In Silico Evaluation of Paxlovid’s Pharmacometrics for SARS-CoV-2: A Multiscale Approach
Abstract
:1. Introduction
2. Methods
2.1. The Hybrid PDE-ABM Model
2.1.1. Epithelial Cells
- All living uninfected cells are susceptible target cells to virus infection;
- Since the time frame of infection is relatively short, cell birth and cell division are ignored;
- Infection is not reversible: an infected cell can not become a healthily functioning uninfected cell again;
- Viral infection itself is the only reason for cell death, i.e., death related to any other natural cause is not accounted for (considering the 17-month half-life of lung epithelial cells [13], natural apoptosis may be ignored over the course of a 5-day Paxlovid treatment);
- The uninfected→infected state change: a target cell may become infected depending on the local virus concentration at the given cell. Infection itself is randomized and it occurs with a probability of (for more details, see [6]);
- The infected→dead state change: Analogously to infection, death is governed by a stochastic model with probability .
2.1.2. Virus Concentration
- (i)
- Virus particles spread across the domain primarily via diffusion.
- (ii)
- A non-specific, non-adaptive, simplified immune system is assumed which clears virions with a constant rate.
- (iii)
- A local nirmatrelvir concentration of reduces virus production from infected cells by a ratio of for more details, see Section 2.1.3.
- (iv)
- Infected cells generate new virus particles in a process that is formally described by the source functions:
2.1.3. Drug Concentration
2.2. Parametrization
- Drug removal rates: . As we do not have direct information on the coefficients, we deduce them indirectly by using frequently measured nirmatrelvir blood concentration values communicated in [20]. Of course, this argument raises the question of whether it is reasonable to use blood concentration values to estimate local drug concentrations in the lung—the validity of this approach is reassured by the results of [22]. The and coefficients were set using Wolfram Mathematica [23]. We note that in the process, we also benefited from a priori information on ritonavir from [20,24]: we used that in non-ritonavir-boosted cases, the active component is apparently metabolized 3–4 times faster. The Wolfram Mathematica notebook is available in our public Github repository [25].
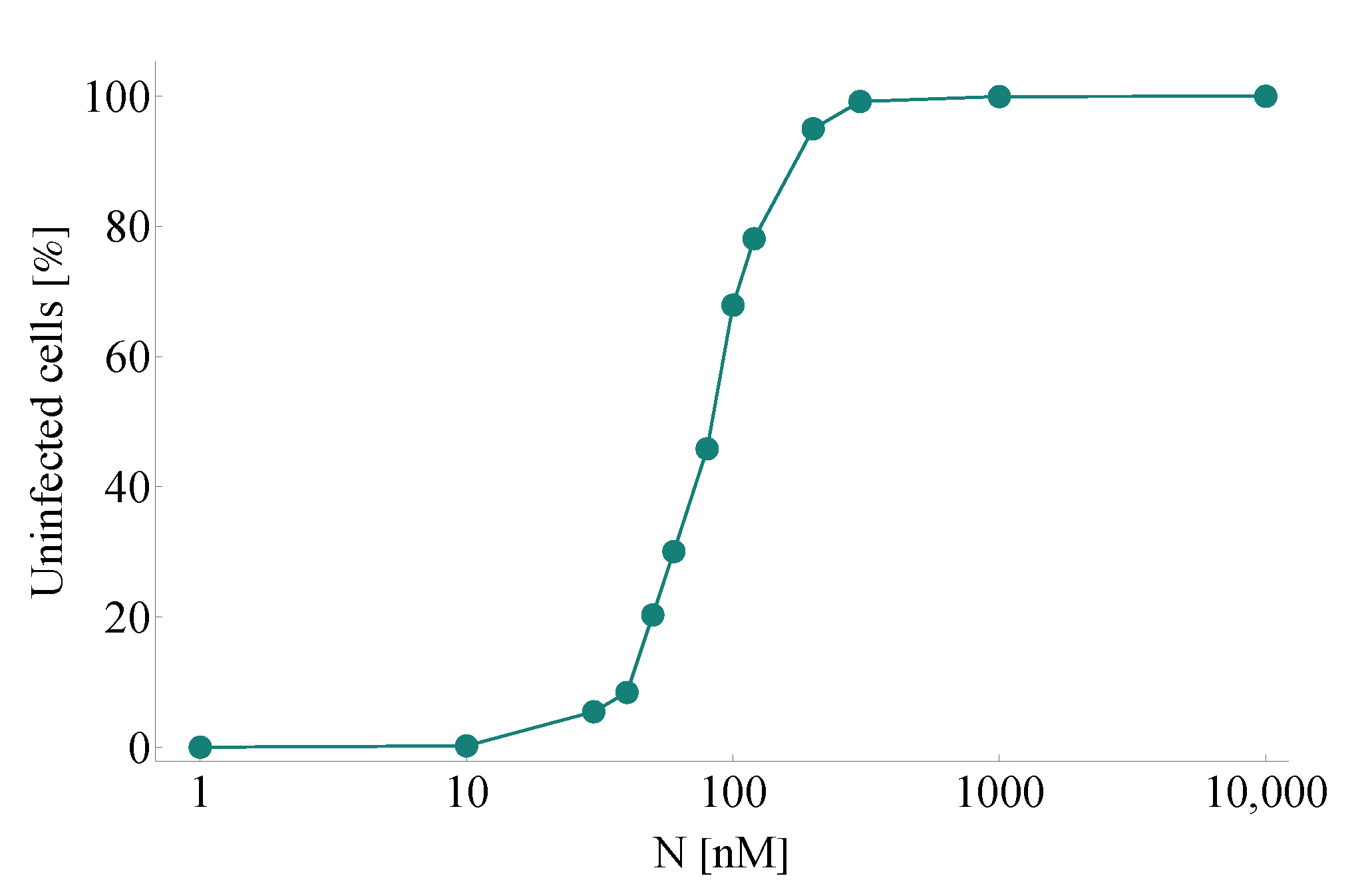
- Efficacy: . In our model, is defined by means of a Hill function—the parameters of the latter are set precisely to obtain an efficacy of when the drug concentration takes the value of EC for nirmatrelvir w.r.t. SARS-CoV-2 (the latter parameter is approximately 62 nM according to [3]). Formally, is defined as
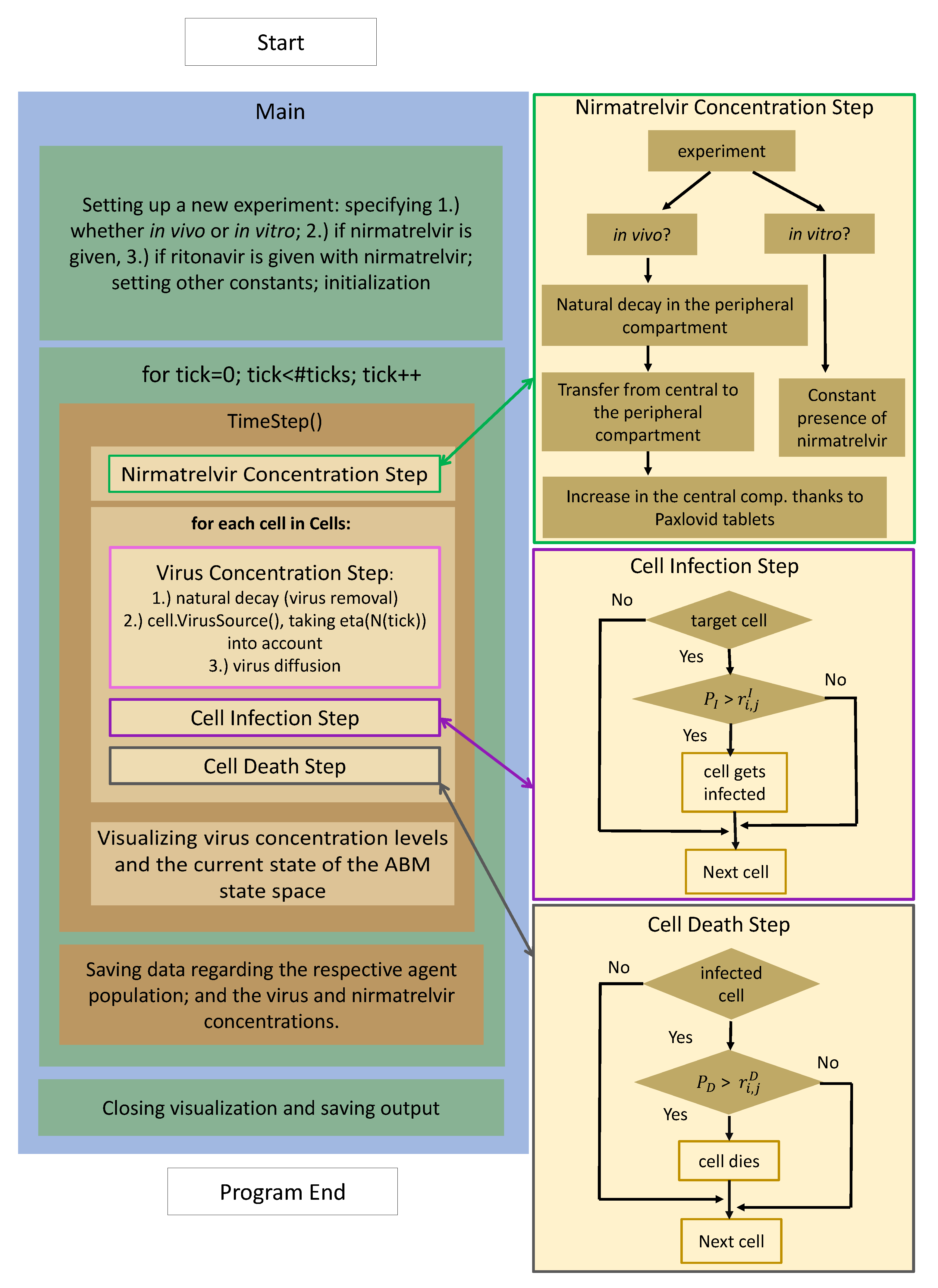
2.3. Implementation
- 1.
- Nirmatrelvir concentration is calculated first (highlighted with a green frame): the most fundamental details of the process are given on the right-hand side, highlighted with an identical color. There is a clear distinction between in vitro and in vivo cases: while the first scenario is implemented assuming a constant drug concentration, the latter utilizes a two-compartment PK system.
- 2.
- Updating virus concentration values entails recalculating the current values considering both natural decay and inflow from infected cells taking virus diffusion into account.
- 3.
- After the new drug and virus concentration values have been obtained at each cell, we are ready to update the cell states, i.e., consider potential cell infection and cell death—the former is highlighted in purple, the latter in gray. The schematic details are shown on the right with corresponding colors; we emphasize that the respective stochastic cores of these processes are very similar to each other. In both cases, the algorithm calculates the probability of either infection or death.
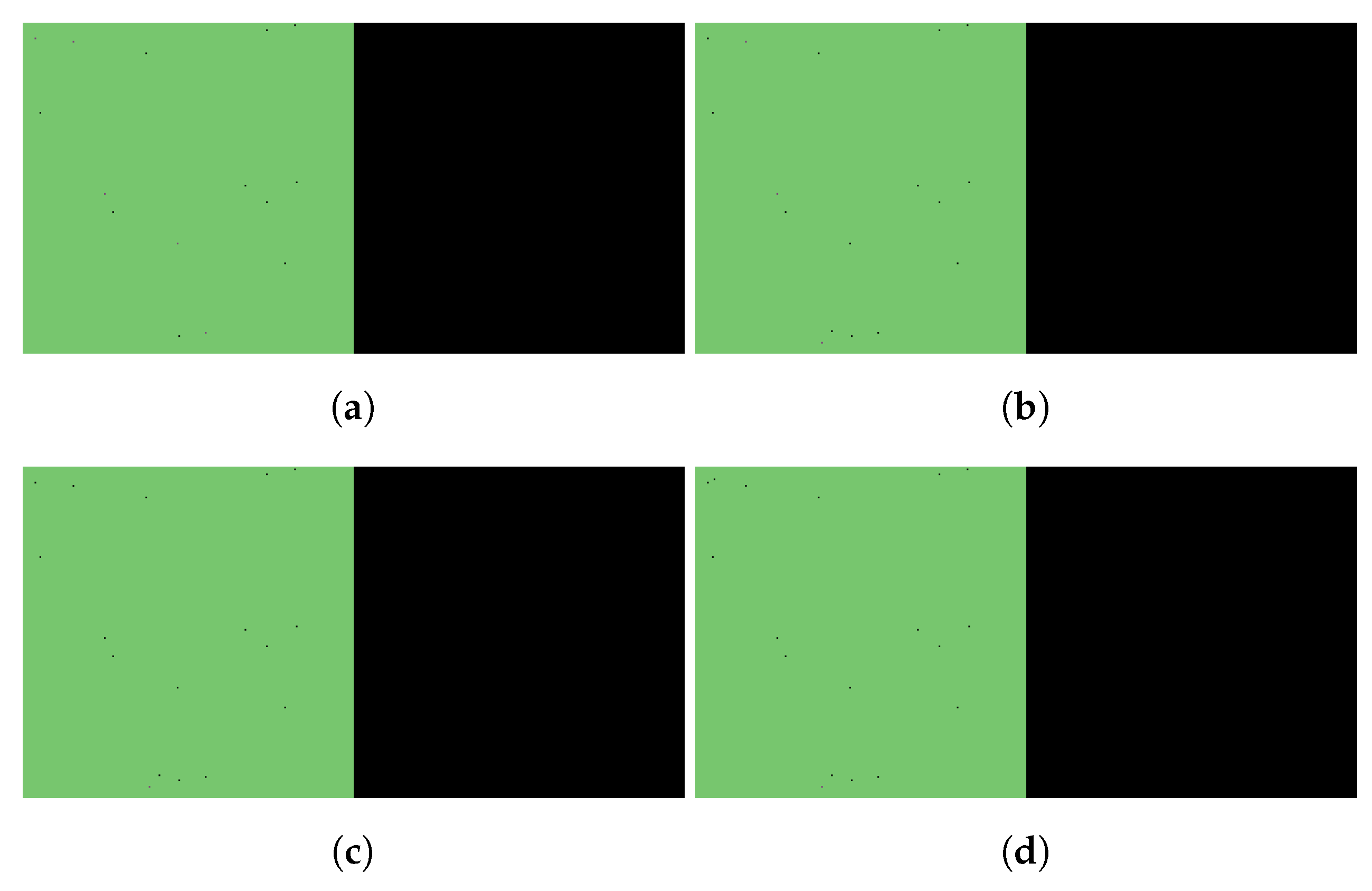
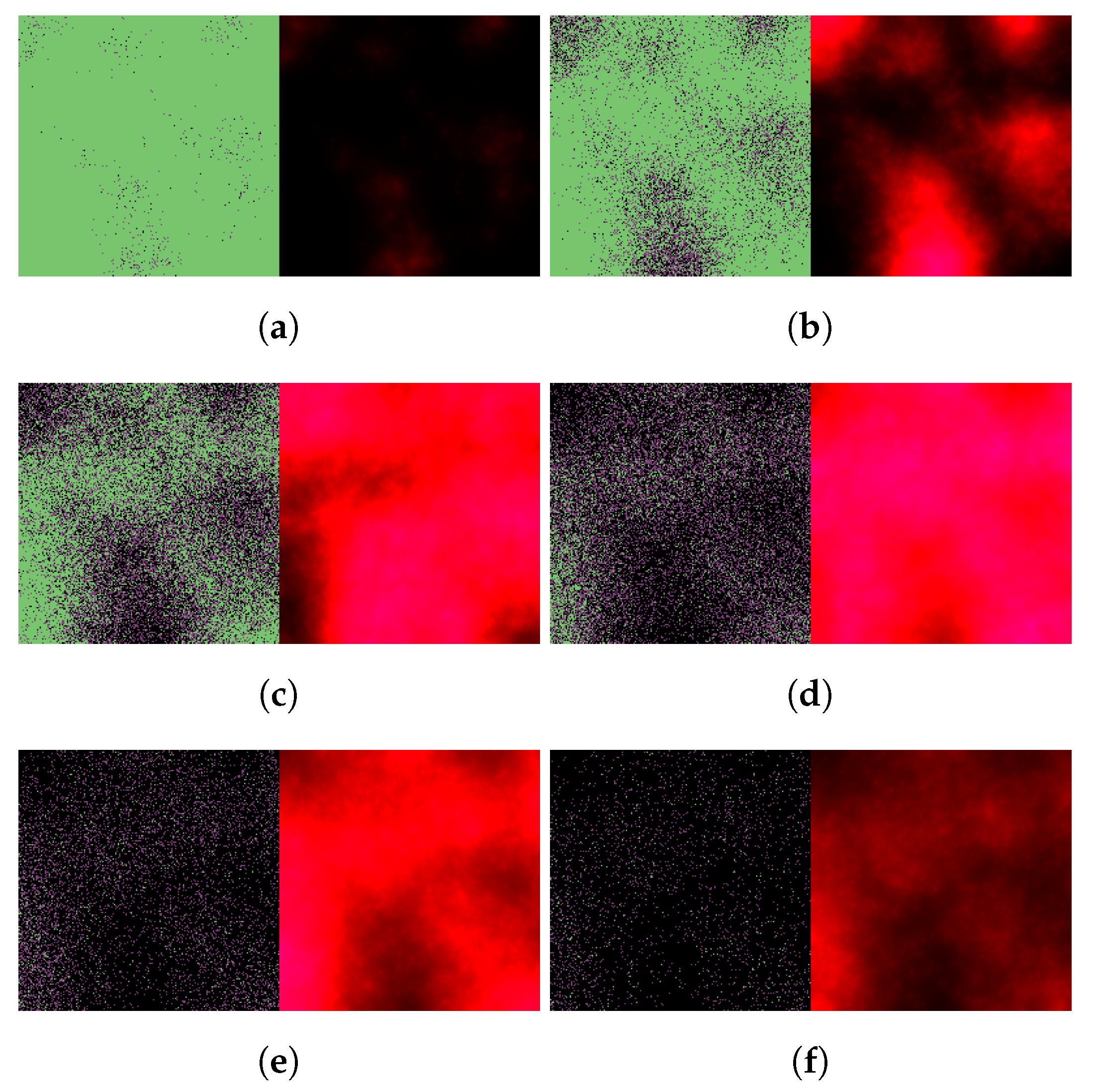
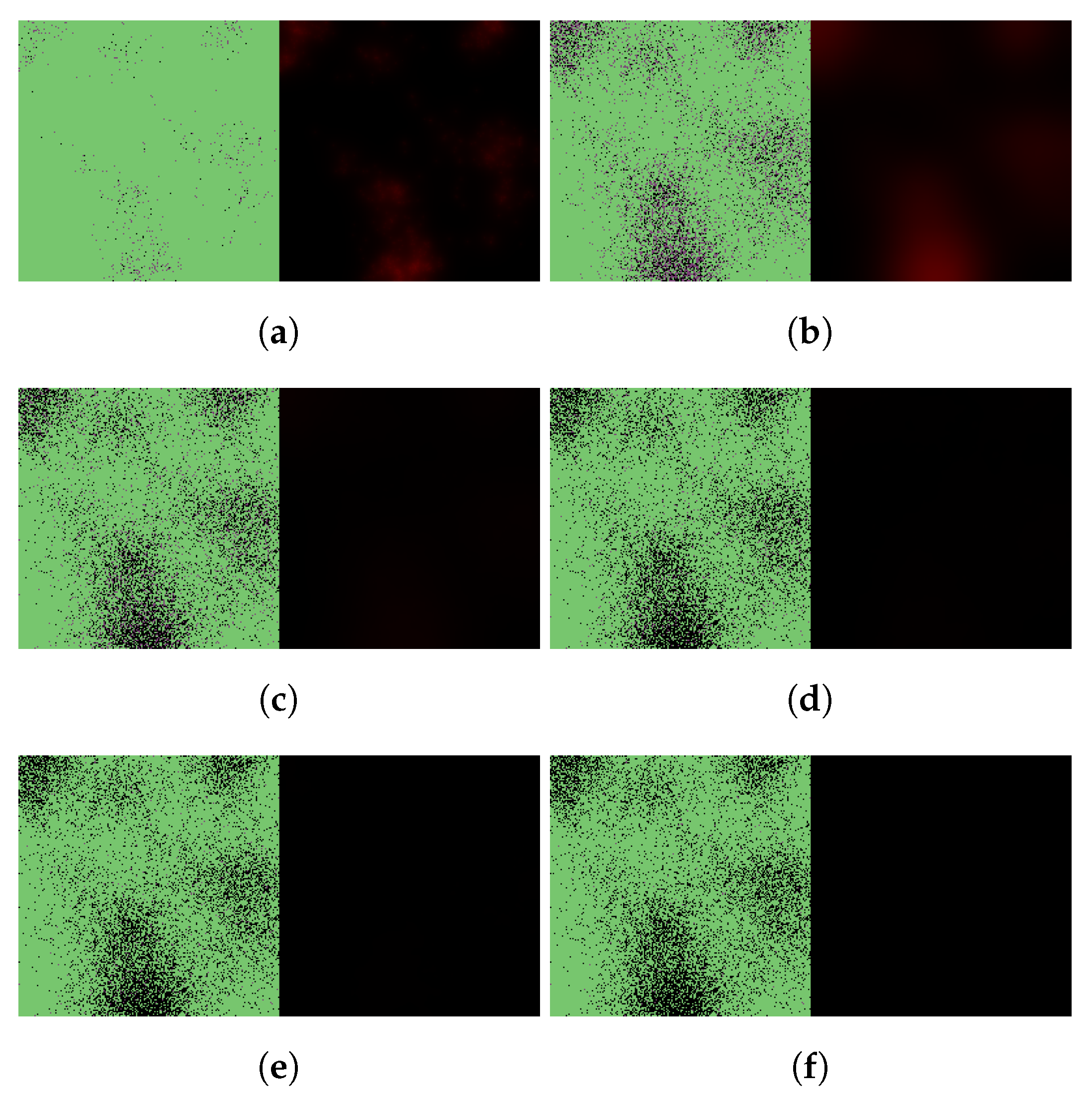
3. Results
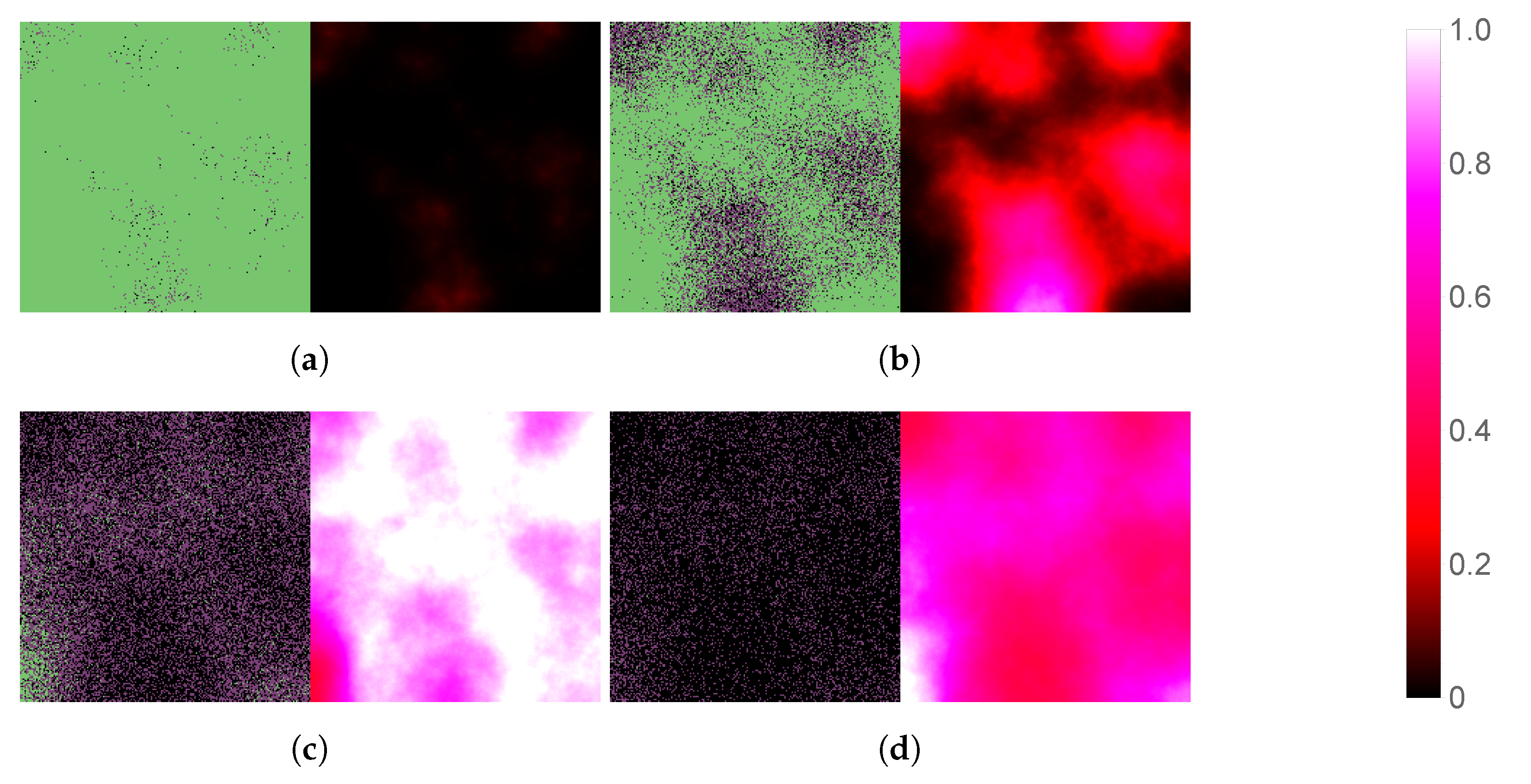
3.1. Replication of In Vitro Pharmacometrics of Paxlovid
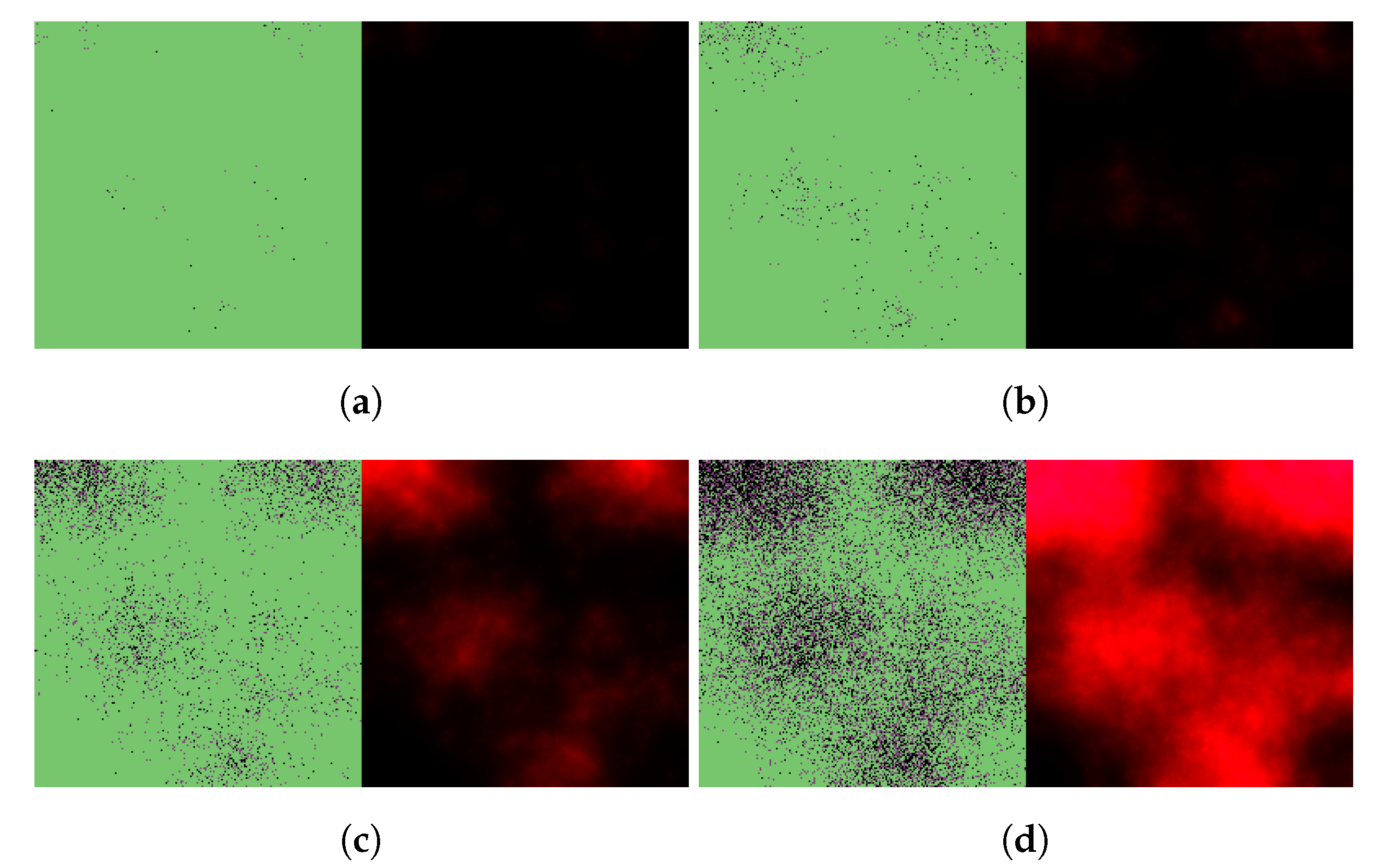
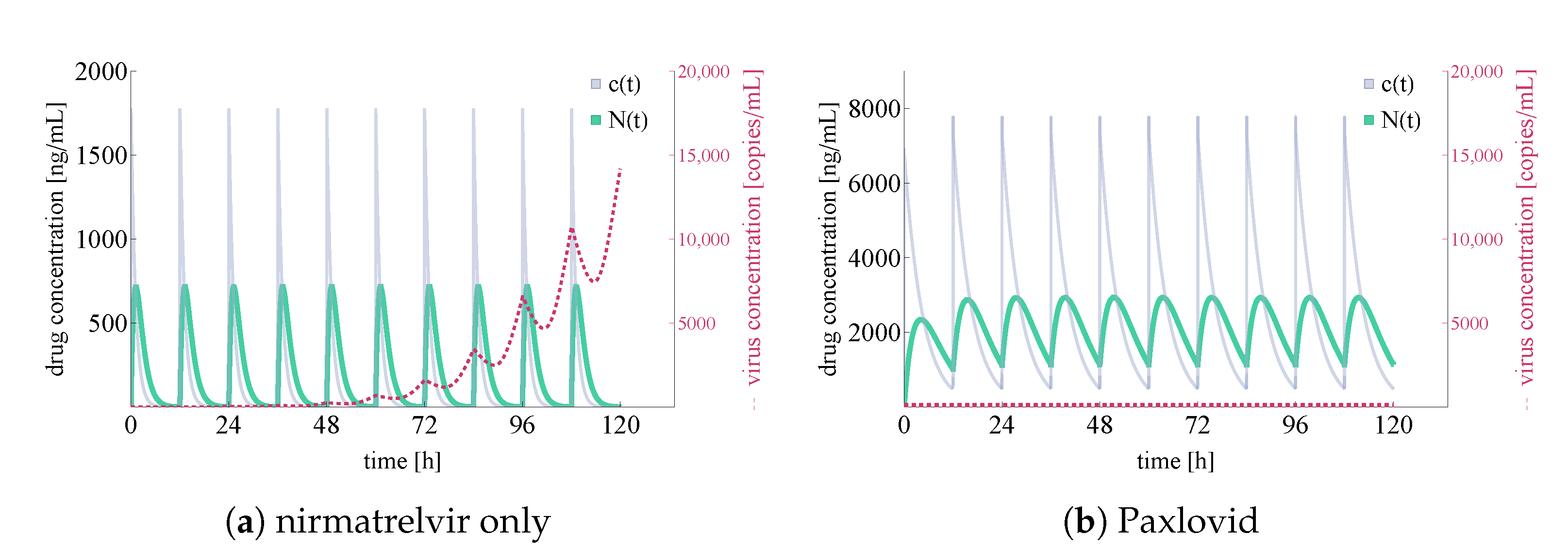
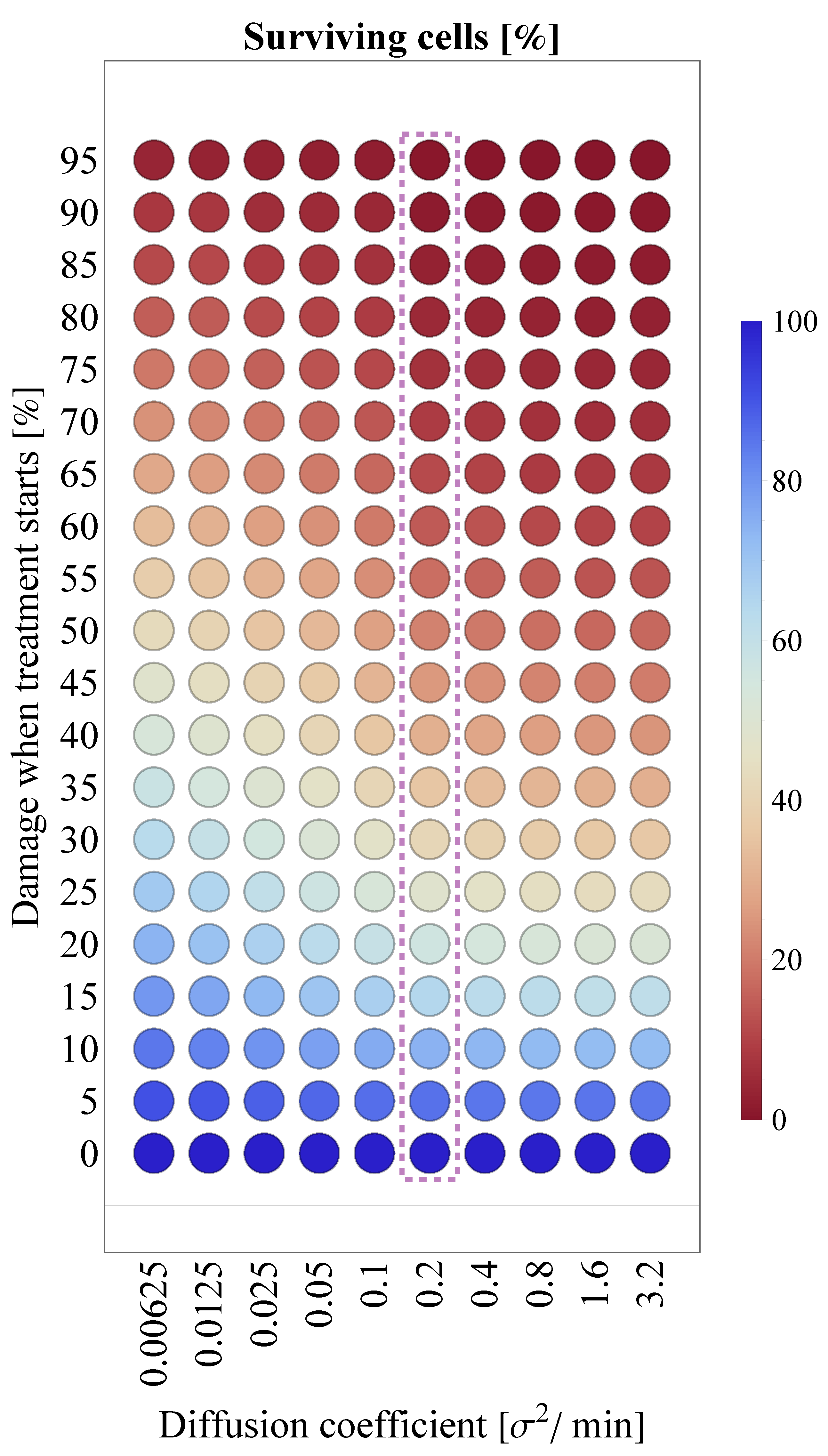
3.2. Exploring In Vivo Pharmacometrics of Paxlovid
3.2.1. In Silico Testing of Immediate Paxlovid-Based Intervention
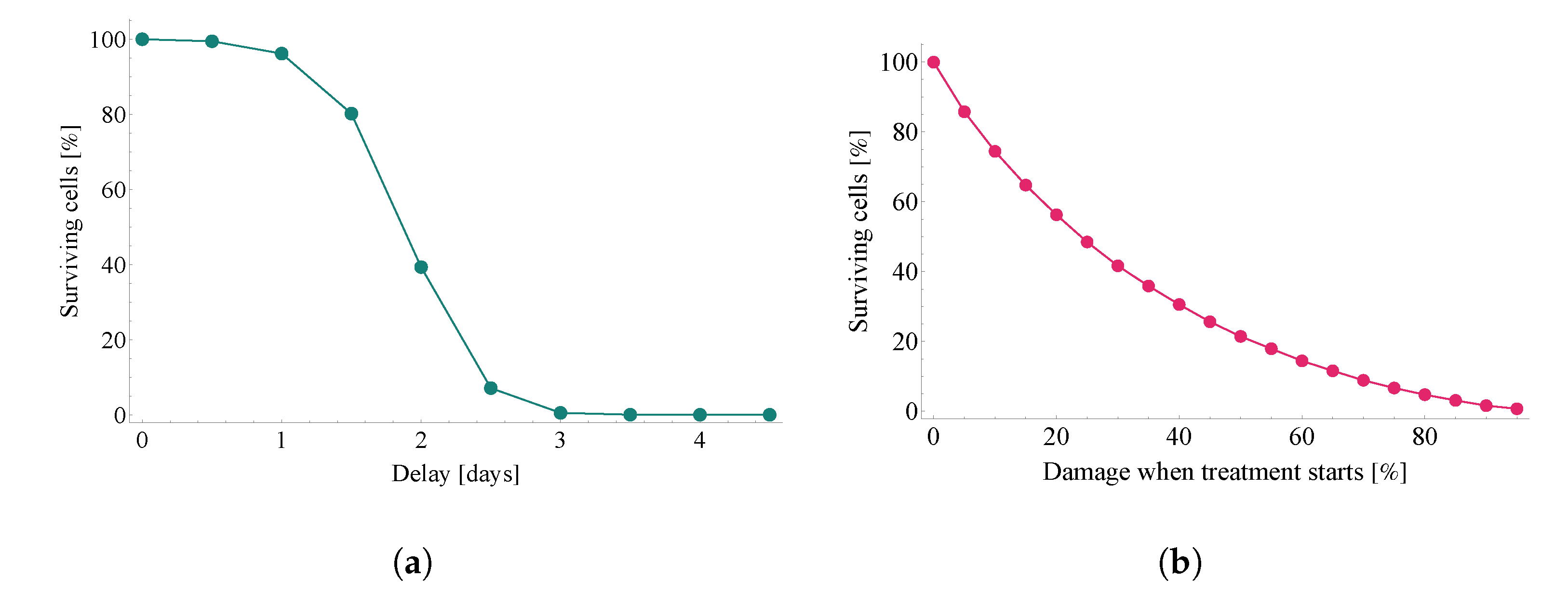
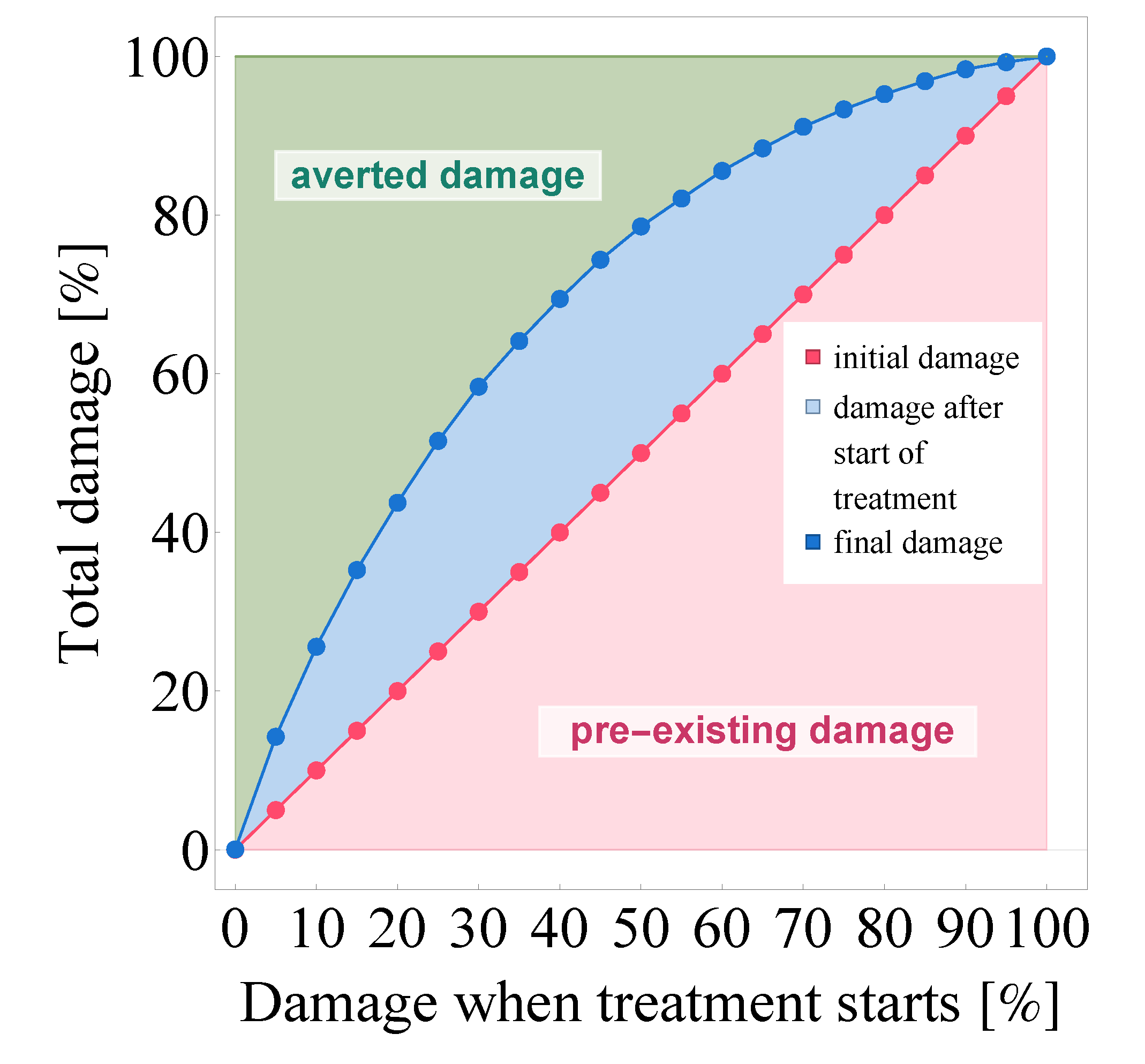
3.2.2. Evaluating the Effects of Treatment Delay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pfizer. PAXLOVID™ (Nirmatrelvir Tablers; Ritonavir Tablets). Pfizer Medical Information. 2022. Available online: https://www.pfizermedicalinformation.com/en-us/nirmatrelvir-tablets-ritonavir-tablets/clinical-pharmacology (accessed on 21 April 2022).
- Owen, D.R.; Allerton, C.M.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- FDA. Fact Sheet For Healthcare Providers: Emergency Use Authoriziation for Paxlovid™. United States Food and Drug Administration. 2022. Available online: https://www.fda.gov/media/155050/download (accessed on 21 April 2022).
- EMA. Annex I—Conditions of Use, Conditions for Distribution and Patients Targeted and Conditions for Safety Monitoring Addressed to Member States—for Unauthorised Product—Paxlovid (PF-07321332 150 mg and ritonavir 100 mg)—Available for Use. European Medicines Agency. 2022. Available online: https://www.ema.europa.eu/en/documents/referral/paxlovid-pf-07321332-ritonavir-covid-19-article-53-procedure-conditions-use-conditions-distribution_en.pdf (accessed on 21 April 2022).
- Perelson, A.S.; Ke, R. Mechanistic modeling of SARS-CoV-2 and other infectious diseases and the effects of therapeutics. Clin. Pharmacol. Ther. 2021, 109, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Marzban, S.; Han, R.; Juhász, N.; Röst, G. A hybrid PDE–ABM model for viral dynamics with application to SARS-CoV-2 and influenza. R Soc. Open Sci. 2021, 8, 210787. [Google Scholar] [CrossRef] [PubMed]
- Gianlupi, J.F.; Mapder, T.; Sego, T.J.; Sluka, J.P.; Quinney, S.K.; Craig, M.; Stratford, R.E., Jr.; Glazier, J.A. Multiscale Model of Antiviral Timing, Potency, and Heterogeneity Effects on an Epithelial Tissue Patch Infected by SARS–CoV–2. Viruses 2022, 14, 605. [Google Scholar] [CrossRef] [PubMed]
- Ghaffarizadeh, A.; Heiland, R.; Friedman, S.H.; Mumenthaler, S.M.; Macklin, P. PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput. Biol. 2018, 14, e1005991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, R.R.; Baratchart, E.; West, J.; Schenck, R.O.; Miller, A.K.; Gallaher, J.; Gatenbee, C.D.; Basanta, D.; Robertson-Tessi, M. Anderson, A.R. Hybrid Automata Library: A flexible platform for hybrid modeling with real-time visualization. PLoS Comput. Biol. 2020, 16, e1007635. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the numbers. eLife 2020, 9, e57309. [Google Scholar] [CrossRef] [PubMed]
- Carcaterra, M.; Caruso, C. Alveolar epithelial cell type II as main target of SARS-CoV-2 virus and COVID-19 development via NF-Kb pathway deregulation: A physio-pathological theory. Med. Hypotheses 2021, 146, 110412. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.J. Biology of alveolar type II cells. Respirology 2006, 11, S12–S15. [Google Scholar] [CrossRef] [PubMed]
- Rawlins, E.L.; Hogan, B.L.M. Ciliated epithelial cell lifespan in the mouse trachea and lung. Am. J. Physiol. Lung Cell Mol. Physiol. 2008, 295, L231–L234. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, C.; Forrest, S.; Koster, F.T. Modeling Influenza Viral Dynamics in Tissue. In Artificial Immune Systems; ICARIS, Lecture Notes in Computer Science; Bersini, H., Carneiro, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4163. [Google Scholar]
- Lord, J.S.; Bonsall, M.B. The evolutionary dynamics of viruses: Virion release strategies, time delays and fitness minima. Virus Evol. 2021, 7, veab039. [Google Scholar] [CrossRef] [PubMed]
- Perelson, A.S. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2002, 2, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Vargas, E.A.; Velasco-Hernandez, J.X. In-host mathematical modelling of COVID-19 in humans. Annu. Rev. Control 2020, 50, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G.J.; Shapiro, S.D. (Eds.) Encyclopedia of Respiratory Medicine, 1st ed.; Academic Press: Cambridge, MA, USA, 2006; Volume 3. [Google Scholar]
- Willführ, A.; Brandenberger, C.; Piatkowski, T.; Grothausmann, R.; Nyengaard, J.R.; Ochs, M.; Mühlfeld, C. Estimation of the number of alveolar capillaries by the Euler number (Euler-Poincaré characteristic). Am. J. Physiol. Lung Cell Mol. Physiol. 2015, 309, L1286–L1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.S.; Toussi, S.S.; Hackman, F.; Chan, P.L.; Rao, R.; Allen, R.; Van Eyck, L.; Pawlak, S.; Kadar, E.P.; Clark, F.; et al. Innovative Randomized Phase 1 Study and Dosing Regimen Selection to Accelerate and Inform Pivotal COVID-19 Trial of Nirmatrelvir. Clin. Pharmacol. Ther. 2022, cpt.2603. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Cardozo-Ojeda, E.F.; Schiffer, J.T. Potency and timing of antiviral therapy as determinants of duration of SARS-CoV-2 shedding and intensity of inflammatory response. Sci. Adv. 2020, 6, eabc7112. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.J.; Chang, L.; Yang, Y.; Wang, X.; Xie, Y.C.; Shen, J.S.; Tan, B.; Liu, J. Pharmacokinetics and tissue distribution of remdesivir and its metabolites nucleotide monophosphate, nucleotide triphosphate, and nucleoside in mice. Acta Pharmacol. Sin. 2021, 42, 1195–1200. [Google Scholar] [CrossRef] [PubMed]
- Mathematica. Version 13.0.0; Wolfram Research, Inc.: Champaign, IL, USA, 2021. Available online: https://www.wolfram.com/mathematica (accessed on 13 December 2021).
- Olkkola, K.T.; Palkama, V.J.; Neuvonen, P.J. Ritonavir’s role in reducing fentanyl clearance and prolonging its half-life. Anesthesiology 1999, 91, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Bartha, F.A.; Juhász, N.; Marzban, S.; Han, R.; Röst, G. Supplementary Codes for In Silico Evaluation of Paxlovid’s Pharmacometrics for SARS–CoV–2: A Multiscale Approach. Github 2022. Available online: https://github.com/epidelay/paxlovid-x-sars-cov-2 (accessed on 21 April 2022).

| Symbol | Parameter | Unit | Ritonavir–Boosted | Value |
|---|---|---|---|---|
| drug removal rate in the stomach | false | 0.015 | ||
| true | 0.005 | |||
| drug removal rate in the lung | false | 0.013 | ||
| true | 0.004 | |||
| drug source in the stomach | false | 1800 | ||
| true | 6800 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartha, F.A.; Juhász, N.; Marzban, S.; Han, R.; Röst, G. In Silico Evaluation of Paxlovid’s Pharmacometrics for SARS-CoV-2: A Multiscale Approach. Viruses 2022, 14, 1103. https://doi.org/10.3390/v14051103
Bartha FA, Juhász N, Marzban S, Han R, Röst G. In Silico Evaluation of Paxlovid’s Pharmacometrics for SARS-CoV-2: A Multiscale Approach. Viruses. 2022; 14(5):1103. https://doi.org/10.3390/v14051103
Chicago/Turabian StyleBartha, Ferenc A., Nóra Juhász, Sadegh Marzban, Renji Han, and Gergely Röst. 2022. "In Silico Evaluation of Paxlovid’s Pharmacometrics for SARS-CoV-2: A Multiscale Approach" Viruses 14, no. 5: 1103. https://doi.org/10.3390/v14051103
APA StyleBartha, F. A., Juhász, N., Marzban, S., Han, R., & Röst, G. (2022). In Silico Evaluation of Paxlovid’s Pharmacometrics for SARS-CoV-2: A Multiscale Approach. Viruses, 14(5), 1103. https://doi.org/10.3390/v14051103






