, J.; Green, M.; Sands, E.; Zhai, Y.; Brown, K.; Burberry, A.; Cho, K.R.; Fearon, E.R. Mutant Kras Promotes Hyperplasia and Alters Differentiation in the Colon Epithelium but Does Not Expand the Presumptive Stem Cell Pool. Gastroenterology 2011, 141, 1003–1013. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.B.; Liu, P.Y.; Shi, Y.H.; Li, P. Single-Cell Sequencing and Organoids: A Powerful Combination for Modelling Organ Development and Diseases. Rev. Physiol. Biochem. Pharmacol. 2021, 179, 189–210. [Google Scholar] [CrossRef]
- Sato, T.; Clevers, H. Growing Self-Organizing Mini-Guts from a Single Intestinal Stem Cell: Mechanism and Applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Fu, F.; Guo, S.S.; Wang, H.F.; He, X.J.; Xue, M.; Yin, L.D.; Feng, L.; Liu, P.H. Porcine Intestinal Enteroids: A New Model for Studying Enteric Coronavirus Porcine Epidemic Diarrhea Virus Infection and the Host Innate Response. J. Virol. 2019, 93, e01682-18. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Zheng, J.Y.; Chen, Y.L.; Wang, T.J.; Zhang, Z.N.; Shan, Y.; Xu, J.D.; Yue, M.; Fang, W.H.; Li, X.L. Utility Evaluation of Porcine Enteroids as PDCoV Infection Model in vitro. Front. Microbiol 2020, 11, 821. [Google Scholar] [CrossRef]
- Khalil, H.A.; Lei, N.Y.; Brinkley, G.; Scott, A.; Wang, J.F.; Kar, U.K.; Jabaji, Z.B.; Lewis, M.; Martin, M.G.; Dunn, J.C.Y.; et al. A novel culture system for adult porcine intestinal crypts. Cell Tissue Res. 2016, 365, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, Y.; Zhang, M.; Wang, H.; Cui, A.; Zhao, J.; Ji, W.; Chen, Y.G. Establishment of porcine and monkey colonic organoids for drug toxicity study. Cell Regen. 2021, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.D.; Chen, J.F.; Li, L.; Guo, S.S.; Xue, M.; Zhang, J.L.; Liu, X.; Feng, L.; Liu, P.H. Aminopeptidase N Expression, Not Interferon Responses, Determines the Intestinal Segmental Tropism of Porcine Deltacoronavirus. J. Virol. 2020, 94, e00480-20. [Google Scholar] [CrossRef]
- Yin, L.D.; Liu, X.; Hu, D.M.; Luo, Y.; Zhang, G.Z.; Liu, P.H. Swine Enteric Coronaviruses (PEDV, TGEV, and PDCoV) Induce Divergent Interferon-Stimulated Gene Responses and Antigen Presentation in Porcine Intestinal Enteroids. Front. Immunol. 2022, 12, 5946. [Google Scholar] [CrossRef] [PubMed]
- Vermeire, B.; Gonzalez, L.M.; Jansens, R.J.J.; Cox, E.; Devriendt, B. Porcine small intestinal organoids as a model to explore ETEC-host interactions in the gut. Vet. Res. 2021, 52, 107. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human Intestinal Enteroids: A New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.B.; Bijvelds, M.; Dang, W.; Xu, L.; van der Eijk, A.A.; Knipping, K.; Tuysuz, N.; Dekkers, J.F.; Wang, Y.J.; de Jonge, J.; et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antivir. Res. 2015, 123, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.L.; Duan, X.H.; Yang, L.L.; Nilsson-Payant, B.E.; Wang, P.F.; Duan, F.Y.; Tang, X.M.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [Google Scholar] [CrossRef]
- Xue, B.H.; Li, Y.; He, Y.L.; Wei, R.Y.; Sun, R.Z.; Yin, Z.; Bou, G.; Liu, Z.H. Porcine Pluripotent Stem Cells Derived from IVF Embryos Contribute to Chimeric Development In Vivo. PLoS ONE 2016, 11, e0151737. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.Y.; Yu, T.; Cai, Y.X.; Wang, H.Y. Preserving self-renewal of porcine pluripotent stem cells in serum-free 3i culture condition and independent of LIF and b-FGF cytokines. Cell Death Discov. 2018, 4, 21. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.F.; Nowak-Imialek, M.; Chen, X.; Chen, D.S.; Herrmann, D.; Ruan, D.G.; Chen, A.C.H.; Eckersley-Maslin, M.A.; Ahmad, S.; Lee, Y.L.; et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 2019, 21, 687–699. [Google Scholar] [CrossRef] [Green Version]
- Wilke, G.; Funkhouser-Jones, L.J.; Wang, Y.; Ravindran, S.; Wang, Q.L.; Beatty, W.L.; Baldridge, M.T.; VanDussen, K.L.; Shen, B.; Kuhlenschmidt, M.S.; et al. A Stem-Cell-Derived Platform Enables Complete Cryptosporidium Development In Vitro and Genetic Tractability. Cell Host Microbe 2019, 26, 123–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, S.S.; Tocchi, A.; Holly, M.K.; Parks, W.C.; Smith, J.G. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 2015, 8, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, N.; Chen, J.N.; Huang, X.; Zhang, N.; Yang, S.S.; Liu, G.; Liu, G.L. Next-Generation Porcine Intestinal Organoids: An Apical-Out Organoid Model for Swine Enteric Virus Infection and Immune Response Investigations. J. Virol. 2021, 95, e01006-20. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, H.Y. Porcine enteric coronaviruses: An updated overview of the pathogenesis, prevalence, and diagnosis. Vet. Res. Commun. 2021, 45, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Turlewicz-Podbielska, H.; Pomorska-Mol, M. Porcine Coronaviruses: Overview of the State of the Art. Virol. Sin. 2021, 36, 833–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Li, W.T.; Hulswit, R.J.G.; Kenney, S.P.; Widjaja, I.; Jung, K.; Alhamo, M.A.; van Dieren, B.; van Kuppeveld, F.J.M.; Saif, L.J.; Bosch, B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. USA 2018, 115, E5135–E5143. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Liu, Y.; Ji, C.M.; Yang, Y.L.; Liang, Q.Z.; Zhao, P.; Xu, L.D.; Lei, X.M.; Luo, W.T.; Qin, P.; et al. Porcine Deltacoronavirus Engages the Transmissible Gastroenteritis Virus Functional Receptor Porcine Aminopeptidase N for Infectious Cellular Entry. J. Virol. 2018, 92, e00318-18. [Google Scholar] [CrossRef] [Green Version]
- Wilson, S.S.; Bromme, B.A.; Holly, M.K.; Wiens, M.E.; Gounder, A.P.; Sul, Y.; Smith, J.G. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog. 2017, 13, e1006446. [Google Scholar] [CrossRef]
- Doyle, M.P.; Erickson, M.C. Reducing the carriage of foodborne pathogens in livestock and poultry. Poult. Sci. 2006, 85, 960–973. [Google Scholar] [CrossRef]
- Melkebeek, V.; Goddeeris, B.M.; Cox, E. ETEC vaccination in pigs. Vet. Immunol. Immunopathol. 2013, 152, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.Y.; Huang, D.G.; Gao, C.Q.; Yan, H.C.; Zou, S.G.; Wang, X.Q. Heat-stable enterotoxin inhibits intestinal stem cell expansion to disrupt the intestinal integrity by downregulating the Wnt/beta-catenin pathway. Stem Cells 2021, 39, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Li, X.G.; Zhu, M.; Chen, M.X.; Fan, H.B.; Fu, H.L.; Zhou, J.Y.; Zhai, Z.Y.; Gao, C.Q.; Yan, H.C.; Wang, X.Q. Acute exposure to deoxynivalenol inhibits porcine enteroid activity via suppression of the Wnt/beta-catenin pathway. Toxicol. Lett. 2019, 305, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, P.; Schnepel, N.; Langeheine, M.; Kunnemann, K.; Grassl, G.A.; Brehm, R.; Seeger, B.; Mazzuoli-Weber, G.; Breves, G. Intestinal organoid-based 2D monolayers mimic physiological and pathophysiological properties of the pig intestine. PLoS ONE 2021, 16, e0256143. [Google Scholar] [CrossRef]
- Leatham, M.P.; Banerjee, S.; Autieri, S.M.; Mercado-Lubo, R.; Conway, T.; Cohen, P.S. Precolonized Human Commensal Escherichia coli Strains Serve as a Barrier to E. coli O157:H7 Growth in the Streptomycin-Treated Mouse Intestine. Infect. Immun. 2009, 77, 2876–2886. [Google Scholar] [CrossRef] [Green Version]
- Klotz, C.; Aebischer, T.; Seeber, F. Stem cell-derived cell cultures and organoids for protozoan parasite propagation and studying host-parasite interaction. Int. J. Med. Microbiol. 2012, 302, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Betancourt, E.D.; Hamid, B.; Fabian, B.T.; Klotz, C.; Hartmann, S.; Seeber, F. From Entry to Early Dissemination-Toxoplasma gondii’s Initial Encounter With Its Host. Front. Cell. Infect. Microbiol. 2019, 9, 46. [Google Scholar] [CrossRef]
- Clough, B.; Frickel, E.M. The Toxoplasma Parasitophorous Vacuole: An Evolving Host-Parasite Frontier. Trends Parasitol. 2017, 33, 473–488. [Google Scholar] [CrossRef]
- Dubey, J.P.; Jones, J.L. Toxoplasma gondii infection in humans and animals in the United States. Int. J. Parasitol. 2008, 38, 1257–1278. [Google Scholar] [CrossRef]
- Torgerson, P.R.; Mastroiacovo, P. The global burden of congenital toxoplasmosis: A systematic review. Bull. World Health Organ. 2013, 91, 501–508. [Google Scholar] [CrossRef]
- Wallander, C.; Frossling, J.; Dorea, F.C.; Uggla, A.; Vagsholm, I.; Lunden, A. Pasture is a risk factor for Toxoplasma gondii infection in fattening pigs. Vet. Parasitol. 2016, 224, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Dutta, D.; Schaefer, D.A.; Iakobachvili, N.; Artegiani, B.; Sachs, N.; Boonekamp, K.E.; Bowden, G.; Hendrickx, A.P.A.; Willems, R.J.L.; et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018, 3, 814–823. [Google Scholar] [CrossRef]
- Luu, L.; Johnston, L.J.; Derricott, H.; Armstrong, S.D.; Randle, N.; Hartley, C.S.; Duckworth, C.A.; Campbell, B.J.; Wastling, J.M.; Coombes, J.L. An Open-Format Enteroid Culture System for Interrogation of Interactions Between Toxoplasma gondii and the Intestinal Epithelium. Front. Cell. Infect. Microbiol. 2019, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Holthaus, D.; Kraft, M.R.; Krug, S.M.; Wolf, S.; Muller, A.; Delgado Betancourt, E.; Schorr, M.; Holland, G.; Knauf, F.; Schulzke, J.D.; et al. Dissection of Barrier Dysfunction in Organoid-Derived Human Intestinal Epithelia Induced by Giardia duodenalis. Gastroenterology 2022, 162, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Duque-Correa, M.A.; Schreiber, F.; Rodgers, F.H.; Goulding, D.; Forrest, S.; White, R.; Buck, A.; Grencis, R.K.; Berriman, M. Development of caecaloids to study host-pathogen interactions: New insights into immunoregulatory functions of Trichuris muris extracellular vesicles in the caecum. Int. J. Parasitol. 2020, 50, 707–718. [Google Scholar] [CrossRef]
- Gonzalez, L.M.; Moeser, A.J.; Blikslager, A.T. Porcine models of digestive disease: The future of large animal translational research. Transl. Res. 2015, 166, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Tsai, Y.H.; Tseng, S.H. Selenite Stimulates the Proliferation of Intestinal Stem Cells With Elevated Antioxidative Activity. Transplant. Proc. 2016, 48, 507–511. [Google Scholar] [CrossRef]
- Chen, Y.; Tseng, S.H.; Yao, C.L.; Li, C.; Tsai, Y.H. Distinct Effects of Growth Hormone and Glutamine on Activation of Intestinal Stem Cells. J. Parenter. Enter. Nutr. 2018, 42, 642–651. [Google Scholar] [CrossRef]
- Odle, J.; Lin, X.; Jacobi, S.K.; Kim, S.W.; Stahl, C.H. The Suckling Piglet as an Agrimedical Model for the Study of Pediatric Nutrition and Metabolism. Annu. Rev. Anim. Biosci. 2014, 2, 419–444. [Google Scholar] [CrossRef]
- Che, L.Q.; Xuan, Y.; Hu, L.; Liu, Y.; Xu, Q.; Fang, Z.F.; Lin, Y.; Xu, S.Y.; Wu, D.; Zhang, K.Y.; et al. Effect of Postnatal Nutrition Restriction on the Oxidative Status of Neonates with Intrauterine Growth Restriction in a Pig Model. Neonatology 2015, 107, 93–99. [Google Scholar] [CrossRef]
- Heinritz, S.N.; Weiss, E.; Eklund, M.; Aumiller, T.; Louis, S.; Rings, A.; Messner, S.; Camarinha-Silva, A.; Seifert, J.; Bischoff, S.C.; et al. Intestinal Microbiota and Microbial Metabolites Are Changed in a Pig Model Fed a High-Fat/Low-Fiber or a Low-Fat/High-Fiber Diet. PLoS ONE 2016, 11, e0154329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.S.; Xiong, X.; Wang, X.C.; Li, T.J.; Yin, Y.L. Effects of weaning on intestinal crypt epithelial cells in piglets. Sci. Rep. 2016, 6, 36939. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.B.; Li, J.; Wang, Y.; Wang, L.; Yin, Y.B.; Yin, L.M.; Yang, H.S.; Yin, Y.L. Dietary vitamin A affects growth performance, intestinal development, and functions in weaned piglets by affecting intestinal stem cells. J. Anim. Sci. 2020, 98, skaa020. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Qin, Y.C.; Gao, C.Q.; Yan, H.C.; Wang, X.Q. l-Glutamate drives porcine intestinal epithelial renewal by increasing stem cell activity via upregulation of the EGFR-ERK-mTORC1 pathway. Food Funct. 2020, 11, 2714–2724. [Google Scholar] [CrossRef]
- Miyoshi, H.; Stappenbeck, T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 2013, 8, 2471–2482. [Google Scholar] [CrossRef] [PubMed]
- de Jeude, J.F.V.; Vermeulen, J.L.M.; Montenegro-Miranda, P.S.; Van den Brink, G.R.; Heijmans, J. A Protocol for Lentiviral Transduction and Downstream Analysis of Intestinal Organoids. J. Vis. Exp. 2015, 98, 52531. [Google Scholar] [CrossRef] [Green Version]
- Driehuis, E.; Clevers, H. CRISPR/Cas 9 genome editing and its applications in organoids. Am. J. Physiol. Liver Physiol. 2017, 312, G257–G265. [Google Scholar] [CrossRef]
- Morizane, R.; Lam, A.Q.; Freedman, B.S.; Kishi, S.; Valerius, M.T.; Bonventre, J.V. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 2015, 33, 1193–1200. [Google Scholar] [CrossRef] [Green Version]
- Devarasetty, M.; Forsythe, S.; Shupe, T.D.; Soker, S.; Bishop, C.E.; Atala, A.; Skardal, A. Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids. Biosensors 2017, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Sgodda, M.; Dai, Z.; Zweigerdt, R.; Sharma, A.D.; Ott, M.; Cantz, T. A Scalable Approach for the Generation of Human Pluripotent Stem Cell-Derived Hepatic Organoids with Sensitive Hepatotoxicity Features. Stem Cells Dev. 2017, 26, 1490–1504. [Google Scholar] [CrossRef]
- Muller, T.; Hess, M.W.; Schiefermeier, N.; Pfaller, K.; Ebner, H.L.; Heinz-Erian, P.; Ponstingl, H.; Partsch, J.; Rollinghoff, B.; Kohler, H.; et al. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat. Genet. 2008, 40, 1163–1165. [Google Scholar] [CrossRef] [PubMed]
- Ruemmele, F.M.; Muller, T.; Schiefermeier, N.; Ebner, H.L.; Lechner, S.; Pfaller, K.; Thoni, C.E.; Goulet, O.; Lacaille, F.; Schmitz, J.; et al. Loss-of-Function of MYO5B is the Main Cause of Microvillus Inclusion Disease: 15 Novel Mutations and a CaCo-2 RNAi Cell Model. Hum. Mutat. 2010, 31, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Erickson, R.P.; Larson-Thome, K.; Valenzuela, R.K.; Whitaker, S.E.; Shub, M.D. Navajo Microvillous Inclusion Disease Is Due to a Mutation in MYO5B. Am. J. Med. Genet. A 2008, 146A, 3117–3119. [Google Scholar] [CrossRef]
- Al-Sinani, S.; Sharef, S.W.; Lakhtakia, R.; Abdellatif, M. Diagnosis of microvillous inclusion disease: A case report and literature review with significance for oman. Oman Med. J. 2012, 27, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Engevik, A.C.; Coutts, A.W.; Kaji, I.; Rodriguez, P.; Ongaratto, F.; Saqui-Salces, M.; Medida, R.L.; Meyer, A.R.; Kolobova, E.; Engevik, M.A.; et al. Editing Myosin VB Gene to Create Porcine Model of Microvillus Inclusion Disease, With Microvillus-Lined Inclusions and Alterations in Sodium Transporters. Gastroenterology 2020, 158, 2236–2249.e9. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Nanki, K.; Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 2015, 10, 1474–1485. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, P.; Fang, P.; Ren, T.; Fang, L.; Xiao, S. Porcine Intestinal Organoids: Overview of the State of the Art. Viruses 2022, 14, 1110. https://doi.org/10.3390/v14051110
Ma P, Fang P, Ren T, Fang L, Xiao S. Porcine Intestinal Organoids: Overview of the State of the Art. Viruses. 2022; 14(5):1110. https://doi.org/10.3390/v14051110
Chicago/Turabian StyleMa, Panpan, Puxian Fang, Tianze Ren, Liurong Fang, and Shaobo Xiao. 2022. "Porcine Intestinal Organoids: Overview of the State of the Art" Viruses 14, no. 5: 1110. https://doi.org/10.3390/v14051110
APA StyleMa, P., Fang, P., Ren, T., Fang, L., & Xiao, S. (2022). Porcine Intestinal Organoids: Overview of the State of the Art. Viruses, 14(5), 1110. https://doi.org/10.3390/v14051110





