In Vitro and In Vivo Assessments of Two Newly Isolated Bacteriophages against an ST13 Urinary Tract Infection Klebsiella pneumoniae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Bacteriophage Isolation
2.3. Bacteriophages Genome Sequencing
2.4. Temperatures and pH Stability
2.5. Host Range and Efficiency of Plating
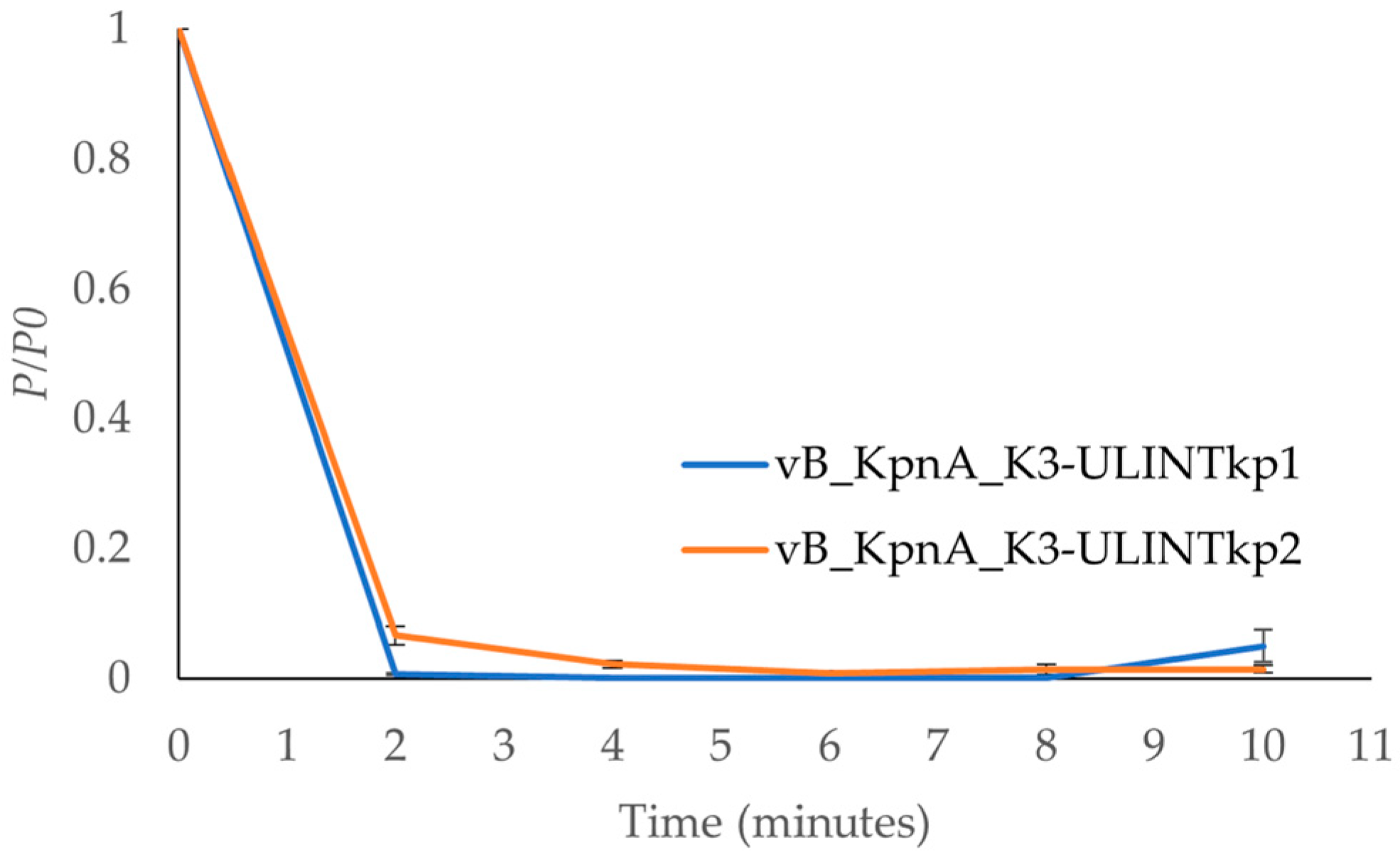
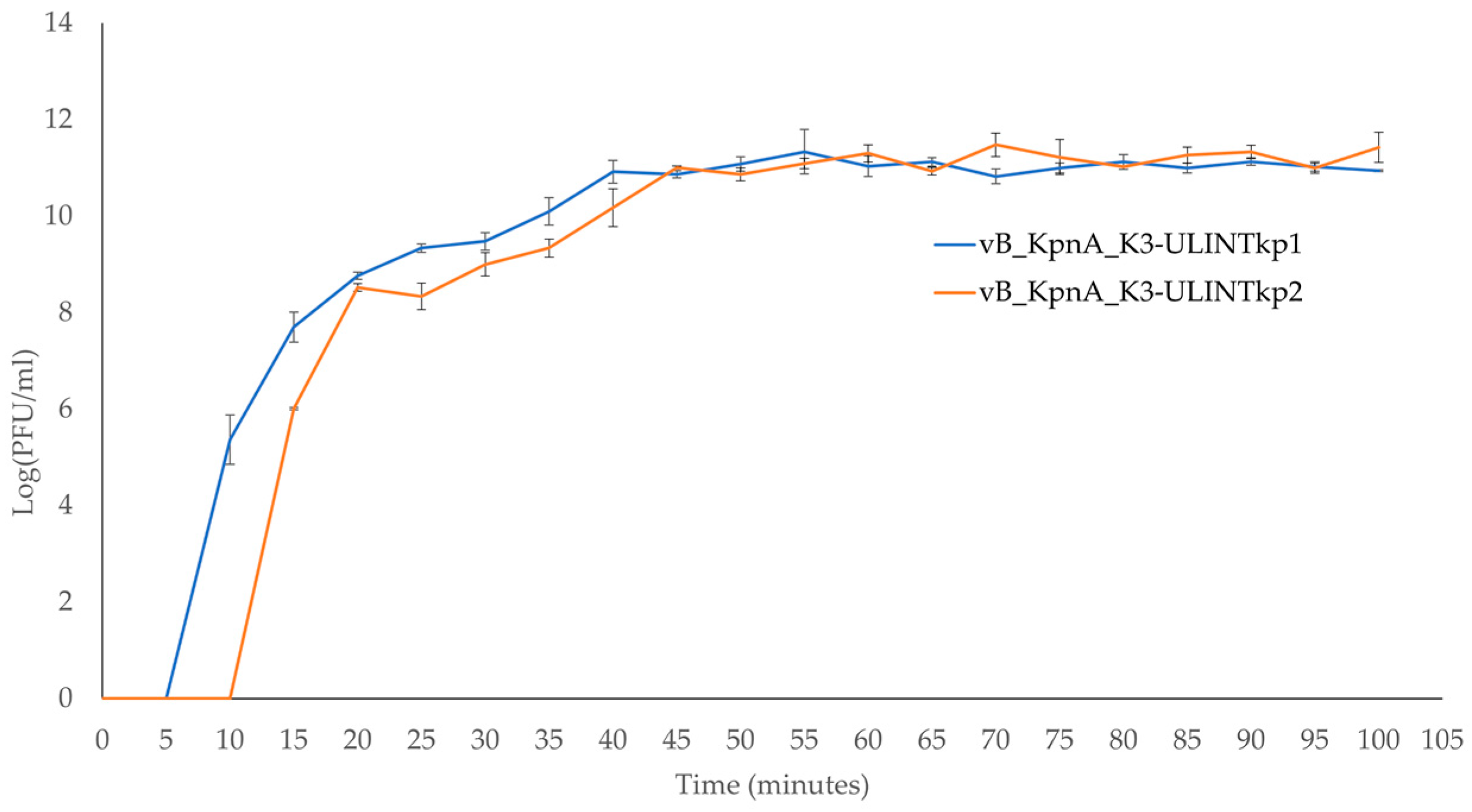
2.6. Adsorption Times and Low MOI Kinetic Curves
2.7. In Vitro Bacteriophage Activity
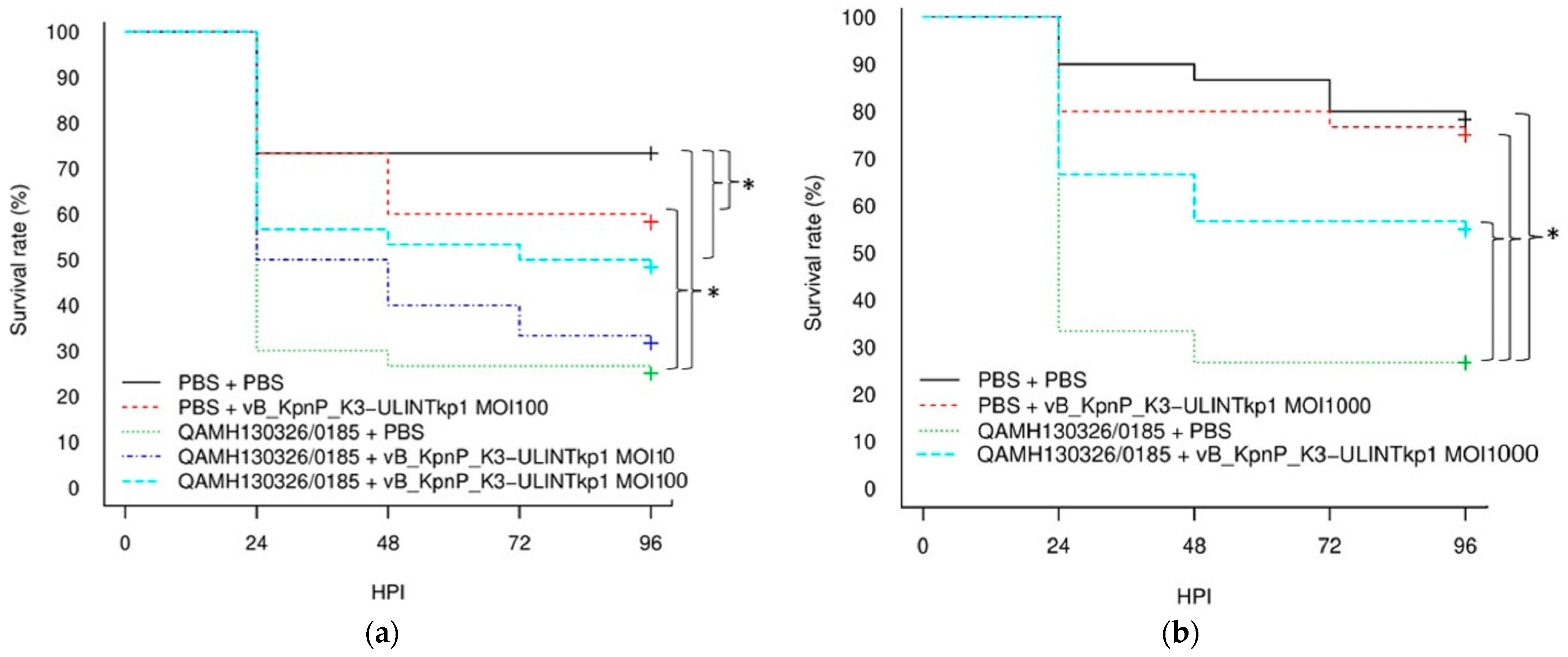
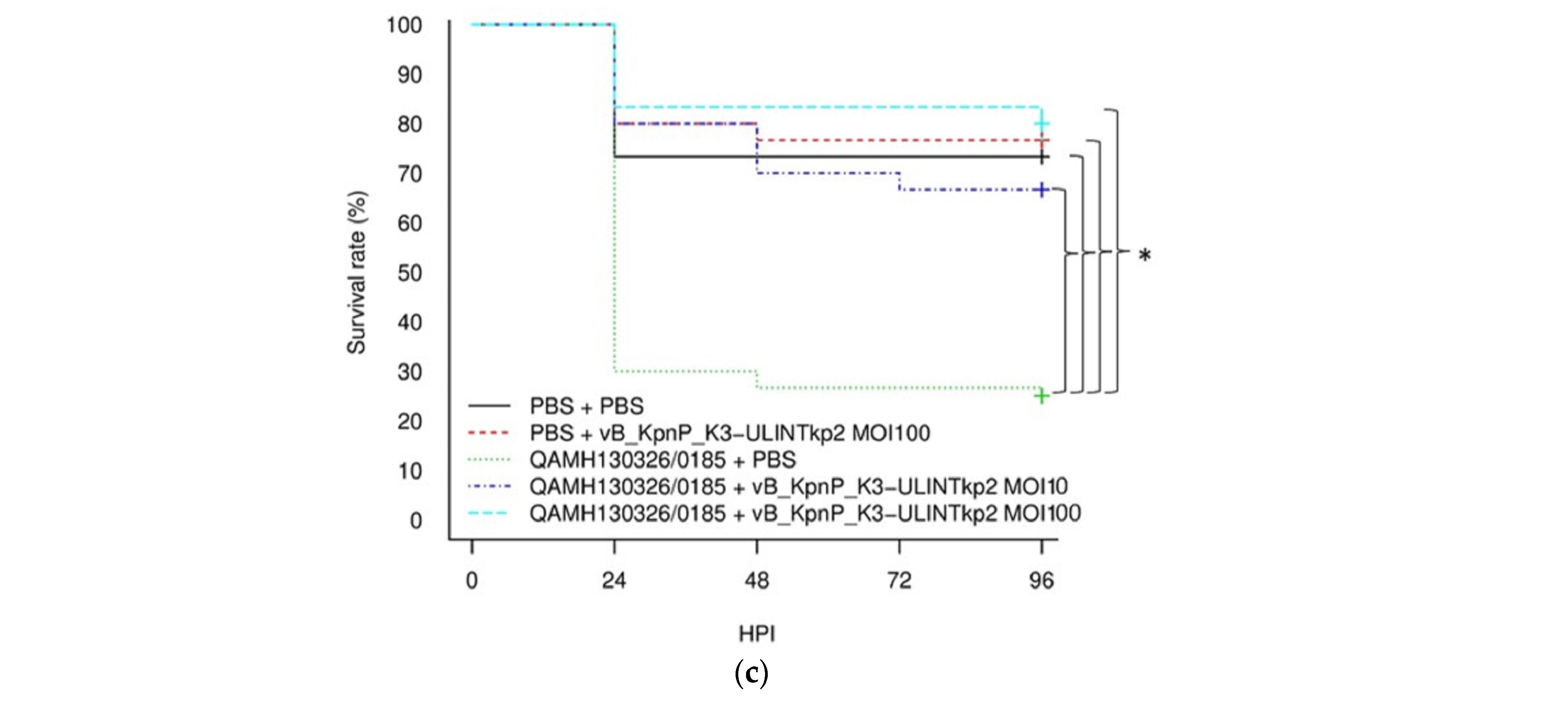
2.8. Survival Rate in Galleria mellonella Larvae
2.9. Bacterial Load and Bacteriophage Replication in Galleria mellonella Larvae
3. Results
3.1. Bacterial Strain
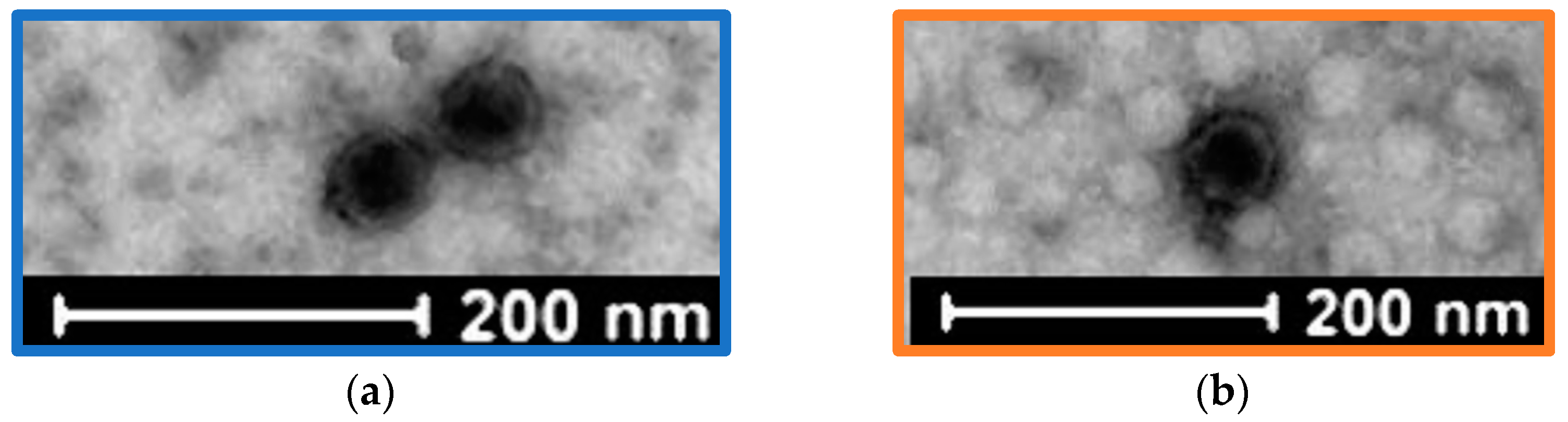
3.2. Bacteriophage Isolation
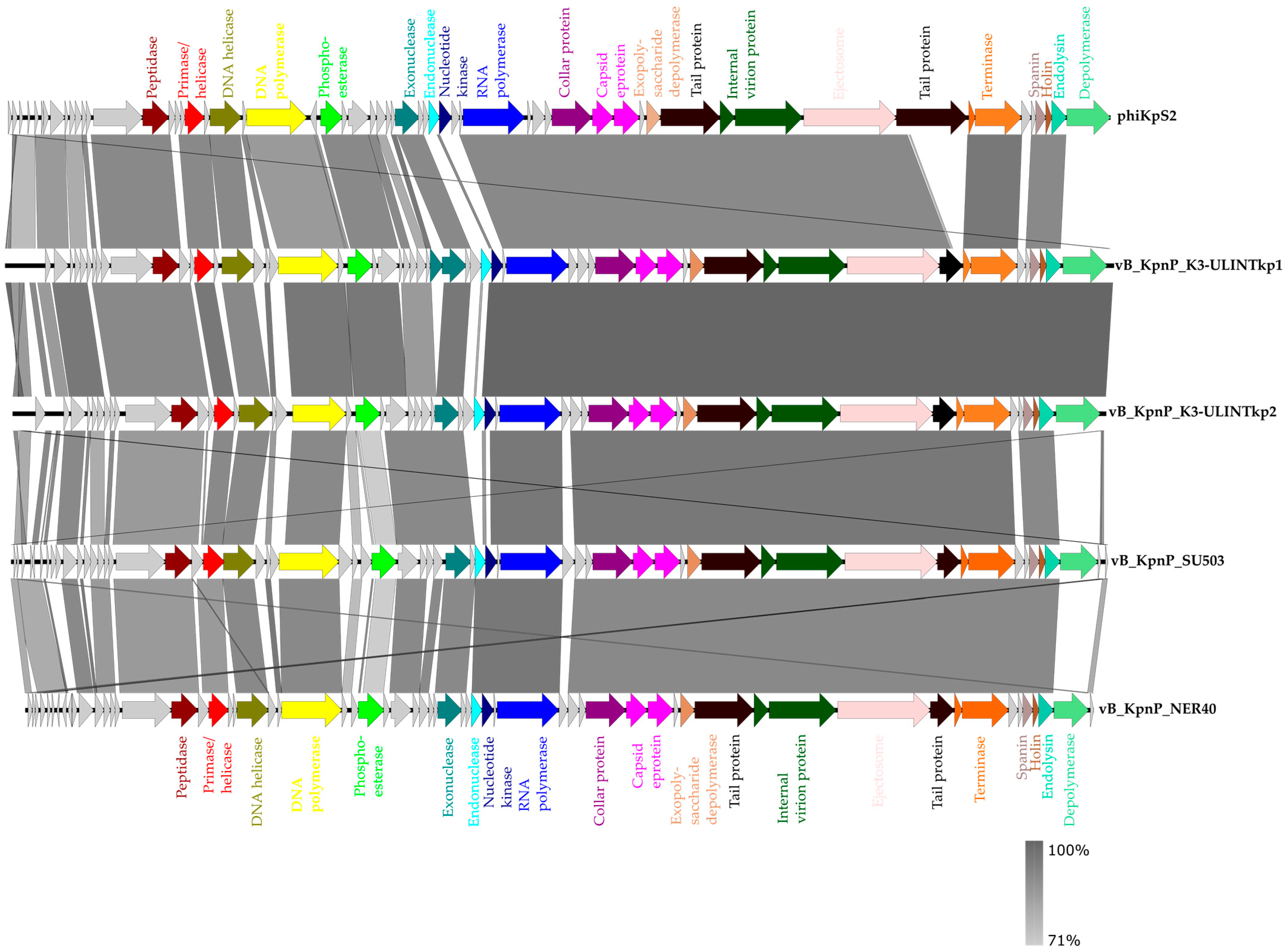
3.3. Bacteriophage Genome Sequencing
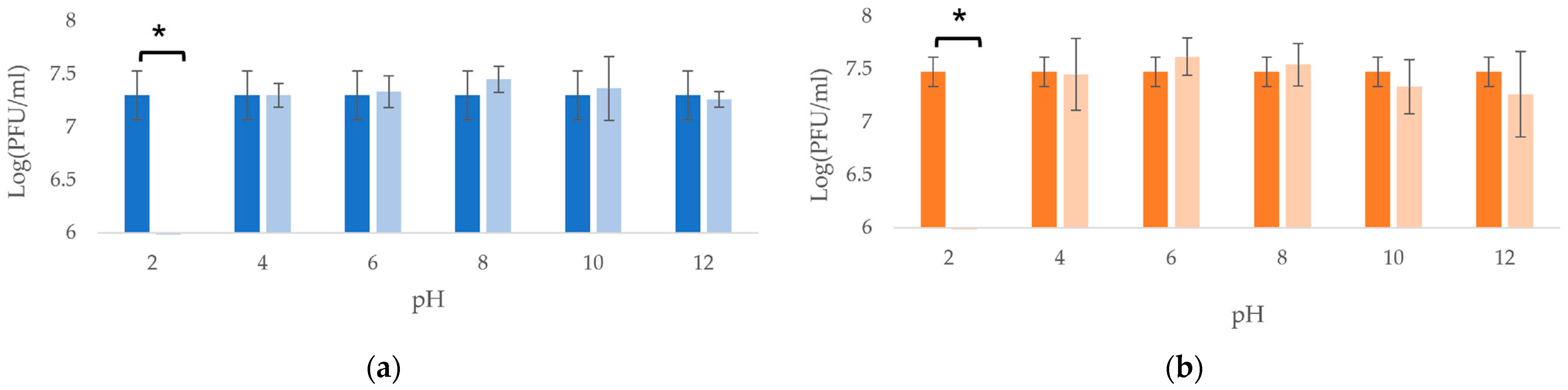
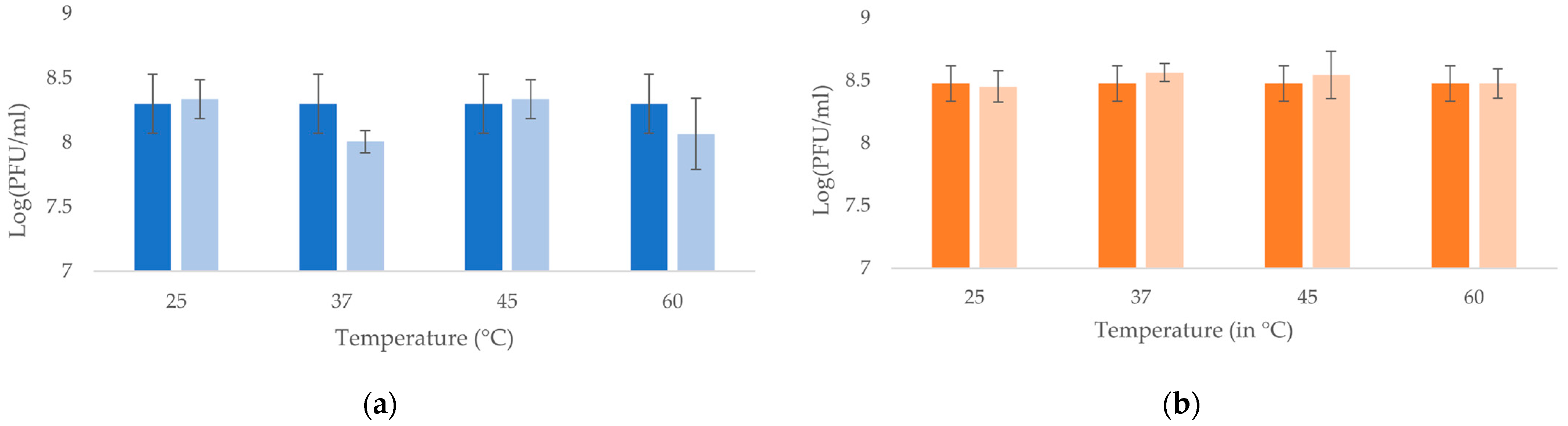
3.4. Temperatures and pH Stability
3.5. Host Range and Efficiency of Plating
3.6. Adsorption Times and Low MOI Kinetic Curves
3.7. In Vitro Bacteriophage Activity
3.8. Survival Rate in Galleria mellonella Larvae
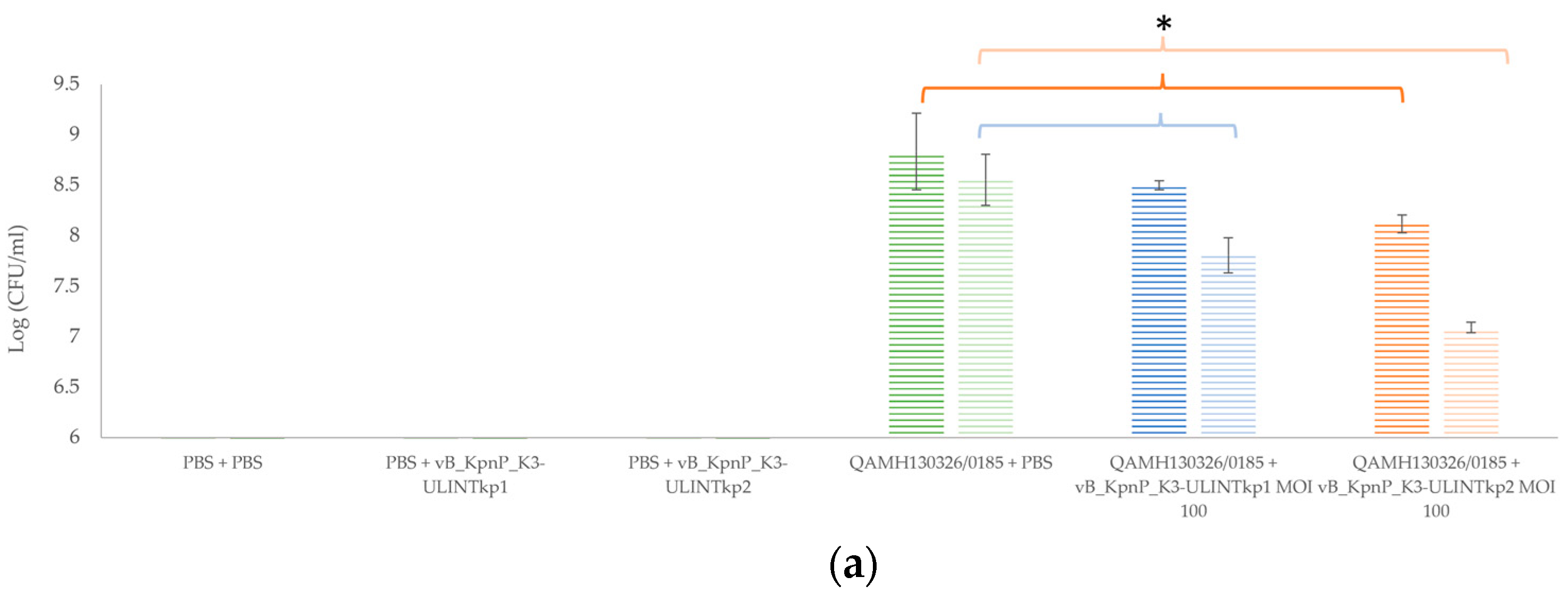
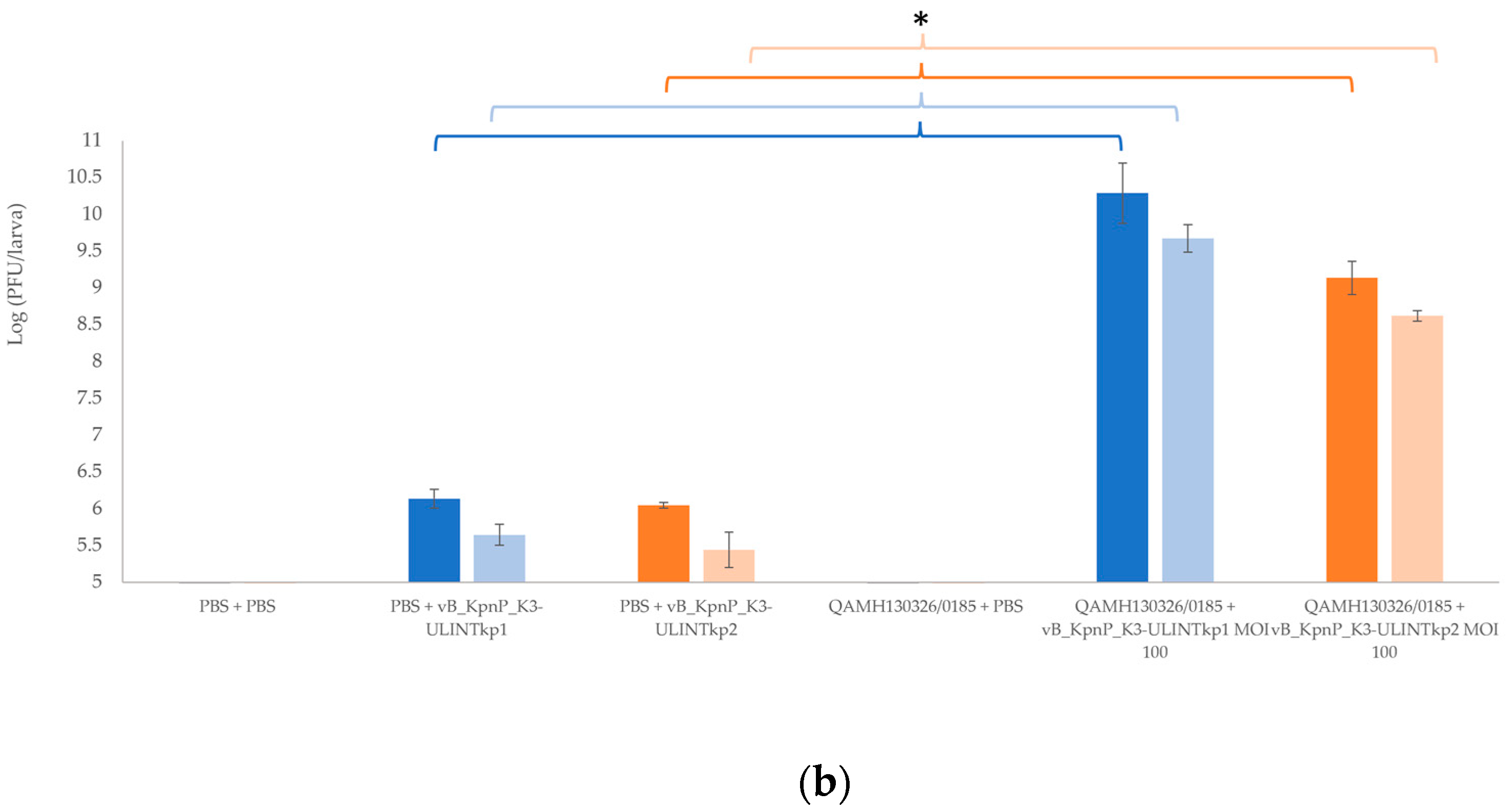
3.9. Bacterial Load and Bacteriophage Replication in Galleria mellonella Larvae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shaidullina, E.; Shelenkov, A.; Yanushevich, Y.; Mikhaylova, Y.; Shagin, D.; Alexandrova, I.; Ershova, O.; Akimkin, V.; Kozlov, R.; Edelstein, M. Antimicrobial Resistance and Genomic Characterization of OXA-48- and CTX-M-15-Co-Producing Hypervirulent Klebsiella pneumoniae ST23 Recovered from Nosocomial Outbreak. Antibiotics 2020, 9, 862. [Google Scholar] [CrossRef] [PubMed]
- Health First Europe. Available online: https://healthfirsteurope.eu/topic/amr-patient-group/ (accessed on 4 March 2022).
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- De Freitas, L.C. WHO (2017) Global Priority List of Antibiotic-resistant Bacteria to guide Research, Discovery, and Development of New Antibiotics. Cad. Pesqui. 2017, 43, 348–365. [Google Scholar]
- Brisse, S.; Passet, V.; Haugaard, A.B.; Babosan, A.; Kassis-Chikhani, N.; Struve, C.; Decre, D. Wzi Gene Sequencing, a Rapid Method for Determination of Capsular Type for Klebsiella strains. J. Clin. Microbiol. 2013, 51, 4073–4078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brisse, S.; Passet, V.; Grimont, P.A.D. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int. J. Syst. Evol. Microbiol. 2014, 64, 3146–3152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drancourt, M.; Bollet, C.; Carta, A.; Rousselier, P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 925–932. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martínez, L.; Silva, J.; Martínez-Romero, E. Klebsiella variicola, A Novel Species with Clinical and Plant-Associated Isolates. Syst. Appl. Microbiol. 2004, 27, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Saha, R.; Farrance, C.E.; Verghese, B.; Hong, S.; Donofrio, R.S. Klebsiella michiganensis sp. nov., a New Bacterium Isolated from a Tooth Brush Holder. Curr. Microbiol. 2013, 66, 72–78. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Ronald, A.; Ludwig, E. Urinary tract infections in adults with diabetes. Int. J. Antimicrob. Agents 2001, 17, 287–292. [Google Scholar] [CrossRef]
- Ronald, A. The Etiology of Urinary Tract Infection: Traditional and Emerging Pathogens. Disease-a-Month 2003, 49, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Belgian Antibiotic Policy Coordination Commission. Available online: https://www.health.belgium.be/sites/default/files/uploads/fields/fpshealth_theme_file/guide_belge_bapcoc_fr_2021_a4.pdf (accessed on 4 March 2022).
- Liu, H.Y.; Lin, H.C.; Lin, Y.C.; Yu, S.H.; Wu, W.H.; Lee, Y.J. Antimicrobial susceptibilities of urinary extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae to fosfomycin and nitrofurantoin in a teaching hospital in Taiwan. J. Microbiol. Immunol. Infect. 2011, 44, 364–368. [Google Scholar] [CrossRef] [Green Version]
- Deresinski, S. Bacteriophage Therapy: Exploiting Smaller Fleas. Clin. Infect. Dis. 2009, 48, 1096–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aragón, I.M.; Herrera-Imbroda, B.; Queipo-Ortuño, M.I.; Castillo, E.; Del Moral, J.S.G.; Gómez-Millán, J.; Yucel, G.; Lara, M.F. The Urinary Tract Microbiome in Health and Disease. Eur. Urol. Focus 2018, 4, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Garretto, A.; Miller-Ensminger, T.; Wolfe, A.J.; Putonti, C. Bacteriophages of the lower urinary tract. Nat. Rev. Urol. 2019, 16, 422–432. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Vesal, S.; Moradabadi, A.; Hashemi, A.; Shariati, A. Bacteriophage therapy for inhibition of multi drug-resistant uropathogenic bacteria: A narrative review. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 30. [Google Scholar] [CrossRef]
- Loubet, P.; Ranfaing, J.; Dinh, A.; Dunyach-Remy, C.; Bernard, L.; Bruyère, F.; Lavigne, J.P.; Sotto, A. Alternative Therapeutic Options to Antibiotics for the Treatment of Urinary Tract Infections. Front. Microbiol. 2020, 11, 1509. [Google Scholar] [CrossRef]
- Malik, S.; Sidhu, P.K.; Rana, J.S.; Nehra, K. Managing urinary tract infections through phage therapy: A novel approach. Folia Microbiol. 2020, 65, 217–231. [Google Scholar] [CrossRef]
- Cieślik, M.; Bagińska, N.; Górski, A.; Jończyk-Matysiak, E. Animal Models in the Evaluation of the Effectiveness of Phage Therapy for Infections Caused by Gram-Negative Bacteria from the ESKAPE Group and the Reliability of Its Use in Humans. Microorganisms 2021, 9, 206. [Google Scholar] [CrossRef]
- Anand, T.; Virmani, N.; Kumar, S.; Mohanty, A.K.; Pavulraj, S.; Bera, B.C.; Vaid, R.K.; Ahlawat, U.; Tripathi, B.N. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J. Glob. Antimicrob. Resist. 2020, 21, 34–41. [Google Scholar] [CrossRef]
- Wintachai, P.; Naknaen, A.; Thammaphet, J.; Pomwised, R.; Phaonakrop, N.; Roytrakul, S.; Smith, D.R. Characterization of extended-spectrum-β-lactamase producing Klebsiella pneumoniae phage KP1801 and evaluation of therapeutic efficacy in vitro and in vivo. Sci. Rep. 2020, 10, 11803. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Nachimuthu, R. Therapeutic Characterization and Efficacy of Bacteriophage Cocktails Infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter Species. Front. Microbiol. 2019, 10, 574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiry, D.; Passet, V.; Danis-Wlodarczyk, K.; Lood, C.; Wagemans, J.; De Sordi, L.; van Noort, V.; Dufour, N.; Debarbieux, L.; Mainil, J.G.; et al. New Bacteriophages against Emerging Lineages ST23 and ST258 of Klebsiella pneumoniae and Efficacy Assessment in Galleria mellonella Larvae. Viruses 2019, 11, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiry, D.; Berrah, A.; Evrard, J.; Duprez, J.N.; Mainil, J.G.; Saulmont, M. Assessment of two selective agar media to isolate colistin-resistant bovine Escherichia coli: Correlation with minimal inhibitory concentration and presence of mcr genes. J. Microbiol. Methods 2019, 159, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Argimón, S.; David, S.; Underwood, A.; Abrudan, M.; Wheeler, N.E.; Kekre, M.; Abudahab, K.; Yeats, C.A.; Goater, R.; Taylor, B.; et al. Rapid Genomic Characterization and Global Surveillance of Klebsiella Using Pathogenwatch. Clin. Infect. Dis. 2021, 73, S325–S335. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI—Bacteriophage Life Cycle Recognition with Machine Learning and Natural Language Processing. bioRxiv 2020. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/fr/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 8 May 2022).
- Antoine, C.; Laforêt, F.; Blasdel, B.; Fall, A.; Duprez, J.N.; Mainil, J.; Delcenserie, V.; Thiry, D. In Vitro Characterization and In Vivo Efficacy Assessment in Galleria mellonella Larvae of Newly Isolated Bacteriophages against Escherichia coli k1. Viruses 2021, 13, 2005. [Google Scholar] [CrossRef]
- Kropinski, A.M. Measurement of the Rate of Attachment of Bacteriophage to Cells. Methods Mol. Biol. 2009, 501, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Antoine, C.; Laforêt, F.; Blasdel, B.; Glonti, T.; Kutter, E.; Pirnay, J.P.; Mainil, J.; Delcenserie, V.; Thiry, D. Efficacy assessment of PEV2 phage on Galleria mellonella larvae infected with a Pseudomonas aeruginosa dog otitis isolate. Res. Vet. Sci. 2021, 136, 598–601. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.A.; Sutherland, I.W.; Clark, J.; Jones, M.V. Bacteriophage and associated polysaccharide depolymerases—Novel tools for study of bacterial biofilms. J. Appl. Microbiol. 1998, 85, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.U.; Wang, W.; Sun, Q.; Shah, J.A.; Li, C.; Sun, Y.; Li, Y.; Zhang, B.; Chen, W.; Wang, S. Endolysin, a Promising Solution against Antimicrobial Resistance. Antibiotics 2021, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Keen, E.C.; Bliskovsky, V.V.; Malagon, F.; Baker, J.D.; Prince, J.S.; Klaus, J.S.; Adhya, S.L. Novel “Superspreader” Bacteriophages Promote Horizontal Gene Transfer by Transformation. MBio 2017, 8, e02115-16. [Google Scholar] [CrossRef] [Green Version]
- Dutton, G.G.S.; Parolis, H.; Joseleau, J.P.; Marais, M.F. The use of bacteriophage depolymerization in the structural investigation of the capsular polysaccharide from Klebsiella serotype K3. Carbohydr. Res. 1986, 149, 411–423. [Google Scholar] [CrossRef]
- Zamze, S.; Martinez-Pomares, L.; Jones, H.; Taylor, P.R.; Stillion, R.J.; Gordon, S.; Wong, S.Y.C. Recognition of Bacterial Capsular Polysaccharides and Lipopolysaccharides by the Macrophage Mannose Receptor. J. Biol. Chem. 2002, 277, 41613–41623. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, L.; Li, R.; Hou, P.; Zhang, Y.; Fang, M.; Hu, B. Presence and characterization of Klebsiella pneumoniae from the intestinal tract of diarrhoea patients. Lett. Appl. Microbiol. 2018, 66, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Brisse, S.; Fevre, C.; Passet, V.; Issenhuth-Jeanjean, S.; Tournebize, R.; Diancourt, L.; Grimont, P. Virulent clones of Klebsiella pneumoniae: Identification and Evolutionary Scenario Based on Genomic and Phenotypic Characterization. PLoS ONE 2009, 4, e4982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcántar-Curiel, M.D.; Blackburn, D.; Saldaña, Z.; Gayosso-Vázquez, C.; Iovine, N.; De la Cruz, M.A.; Girón, J.A. Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 2013, 4, 129–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Govindarajan, D.K.; Kandaswamy, K. Virulence factors of uropathogens and their role in host pathogen interactions. Cell Surf. 2022, 8, 100075. [Google Scholar] [CrossRef]
- Mendes, G.; Ramalho, J.F.; Bruschy-fonseca, A.; Lito, L.; Duarte, A.; Melo-cristino, J.; Caneiras, C. Whole-Genome Sequencing Enables Molecular Characterization of Non-Clonal Group 258 High-Risk Clones (ST13, ST17, ST147 and ST307) Among Carbapenem-Resistant Klebsiella pneumoniae From a Tertiary University Hospital Centre in Portugal. Microorganisms 2022, 10, 416. [Google Scholar] [CrossRef]
- Marcade, G.; Brisse, S.; Bialek, S.; Marcon, E.; Leflon-Guibout, V.; Passet, V.; Moreau, R.; Nicolas-Chanoine, M.H. The emergence of multidrug-resistant Klebsiella pneumoniae of international clones ST13, ST16, ST35, ST48 and ST101 in a teaching hospital in the Paris region. Epidemiol. Infect. 2013, 141, 1705–1712. [Google Scholar] [CrossRef]
- Ciacci, N.; D’andrea, M.M.; Marmo, P.; Demattè, E.; Amisano, F.; Di Pilato, V.; Fraziano, M.; Lupetti, P.; Rossolini, G.M.; Thaller, M.C. Characterization of vB_Kpn_F48, a Newly Discovered Lytic Bacteriophage for Klebsiella pneumoniae of Sequence Type 101. Viruses 2018, 10, 482. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.R.; Lin, T.L.; Pan, Y.J.; Hsieh, P.F.; Wang, J.T. Isolation of a Bacteriophage Specific for a New Capsular Type of Klebsiella pneumoniae and Characterization of Its Polysaccharide Depolymerase. PLoS ONE 2013, 8, e70092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Sheng, Y.; Ma, R.; Xu, M.; Liu, F.; Qin, R.; Zhu, M.; Zhu, X.; He, P. Identification of a Depolymerase Specific for k64-serotype Klebsiella pneumoniae: Potential Applications in Capsular Typing and Treatment. Antibiotics 2021, 10, 144. [Google Scholar] [CrossRef]
- Lin, T.L.; Hsieh, P.F.; Huang, Y.T.; Lee, W.C.; Tsai, Y.T.; Su, P.A.; Pan, Y.J.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Isolation of a Bacteriophage and Its Depolymerase Specific for K1 capsule of Klebsiella pneumoniae: Implication in Typing and Treatment. J. Infect. Dis. 2014, 210, 1734–1744. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.J.; Lin, T.L.; Chen, C.C.; Tsai, Y.T.; Cheng, Y.H.; Chen, Y.Y.; Hsieh, P.F.; Lin, Y.T.; Wang, J.T. Klebsiella Phage ΦK64-1 Encodes Multiple Depolymerases for Multiple Host Capsular Types. J. Virol. 2017, 91, e02457-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, J.; Fu, H.; Mu, Y.; Sun, Y.; Xu, Y.; Xiu, Z. A Klebsiella pneumoniae bacteriophage and its effect on 1,3-propanediol fermentation. Process Biochem. 2016, 51, 1323–1330. [Google Scholar] [CrossRef]
- Gorodnichev, R.; Kornienko, M.; Kupstov, N.S.; Malakhova, M.; Bespiatykh, D.; Veselovky, V.; Shitikov, E.A.; Ilina, E. Molecular Genetic Characterization Of Three New Klebsiella pneumoniae Bacteriophages Suitable For Phage Therapy. Med. Extrem. Situations 2021, 3, 84–90. [Google Scholar] [CrossRef]
- Eriksson, H.; Maciejewska, B.; Latka, A.; Majkowska-Skrobek, G.; Hellstrand, M.; Melefors, Ö.; Wang, J.T.; Kropinski, A.M.; Drulis-Kawa, Z.; Nilsson, A.S. A Suggested New Bacteriophage Genus, “Kp34likevirus”, within the Autographivirinae Subfamily of Podoviridae. Viruses 2015, 7, 1804–1822. [Google Scholar] [CrossRef] [PubMed]
- Brolund, A.; Hæggman, S.; Edquist, P.J.; Gezelius, L.; Olsson-Liljequist, B.; Wisell, K.T.; Giske, C.G. The DiversiLab system versus pulsed-field gel electrophoresis: Characterisation of extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae. J. Microbiol. Methods 2010, 83, 224–230. [Google Scholar] [CrossRef]
| 1st Injection (10 µL) | 2nd Injection (10 µL) | ||
|---|---|---|---|
| Experiment with MOI 100 and MOI 10 | 1 | PBS | PBS |
| 2 | QAMH130326/0185: 104 (CFU) | PBS | |
| 3 | QAMH130326/0185: 104 (CFU) | vB_KpnA_K3-ULINTkp1 (or kp2): 105 (PFUl) | |
| 4 | QAMH130326/0185: 104 (CFU) | vB_KpnA_K3-ULINTkp1 (or kp2): 106 (PFU) | |
| 5 | PBS | vB_KpnA_K3-ULINTkp1 (or kp2): 106 (PFU) | |
| Experiment with MOI 1000 | 1 | PBS | PBS |
| 2 | QAMH130326/0185: 104 (CFU) | PBS | |
| 3 | QAMH130326/0185: 104 (CFU) | vB_KpnA_K3-ULINTkp1: 107 (PFU/10 µL) | |
| 4 | PBS | vB_KpnA_K3-ULINTkp1: 107 (PFU) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laforêt, F.; Antoine, C.; Blasdel Reuter, B.; Detilleux, J.; Pirnay, J.-P.; Brisse, S.; Fall, A.; Duprez, J.-N.; Delcenserie, V.; Thiry, D. In Vitro and In Vivo Assessments of Two Newly Isolated Bacteriophages against an ST13 Urinary Tract Infection Klebsiella pneumoniae. Viruses 2022, 14, 1079. https://doi.org/10.3390/v14051079
Laforêt F, Antoine C, Blasdel Reuter B, Detilleux J, Pirnay J-P, Brisse S, Fall A, Duprez J-N, Delcenserie V, Thiry D. In Vitro and In Vivo Assessments of Two Newly Isolated Bacteriophages against an ST13 Urinary Tract Infection Klebsiella pneumoniae. Viruses. 2022; 14(5):1079. https://doi.org/10.3390/v14051079
Chicago/Turabian StyleLaforêt, Fanny, Céline Antoine, Bob Blasdel Reuter, Johann Detilleux, Jean-Paul Pirnay, Sylvain Brisse, Abdoulaye Fall, Jean-Noël Duprez, Véronique Delcenserie, and Damien Thiry. 2022. "In Vitro and In Vivo Assessments of Two Newly Isolated Bacteriophages against an ST13 Urinary Tract Infection Klebsiella pneumoniae" Viruses 14, no. 5: 1079. https://doi.org/10.3390/v14051079
APA StyleLaforêt, F., Antoine, C., Blasdel Reuter, B., Detilleux, J., Pirnay, J.-P., Brisse, S., Fall, A., Duprez, J.-N., Delcenserie, V., & Thiry, D. (2022). In Vitro and In Vivo Assessments of Two Newly Isolated Bacteriophages against an ST13 Urinary Tract Infection Klebsiella pneumoniae. Viruses, 14(5), 1079. https://doi.org/10.3390/v14051079







