Palmitoylethanolamide (PEA) Inhibits SARS-CoV-2 Entry by Interacting with S Protein and ACE-2 Receptor
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Methodology
2.1.1. Protein Preparation
2.1.2. Molecular Docking Studies
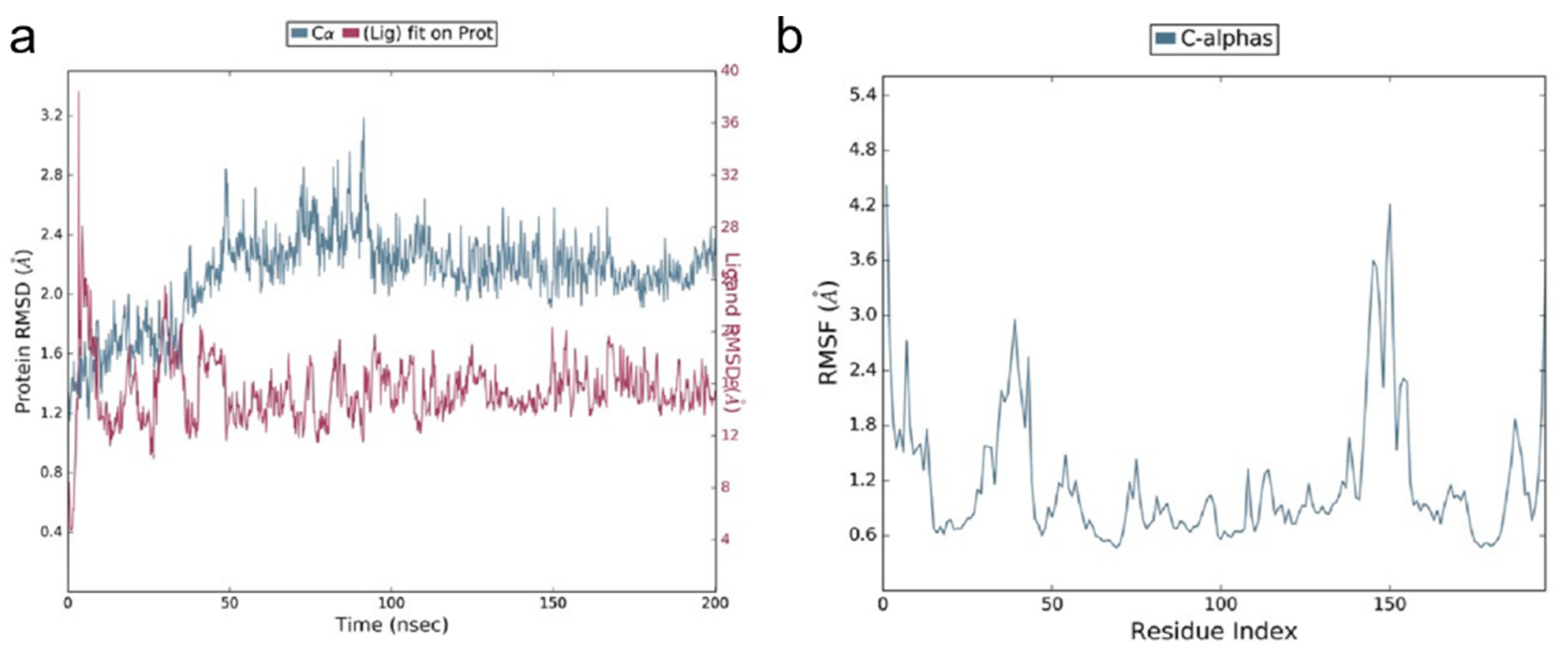
2.1.3. Molecular Dynamic Simulations (MDS)
2.1.4. Cell Culture and Treatments
2.1.5. Infections
2.1.6. Lentiviral Vectors
2.1.7. RNA Extraction and Real-Time PCR
2.1.8. Virus Yield Reduction Assay
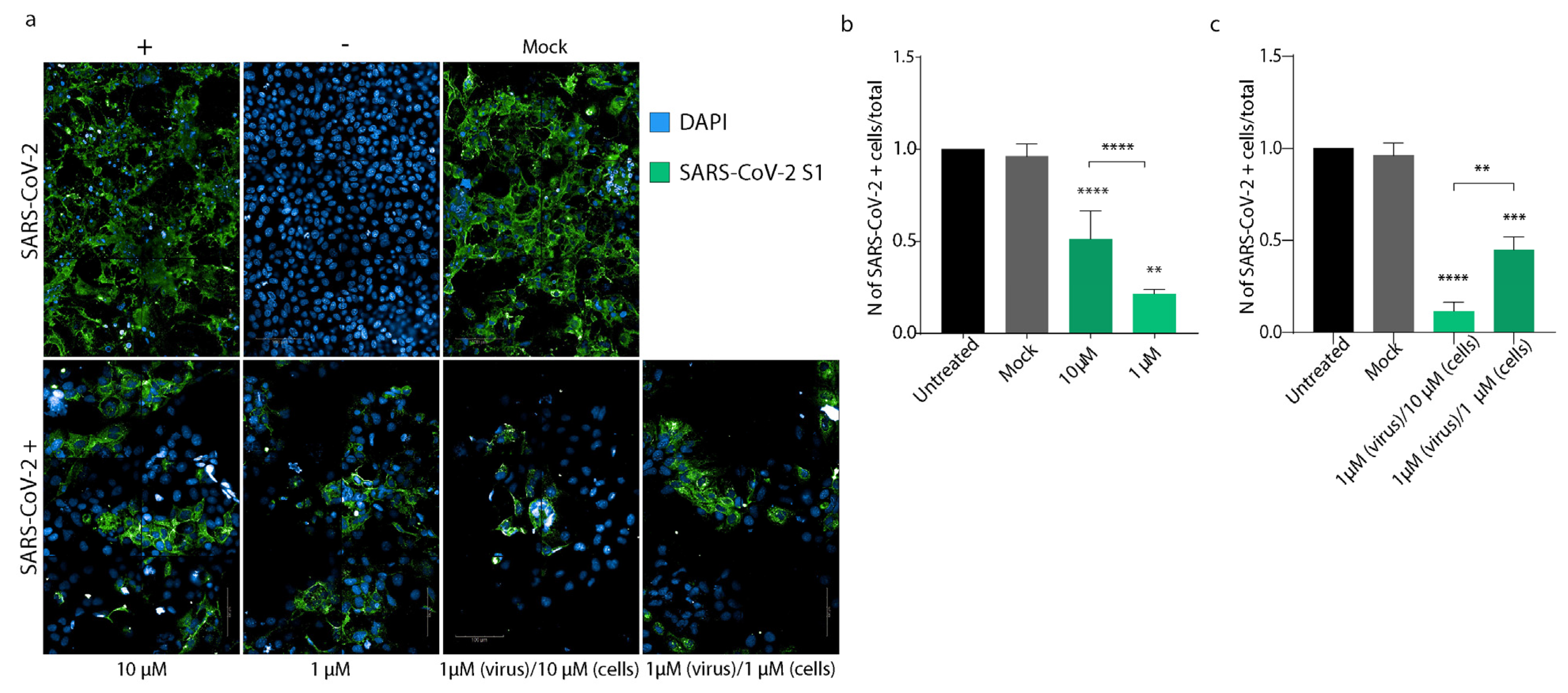
2.1.9. High-Content Confocal Imaging
2.1.10. Western Blot Analysis
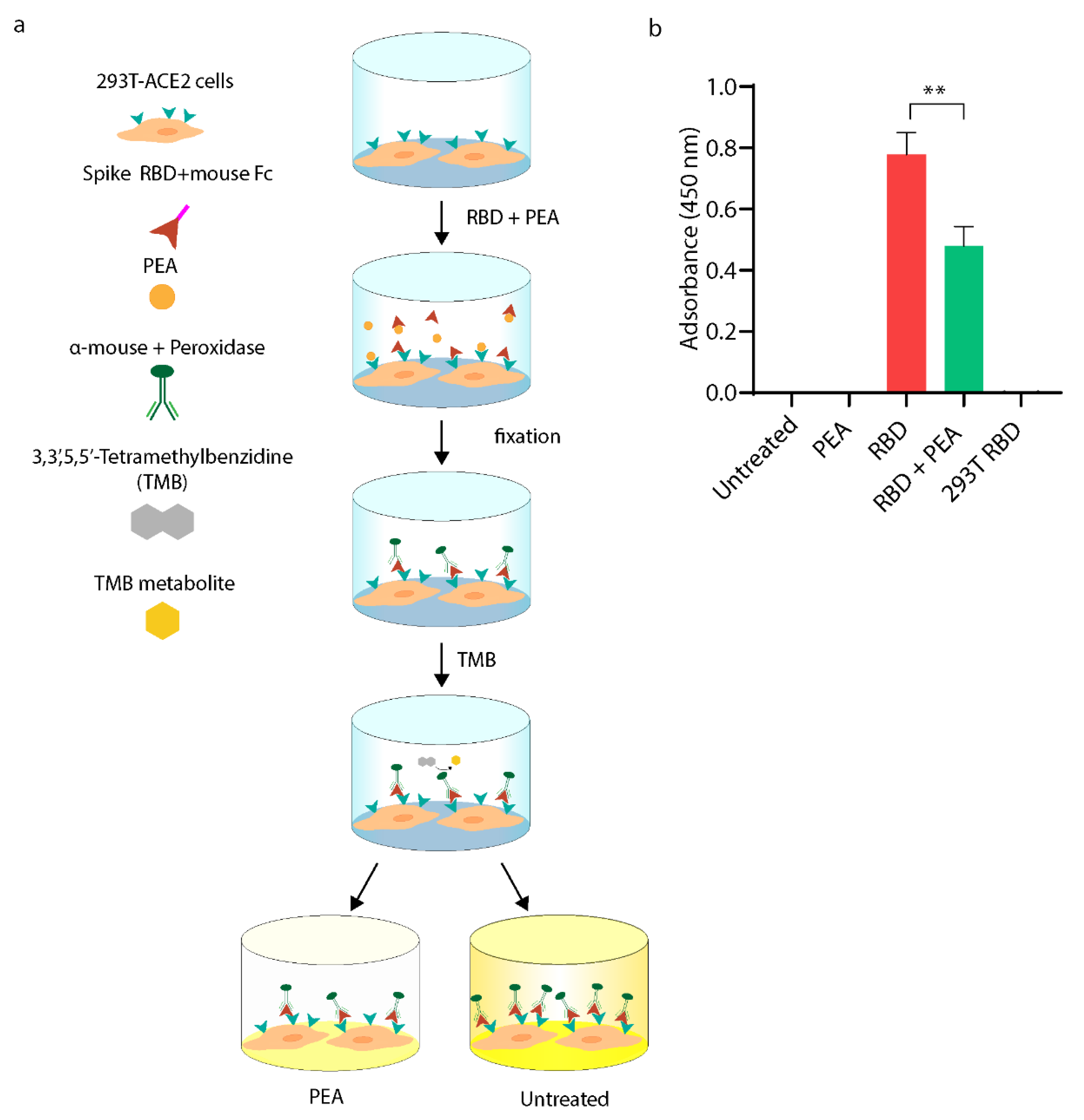
2.1.11. Cell-Based ELISA Assay
2.1.12. Data Analysis
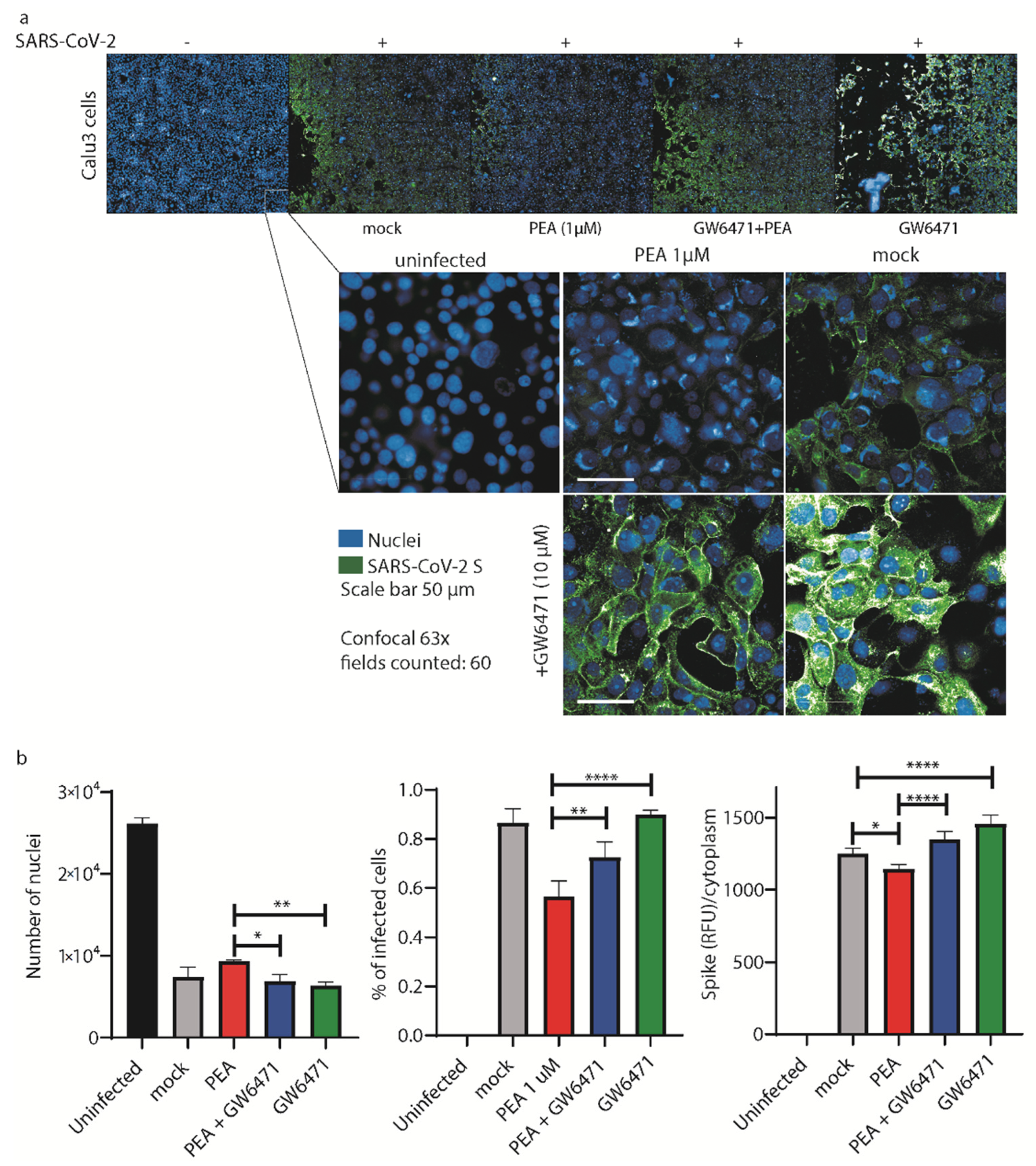
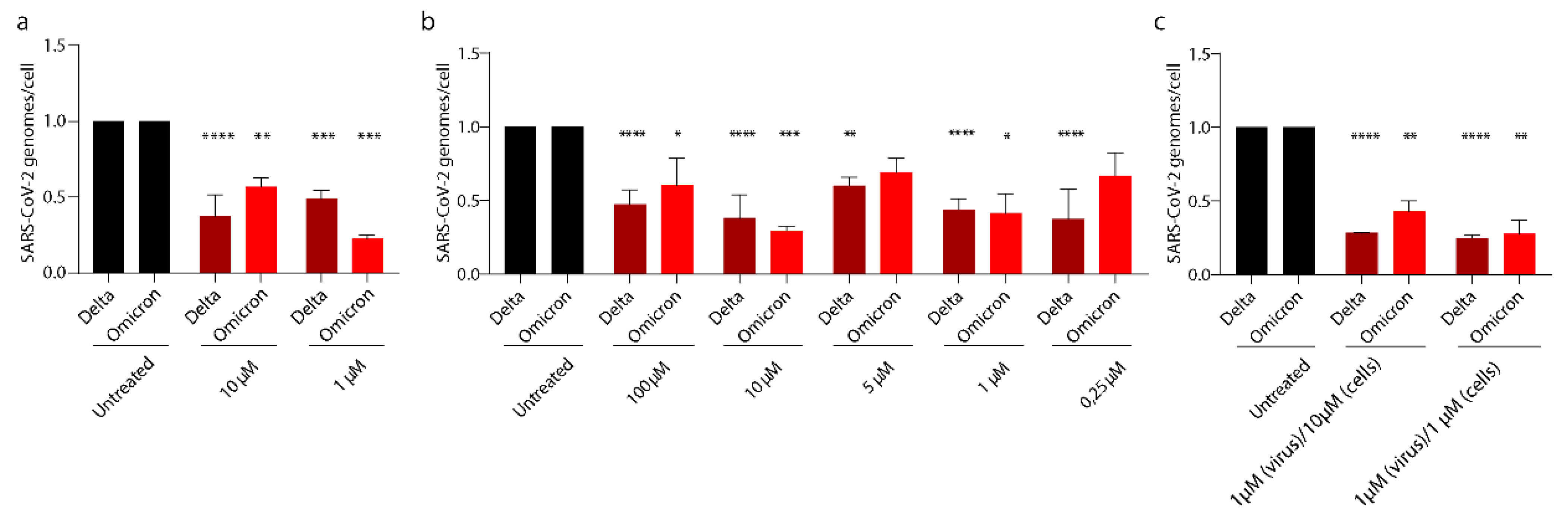
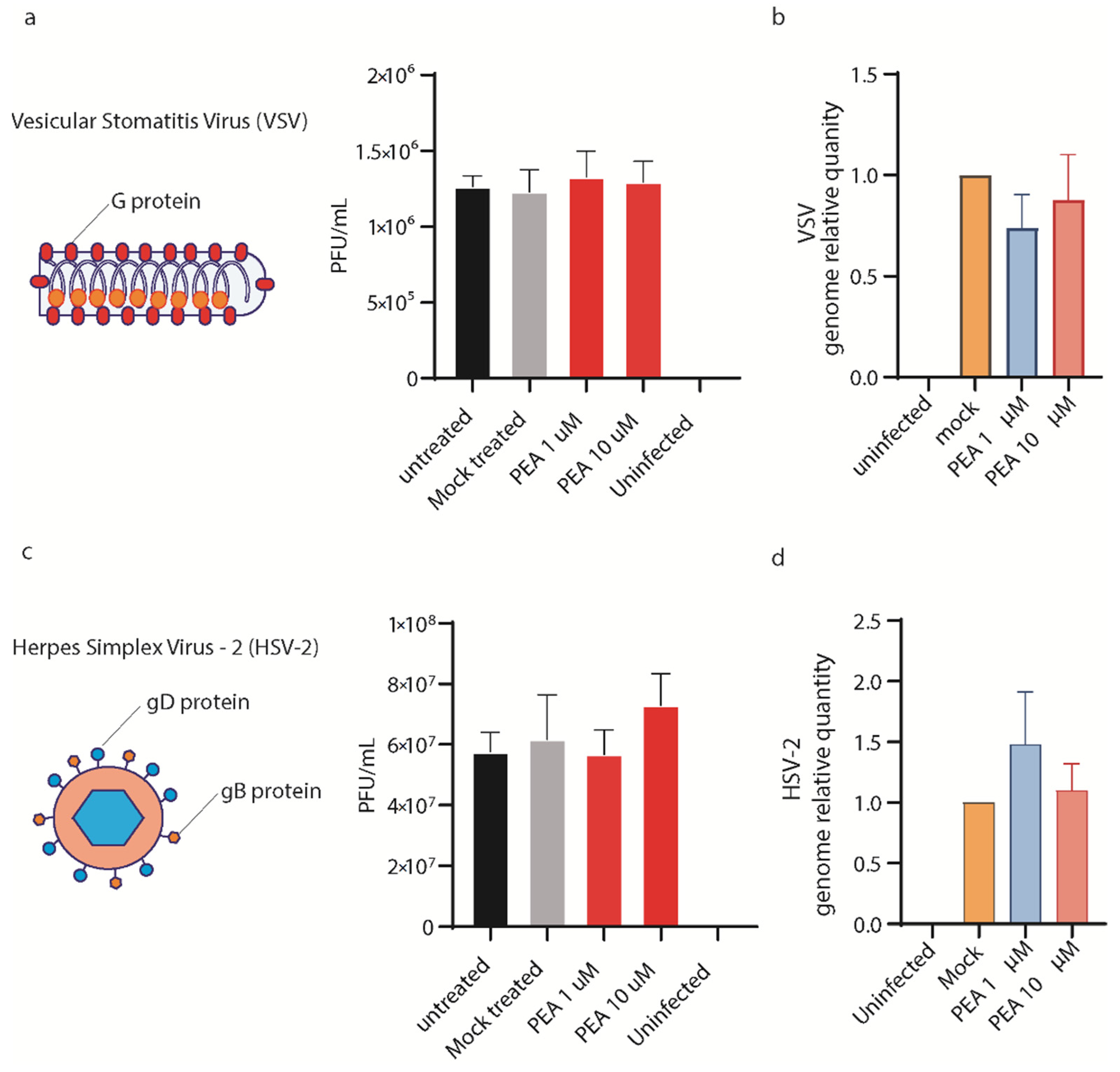
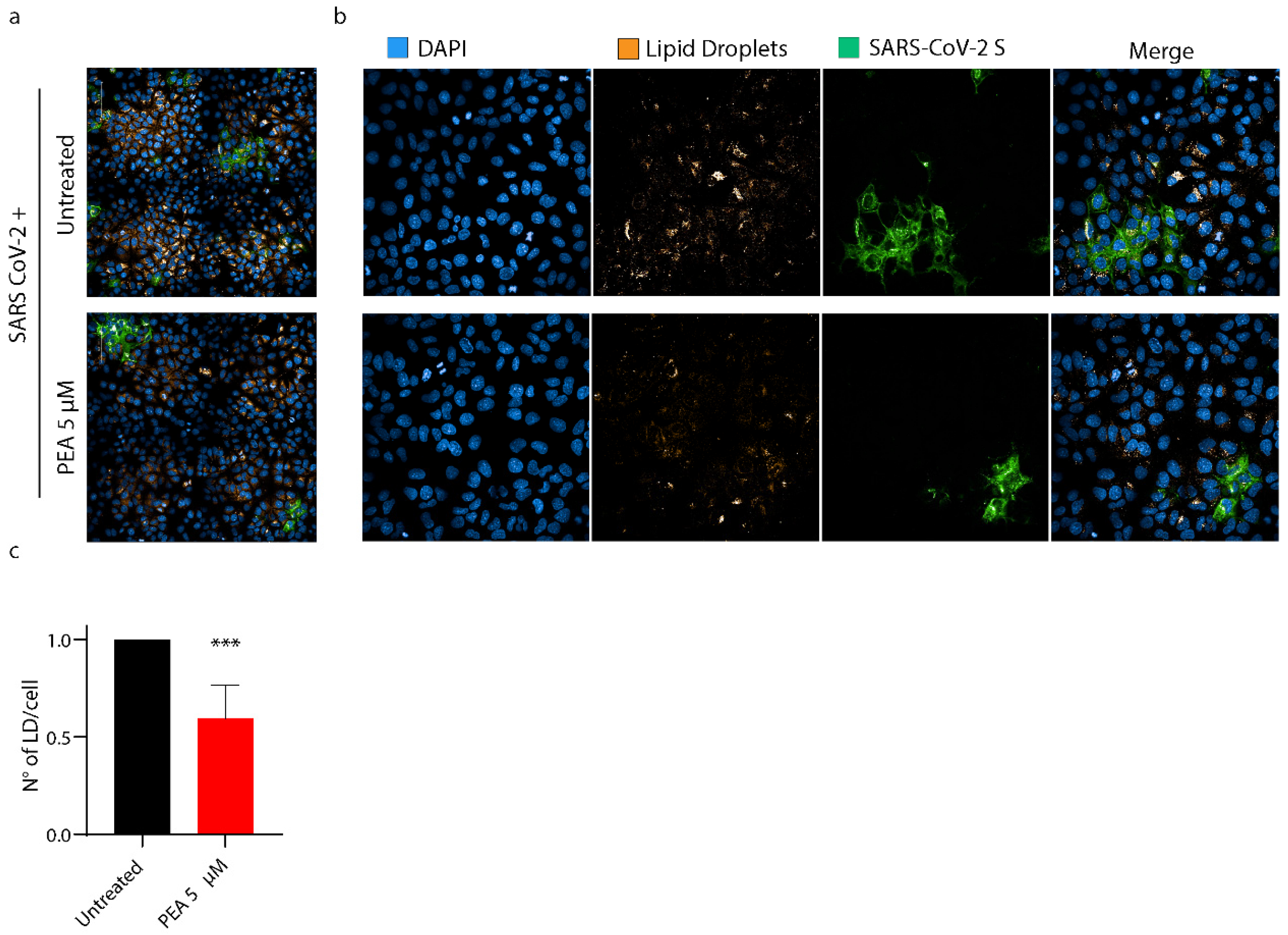
3. Results
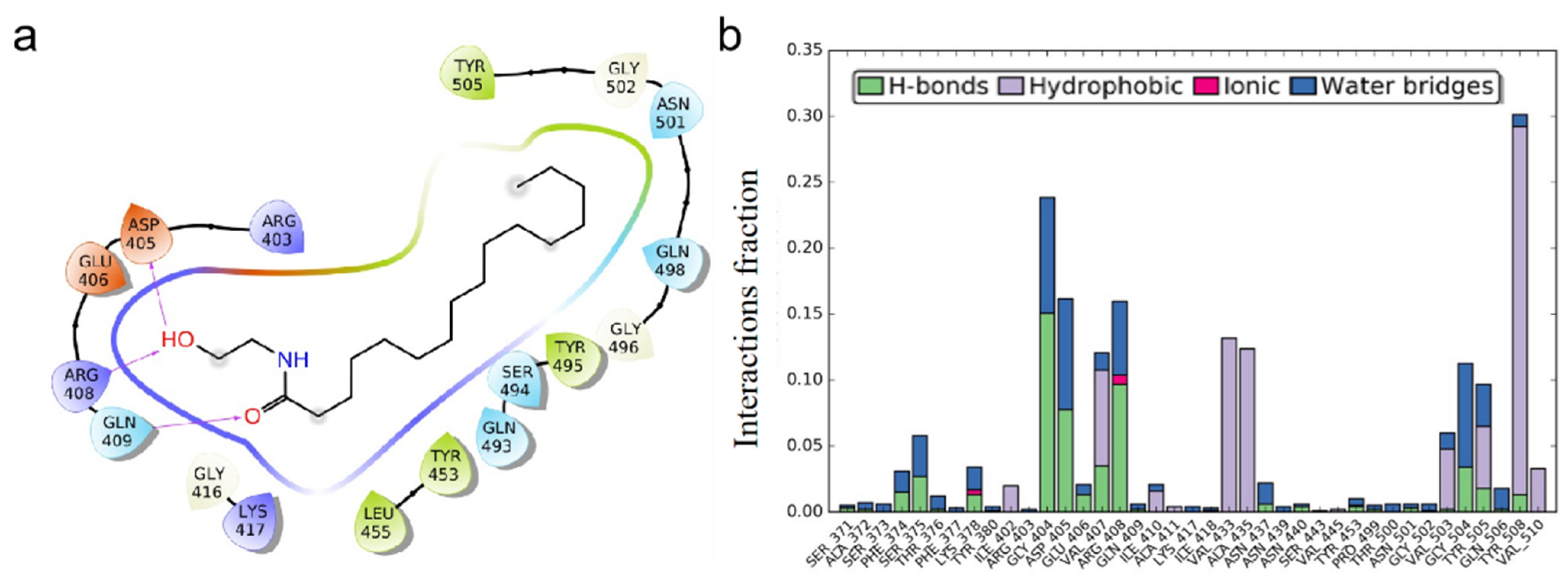
3.1. Molecular Docking Studies of ACE2 Receptor with PEA
3.2. PEA Decreases SARS-CoV-2 RBD Binding to ACE2 Receptor
3.3. PEA Reduces S-Pseudotyped Lentiviral Vector Transduction Efficiency
3.4. PEA Decreases SARS-CoV-2 Infectivity
3.5. PEA Exerts Its Antiviral Activity on SARS-CoV-2 Emerging Variants
3.6. PEA Does Not Block VSV and HSV-2 Entry in Huh-7 Cells
3.7. PEA Decreases the Amount of Intracellular Lipid Droplets
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García, L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020, 11, 1441. [Google Scholar] [CrossRef] [PubMed]
- Mahmudpour, M.; Roozbeh, J.; Keshavarz, M.; Farrokhi, S.; Nabipour, I. COVID-19 Cytokine Storm: The Anger of Inflammation. Cytokine 2020, 133, 155151. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F. The Trinity of COVID-19: Immunity, Inflammation and Intervention. Nat. Rev. Immunol. 2019, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical Course and Outcomes of Critically Ill Patients with SARS-CoV-2 Pneumonia in Wuhan, China: A Single-Centered, Retrospective, Observational Study. Lancet Respir. Med. 2020, 8, P475–P481. [Google Scholar] [CrossRef] [Green Version]
- Grant, R.A.; Morales-Nebreda, L.; Markov, N.S.; Swaminathan, S.; Querrey, M.; Guzman, E.R.; Abbott, D.A.; Donnelly, H.K.; Donayre, A.; Goldberg, I.A.; et al. Circuits between Infected Macrophages and T Cells in SARS-CoV-2 Pneumonia. Nature 2021, 590, 635. [Google Scholar] [CrossRef] [PubMed]
- AminJafari, A.; Ghasemi, S. The Possible of Immunotherapy for COVID-19: A Systematic Review. Int. Immunopharmacol. 2020, 83, 106455. [Google Scholar] [CrossRef]
- Magro, G. COVID-19: Review on Latest Available Drugs and Therapies against SARS-CoV-2. Coagulation and Inflammation Cross-Talking. Virus Res. 2020, 286, 198070. [Google Scholar] [CrossRef]
- Tufan, A.; Avanoğlu Güler, A.; Matucci-Cerinic, M. COVID-19, Immune System Response, Hyperinflammation and Repurposing Antirheumatic Drugs. Turk. J. Med. Sci. 2020, 50, 620–632. [Google Scholar] [CrossRef]
- Mirsaeidi, M.; Quartuccio, L.; Wewers, M.; Mutlu, G.M.; Kimmig, L.M.; Wu, D.; Gold, M.; Pettit, N.N.; Pitrak, D.; Mueller, J.; et al. IL-6 Inhibition in Critically Ill COVID-19 Patients Is Associated with Increased Secondary Infections. Front. Med. 2020, 7, 583897. [Google Scholar] [CrossRef]
- Pettit, N.N.; Nguyen, C.T.; Mutlu, G.M.; Wu, D.; Kimmig, L.; Pitrak, D.; Pursell, K. Late Onset Infectious Complications and Safety of Tocilizumab in the Management of COVID-19. J. Med. Virol. 2021, 93, 1459–1464. [Google Scholar] [CrossRef]
- Butler, M.J.; Barrientos, R.M. The Impact of Nutrition on COVID-19 Susceptibility and Long-Term Consequences. Brain Behav. Immun. 2020, 87, 53–54. [Google Scholar] [CrossRef] [PubMed]
- Zabetakis, I.; Lordan, R.; Norton, C.; Tsoupras, A. COVID-19: The Inflammation Link and the Role of Nutrition in Potential Mitigation. Nutrients 2020, 12, 1466. [Google Scholar] [CrossRef] [PubMed]
- Abedi, V.; Vlasova, A.N.; Cena, H.; Chieppa, M. Coronavirus Disease (COVID-19-SARS-CoV-2) and Nutrition: Is Infection in Italy Suggesting a Connection? Front. Immunol. 2020, 1, 944. [Google Scholar] [CrossRef]
- Stefan, N.; Birkenfeld, A.L.; Schulze, M.B.; Ludwig, D.S. Obesity and Impaired Metabolic Health in Patients with COVID-19. Nat. Rev. Endocrinol. 2020, 16, 341–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muscogiuri, G.; Barrea, L.; Savastano, S.; Colao, A. Nutritional Recommendations for CoVID-19 Quarantine. Eur. J. Clin. Nutr. 2020, 74, 850–851. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.L.R.; Sousa, N.W.A.; Oliveira Carvalho, V. What to Do When the Choice Is No Choice at All? A Critical View on Nutritional Recommendations for CoVID-19 Quarantine. Eur. J. Clin. Nutr. 2020, 74, 1488–1489. [Google Scholar] [CrossRef]
- Sharma, D.; Kunamneni, A. Recent Progress in the Repurposing of Drugs/Molecules for the Management of COVID-19. Expert Rev. Anti-Infective Ther. 2021, 19, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Veeramachaneni, G.K.; Thunuguntla, V.B.S.C.; Bobbillapati, J.; Bondili, J.S. Structural and Simulation Analysis of Hotspot Residues Interactions of SARS-CoV 2 with Human ACE2 Receptor. J. Biomol. Struct. Dyn. 2021, 39, 4015–4025. [Google Scholar] [CrossRef] [PubMed]
- Scherma, M.; Masia, P.; Satta, V.; Fratta, W.; Fadda, P.; Tanda, G. Brain Activity of Anandamide: A Rewarding Bliss? Acta Pharmacol. Sin. 2019, 40, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Sasso, O.; Summa, M.; Armirotti, A.; Pontis, S.; de Mei, C.; Piomelli, D. The N-Acylethanolamine Acid Amidase Inhibitor ARN077 Suppresses Inflammation and Pruritus in a Mouse Model of Allergic Dermatitis. J. Investig. Dermatol. 2018, 138, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Keppel Hesselink, J.M.; De Boer, T.; Witkamp, R.F. Palmitoylethanolamide: A Natural Body-Own Anti-Inflammatory Agent, Effective and Safe against Influenza and Common Cold. Int. J. Inflamm. 2013, 2013, 151028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clayton, P.; Hill, M.; Bogoda, N.; Subah, S.; Venkatesh, R. Palmitoylethanolamide: A Natural Compound for Health Management. Int. J. Mol. Sci. 2021, 22, 5305. [Google Scholar] [CrossRef] [PubMed]
- Pesce, M.; Seguella, L.; Cassarano, S.; Aurino, L.; Sanseverino, W.; Lu, J.; Corpetti, C.; Del Re, A.; Vincenzi, M.; Sarnelli, G.; et al. Phytotherapics in COVID19: Why Palmitoylethanolamide? Phytother. Res. 2021, 35, 2514–2522. [Google Scholar] [CrossRef] [PubMed]
- Noce, A.; Albanese, M.; Marrone, G.; Di Lauro, M.; Pietroboni Zaitseva, A.; Palazzetti, D.; Guerriero, C.; Paolino, A.; Pizzenti, G.; Di Daniele, F.; et al. Ultramicronized Palmitoylethanolamide (Um-PEA): A New Possible Adjuvant Treatment in COVID-19 Patients. Pharmaceuticals 2021, 14, 336. [Google Scholar] [CrossRef]
- Dias, S.S.G.; Soares, V.C.; Ferreira, A.C.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Teixeira, L.; Nunes da Silva, M.A.; Barreto, E.; Mattos, M.; et al. Lipid Droplets Fuel SARS-CoV-2 Replication and Production of Inflammatory Mediators. PLoS Pathog. 2020, 16, e1009127. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [Green Version]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters; Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Lai, M.; Iacono, E.; Spezia, P.G.; Lottini, G.; La Rocca, V.; Quaranta, P.; Pistello, M.; Freer, G. A Low-Cost Simple Test for Weekly Detection of Mycoplasma Hyorhinis and Arginini Contaminations in Cell Cultures and Viral Preparations. J. Virol. Methods 2022, 299, 114327. [Google Scholar] [CrossRef]
- Lai, M.; Amato, R.; La Rocca, V.; Bilgin, M.; Freer, G.; Spezia, P.; Quaranta, P.; Piomelli, D.; Pistello, M. Acid Ceramidase Controls Apoptosis and Increases Autophagy in Human Melanoma Cells Treated with Doxorubicin. Sci. Rep. 2021, 11, 11221. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; La Rocca, V.; Amato, R.; Freer, G.; Costa, M.; Spezia, P.G.; Quaranta, P.; Lombardo, G.; Piomelli, D.; Pistello, M. Ablation of Acid Ceramidase Impairs Autophagy and Mitochondria Activity in Melanoma Cells. Int. J. Mol. Sci. 2021, 22, 3247. [Google Scholar] [CrossRef] [PubMed]
- Freer, G.; Lai, M.; Quaranta, P.; Spezia, P.G.; Pistello, M. Evolution of Viruses and the Emergence of SARS-CoV-2 Variants. N. Microbiol. 2021, 44, 191–204. [Google Scholar]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta Variant of SARS-CoV-2: A Comparative Computational Study of Spike Protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef]
- Chen, X.; Saccon, E.; Appelberg, K.S.; Mikaeloff, F.; Rodriguez, J.E.; Vinhas, B.S.; Frisan, T.; Végvári, Á.; Mirazimi, A.; Neogi, U.; et al. Type-I Interferon Signatures in SARS-CoV-2 Infected Huh7 Cells. Cell Death Discov. 2021, 7, 114. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.-W.; Yuen, T.T.-T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.C.-Y.; Tsang, J.O.-L.; Huang, X.; et al. Comparative Tropism, Replication Kinetics, and Cell Damage Profiling of SARS-CoV-2 and SARS-CoV with Implications for Clinical Manifestations, Transmissibility, and Laboratory Studies of COVID-19: An Observational Study. Lancet Microbe 2020, 1, e14–e23. [Google Scholar] [CrossRef]
- Leier, H.C.; Messer, W.B.; Tafesse, F.G. Lipids and Pathogenic Flaviviruses: An Intimate Union. PLoS Pathog. 2018, 14, e1006952. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, D.; Luongo, L.; Cipriano, M.; Palazzo, E.; Cinelli, M.P.; de Novellis, V.; Maione, S.; Iuvone, T. Palmitoylethanolamide Reduces Granuloma-Induced Hyperalgesia by Modulation of Mast Cell Activation in Rats. Mol. Pain 2011, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Gigli, S.; Seguella, L.; Pesce, M.; Bruzzese, E.; D’Alessandro, A.; Cuomo, R.; Steardo, L.; Sarnelli, G.; Esposito, G. Cannabidiol Restores Intestinal Barrier Dysfunction and Inhibits the Apoptotic Process Induced by Clostridium Difficile Toxin A in Caco-2 Cells. United Eur. Gastroenterol. J. 2017, 5, 1108–1115. [Google Scholar] [CrossRef] [Green Version]
- Matias, I.; Di Marzo, V. Endocannabinoids and the Control of Energy Balance. Trends Endocrinol. Metab. 2007, 18, 27–37. [Google Scholar] [CrossRef]
- Pesce, M.; Esposito, G.; Sarnelli, G. Endocannabinoids in the Treatment of Gasytrointestinal Inflammation and Symptoms. Curr. Opin. Pharmacol. 2018, 43, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Pesce, M.; Seguella, L.; Sanseverino, W.; Lu, J.; Corpetti, C.; Sarnelli, G. The Potential of Cannabidiol in the COVID-19 Pandemic. Br. J. Pharmacol. 2020, 177, 4967–4970. [Google Scholar] [CrossRef] [PubMed]
- Suardíaz, M.; Estivill-Torrús, G.; Goicoechea, C.; Bilbao, A.; Rodríguez de Fonseca, F. Analgesic Properties of Oleoylethanolamide (OEA) in Visceral and Inflammatory Pain. Pain 2007, 133, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.M.; Kirkham, T.C. Anandamide Induces Overeating: Mediation by Central Cannabinoid (CB1) Receptors. Psychopharmacology 1999, 143, 315–317. [Google Scholar] [CrossRef]
- Costiniuk, C.T.; Jenabian, M.A. Acute Inflammation and Pathogenesis of SARS-CoV-2 Infection: Cannabidiol as a Potential Anti-Inflammatory Treatment? Cytokine Growth Factor Rev. 2020, 53, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Onaivi, E.S.; Sharma, V. Cannabis for COVID-19: Can Cannabinoids Quell the Cytokine Storm? Future Sci. OA 2020, 6, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Tavakoli-Yaraki, M.; Salimi, V. Opioids/Cannabinoids as a Potential Therapeutic Approach in COVID-19 Patients. Expert Rev. Respir. Med. 2020, 14, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Das, S. The CBD Dialectic in Mental Health: Benign and Powerful? Psychiatr. Serv. 2020, 71, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, S.; Roshanravan, N.; Tutunchi, H.; Ostadrahimi, A.; Pouraghaei, M.; Kafil, B. Oleoylethanolamide, A Bioactive Lipid Amide, as A Promising Treatment Strategy for Coronavirus/COVID-19. Arch. Med. Res. 2020, 51, 464–467. [Google Scholar] [CrossRef]
- Cabral, G.A.; Ferreira, G.A.; Jamerson, M.J. Endocannabinoids. In Handbook of Experimental Pharmacology; Pertwee, R.G., Ed.; Springer International Publishing: Cham, Switzerland, 2015; Volume 231, ISBN 978-3-319-20824-4. [Google Scholar]
- Petrosino, S.; Iuvone, T.; Di Marzo, V. N-Palmitoyl-Ethanolamine: Biochemistry and New Therapeutic Opportunities. Biochimie 2010, 92, 724–727. [Google Scholar] [CrossRef]
- Heaton, N.S.; Randall, G. Multifaceted Roles for Lipids in Viral Infection. Trends Microbiol. 2011, 19, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Teissier, É.; Pécheur, E.I. Lipids as Modulators of Membrane Fusion Mediated by Viral Fusion Proteins. Eur. Biophys. J. 2007, 36, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Del Re, A.; Corpetti, C.; Pesce, M.; Seguella, L.; Steardo, L.; Palenca, I.; Rurgo, S.; De Conno, B.; Sarnelli, G.; Esposito, G. Ultramicronized Palmitoylethanolamide Inhibits NLRP3 Inflammasome Expression and Pro-Inflammatory Response Activated by SARS-CoV-2 Spike Protein in Cultured Murine Alveolar Macrophages. Metabolites 2021, 11, 592. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonnesu, R.; Thunuguntla, V.B.S.C.; Veeramachaneni, G.K.; Bondili, J.S.; La Rocca, V.; Filipponi, C.; Spezia, P.G.; Sidoti, M.; Plicanti, E.; Quaranta, P.; et al. Palmitoylethanolamide (PEA) Inhibits SARS-CoV-2 Entry by Interacting with S Protein and ACE-2 Receptor. Viruses 2022, 14, 1080. https://doi.org/10.3390/v14051080
Fonnesu R, Thunuguntla VBSC, Veeramachaneni GK, Bondili JS, La Rocca V, Filipponi C, Spezia PG, Sidoti M, Plicanti E, Quaranta P, et al. Palmitoylethanolamide (PEA) Inhibits SARS-CoV-2 Entry by Interacting with S Protein and ACE-2 Receptor. Viruses. 2022; 14(5):1080. https://doi.org/10.3390/v14051080
Chicago/Turabian StyleFonnesu, Rossella, Venkata Bala Sai Chaitanya Thunuguntla, Ganesh Kumar Veeramachaneni, Jayakumar Singh Bondili, Veronica La Rocca, Carolina Filipponi, Pietro Giorgio Spezia, Maria Sidoti, Erika Plicanti, Paola Quaranta, and et al. 2022. "Palmitoylethanolamide (PEA) Inhibits SARS-CoV-2 Entry by Interacting with S Protein and ACE-2 Receptor" Viruses 14, no. 5: 1080. https://doi.org/10.3390/v14051080
APA StyleFonnesu, R., Thunuguntla, V. B. S. C., Veeramachaneni, G. K., Bondili, J. S., La Rocca, V., Filipponi, C., Spezia, P. G., Sidoti, M., Plicanti, E., Quaranta, P., Freer, G., Pistello, M., Mathai, M. L., & Lai, M. (2022). Palmitoylethanolamide (PEA) Inhibits SARS-CoV-2 Entry by Interacting with S Protein and ACE-2 Receptor. Viruses, 14(5), 1080. https://doi.org/10.3390/v14051080






