Abstract
HIV-1 viral particle assembly occurs specifically at the plasma membrane and is driven primarily by the viral polyprotein Gag. Selective association of Gag with the plasma membrane is a key step in the viral assembly pathway, which is traditionally attributed to the MA domain. MA regulates specific plasma membrane binding through two primary mechanisms including: (1) specific interaction of the MA highly basic region (HBR) with the plasma membrane phospholipid phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2], and (2) tRNA binding to the MA HBR, which prevents Gag association with non-PI(4,5)P2 containing membranes. Gag multimerization, driven by both CA–CA inter-protein interactions and NC-RNA binding, also plays an essential role in viral particle assembly, mediating the establishment and growth of the immature Gag lattice on the plasma membrane. In addition to these functions, the multimerization of HIV-1 Gag has also been demonstrated to enhance its membrane binding activity through the MA domain. This review provides an overview of the mechanisms regulating Gag membrane binding through the MA domain and multimerization through the CA and NC domains, and examines how these two functions are intertwined, allowing for multimerization mediated enhancement of Gag membrane binding.
Keywords:
HIV-1; particle assembly; Gag; multimerization; membrane binding; tRNA binding; genomic RNA binding; capsid; nucleocapsid; matrix 1. Introduction
During the HIV-1 replication cycle, the processes of viral particle assembly and release are driven primarily by the structural protein Gag. The 55kDa Gag precursor polyprotein Pr55Gag contains matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains as well as two spacer peptides (SP1 and SP2) (Figure 1A top). Viral assembly begins with translation of the Gag protein from the unspliced viral mRNA in the cytoplasm of the infected cell. After translation, the Gag protein accumulates at the plasma membrane of the host cell, driven by the specific interaction of the MA domain with the plasma membrane phospholipid phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2]. Once Gag binds to the plasma membrane, inter-Gag interactions mediated by CA-CA and NC-RNA binding drive the formation of the immature lattice at the site of viral particle assembly. Here, viral genomic RNA (gRNA) is selectively packaged through the specific interaction of the packaging element (psi) with the NC domain of Gag, ensuring incorporation into the growing viral particle. Through addition of Gag monomers and dimers, this lattice grows, inducing membrane curvature at the site of viral assembly and the budding of the immature viral particle from the surface of the host cell. The host cell ESCRT machinery is recruited to the site of viral assembly by the p6 domain of Gag, which is responsible for the scission of the membranous neck and the release of the immature viral particle (HIV-1 particle assembly is reviewed in [1]). In this review, we will summarize current knowledge regarding the viral and cellular determinants that regulate two essential steps of HIV-1 particle assembly, i.e., Gag membrane binding and multimerization, and then discuss the relationships between the two steps as well as questions that remain to be answered.
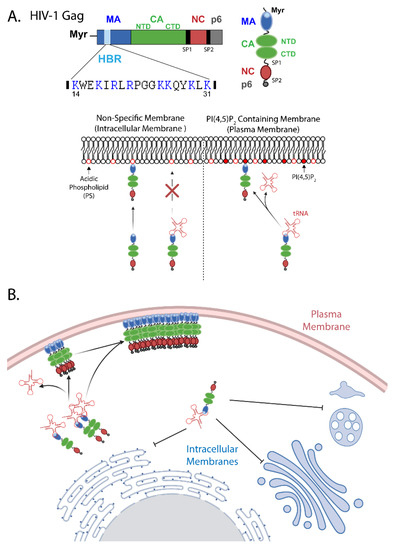
Figure 1.
Plasma membrane-specific binding of HIV-1 Gag. (A) Molecular determinants for Gag membrane binding. Top, the domain organization of HIV-1 Gag. Blue, matrix (MA); green, capsid (CA, N-terminal domain [NTD] and C-terminal domain [CTD]); red, nucleocapsid (NC); gray, p6; and black, spacer peptides 1 and 2 (SP1 and SP2). Additionally, the locations of the myristoyl moiety (Myr) and the highly basic region (HBR, light blue) are shown. The HBR amino acid sequence is shown with basic residues highlighted in blue. Bottom, host molecules that bind the MA HBR. Gag binds membranes through interactions between the HBR and acidic phospholipid headgroups. tRNA binding to the HBR prevents interaction with membranes that contain a prevalent acidic phospholipid, phosphatidylserine (PS), but allows for Gag binding to membranes containing phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2]. For clarity, the myristoyl moiety is not depicted. (B) A model that accounts for specific localization of Gag to the plasma membrane. tRNA-mediated inhibition of membrane binding prevents localization of Gag to intracellular membranes, which lack PI(4,5)P2, but allows for binding to the plasma membrane where PI(4,5)P2 is present.
2. Mechanisms Regulating HIV-1 Gag Membrane Binding
The membrane binding activity of HIV-1 Gag during assembly is attributed to the MA domain. The HIV-1 MA domain contains two major membrane binding signals, an N-terminal myristoylation (Myr) and a conserved stretch of basic amino acids [2,3,4,5,6,7,8,9,10] (Figure 1A top).
2.1. The Role Played by N-Terminal Myristate Moiety
The myristate moiety, which is added to the N-terminal glycine of the MA domain after the removal of the first methionine, contributes to MA membrane binding through hydrophobic interactions with the membrane bilayer. Substitutions of the MA N-terminal residues that prevent the acylation by myristoyl transferase drastically reduces the membrane binding of Gag and the production of viral particles [2,3,5,8,11,12]. While myristate is often found on membrane binding proteins, this modification alone is not expected to drive selective binding to the plasma membrane (reviewed in [13,14]).
2.2. The Role Played by the Highly Basic Region
In addition to myristoylation, early work determined that a polybasic sequence spanning MA residues 14–31 is also involved in membrane binding [9,10,15]. This highly basic region (HBR) forms a positively charged patch on the surface of the MA globular domain [15,16,17,18] and mediates membrane binding through electrostatic interactions with the acidic headgroups of phospholipids such as phosphatidylserine (PS) [5,19,20,21,22].
2.3. PI(4,5)P2 Binding and Plasma Membrane Specificity
HIV-1 particle assembly takes place at the plasma membrane [23,24]. Cell-based and in vitro studies have collectively demonstrated that a specific interaction of the MA HBR with the phospholipid phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2], which is present specifically in the inner leaflet of the plasma membrane, drives the selective binding of Gag to the plasma membrane in the host cell and hence particle assembly at this location [6,23,25,26,27,28,29]. When a phosphatase that depletes PI(4,5)P2 is ectopically expressed in HIV-expressing cells, Gag fails to bind membrane efficiently, resulting in a primarily cytosolic distribution, with a subset of cells displaying mislocalization of Gag to internal compartments instead of the plasma membrane. Consequently, HIV-1 particle release is drastically reduced in these cells. Suppression of host enzymes involved in PI(4,5)P2 generation has a similar effect [30,31]. Interestingly, induction of PI(4,5)P2-enriched endosomal structures also causes a defect in the specific localization, i.e., Gag accumulation at these intracellular vesicles, and a reduction in viral particle release [6].
PI(4,5)P2 is important not only for specific targeting of Gag to the plasma membrane, but also for stable association of Gag with this membrane once bound. Using a system that allows reversible depletion of PI(4,5)P2 at the plasma membrane, Mücksch et al. demonstrated that pre-assembled Gag lattices are detached from the plasma membrane upon depletion of PI(4,5)P2, disassembling back into monomers or lower level oligomers which remain assembly competent [32]. This suggests that even after the establishment of the immature lattice on the membrane, the HBR-PI(4,5)P2 interaction is required for the maintenance of the lattice, preventing the equilibrium of Gag membrane binding from shifting to a cytosolic distribution [32].
In vitro experiments demonstrate that Gag binds more efficiently to membranes containing PI(4,5)P2 than those containing other acidic phospholipids such as PS [25,33,34,35,36,37,38,39,40,41,42]. Increasing the proportion of non-PI(4,5)P2 acidic phospholipids to compensate for the greater negative charge of PI(4,5)P2 does not result in a similar increase in Gag membrane binding, suggesting that Gag recognition of PI(4,5)P2-containing membranes is not simply charge-based but likely involves some recognition of the PI(4,5)P2 headgroup structure [25,38,39,40]. Multiple studies have shown that the interaction of Gag with highly acidic head group of PI(4,5)P2 involves residues within the HBR [25,26,27,43]. This suggests that while MA HBR can mediate membrane binding through interaction with any acidic phospholipid (such as PS), specific recognition of the PI(4,5)P2 headgroup by residues within the HBR preferentially drives high affinity interaction of Gag with PI(4,5)P2-containing membranes.
2.4. RNA-Mediated Suppression of MA-Lipid Binding
In addition to acidic phospholipids, MA has also been shown to bind nucleic acids [27,34,35,44,45,46,47,48,49,50]. RNA binding to the MA domain involves regions that overlap with the site of PI(4,5)P2 interaction within the HBR [44,51,52,53,54]. While the RNA interaction with the HBR inhibits Gag binding to membranes containing non-PI(4,5)P2 acidic phospholipids such as PS, in the absence of RNA, Gag binds these membranes efficiently (Figure 1A bottom). Even in the presence of RNA, Gag retains the ability to bind membranes containing PI(4,5)P2 efficiently, suggesting that PI(4,5)P2 can out-compete RNA for binding to the MA HBR. It is important to note that even in the presence of PI(4,5)P2, the presence of RNA can partially inhibit Gag membrane binding, suggesting that the effect of RNA on Gag-acidic lipid interactions is not binary [25,34,35,55]. Together, these results suggest a hierarchy in which HIV-1 MA HBR binds to PI(4,5)P2 more strongly than RNA, which binds more strongly than other acidic phospholipids such as PS. Thus, while RNA binding to the MA HBR shields it from non-specific interaction with acidic phospholipid containing membranes, PI(4,5)P2 overcomes RNA-mediated inhibition and allows for efficient Gag membrane binding (Figure 1A bottom).
In cells, an RNA crosslinking study showed that tRNA was the major RNA species that binds HIV-1 MA in the cytosol [56]. In vitro experiments demonstrated that tRNA can block Gag binding to non-PI(4,5)P2-containing membranes at concentrations below what is present in the cytosol [42,55]. These observations suggest that in the cellular context tRNA is primarily responsible for the negative regulation of Gag membrane binding, preventing Gag association with intracellular membranes that lack PI(4,5)P2, while allowing for Gag accumulation specifically at the plasma membrane where PI(4,5)P2 is present (Figure 1B). This model is supported by cellular data demonstrating that mutations disrupting MA-RNA binding (K25, K26 and K29, K31 mutants) cause defects in the specific localization of Gag to the plasma membrane [53]. Additionally, HIV-1 Gag chimeras containing MA domains from other retroviruses that are sensitive to RNA-mediated inhibition localize specifically to the plasma membrane, while Gag containing RNA-insensitive MA domains localize promiscuously to intracellular membranes [33,37]. While these studies suggest that tRNA binding to MA HBR spatially regulates Gag membrane binding, it is also conceivable that tRNA imparts some temporal regulation, for example, by preventing pre-mature plasma membrane binding when Gag concentration is still low [57]. Interestingly, although full-length Gag overcomes tRNA-mediated inhibition and binds membranes if they contain PI(4,5)P2 [34], isolated MA domains do not efficiently bind PI(4,5)P2-containing membranes in the presence of tRNA [52]. These observations support the possibility that, in addition to the competition between PI(4,5)P2 and tRNA for binding to the basic residues of the HBR, the Gag domains downstream of MA play a role in regulating tRNA displacement from MA HBR.
3. CA-CA Interactions and Their Role in Immature Gag Lattice Structure
The interactions between adjacent CA domains of the HIV-1 Gag polyprotein are the driving force behind the viral assembly and the organization of the immature hexagonal Gag lattice. HIV-1 CA contains two sub-domains, the N-terminal domain (NTD) and the C-terminal domain (CTD), which play distinct roles in the immature lattice. Both the major homology region (MHR) within the CA CTD [58,59,60,61,62] and a six-helix bundle formed by the region spanning the C terminus of the CA CTD and the N terminus of SP1 contribute to the six-fold interaction interface at the center of the CA hexameric ring [63,64,65,66,67,68,69]. X-ray structures reveal that the cellular co-factor inositol hexakisphosphate (IP6) binds two rings of lysine residues at positions 158 and 227 of the HIV-1 CA domain, stabilizing the six-helix bundle in the immature Gag lattice, and mutations in these residues disrupt CA-interactions at this interface [70,71,72]. In addition to the six-fold interactions, residues within the CTD mediate the two-fold interface between CA domains [60,62,63,73,74]. Mutations within the CA-NTD (such as CA P38E, A42E, E45R) also cause defects in virus assembly [39,73], and residues within this sub-domain contribute to the three-fold CA interaction [62,75]. Together, the CA-NTD, CA-CTD, and SP1 regions cooperate to form an immature Gag lattice consisting of hexametric rings that assemble with one another at their two- and three-fold interfaces (illustrated in Figure 2).
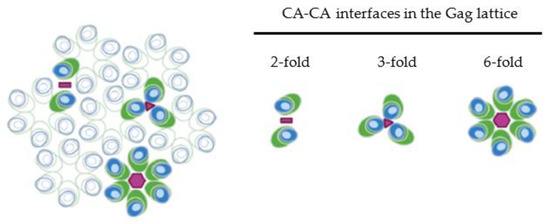
Figure 2.
Organization of MA and CA in the membrane-bound Gag lattice. Gag domains are depicted as in Figure 1 but only MA and CA are shown. Blue, MA; light blue, HBR; and green, CA. Three types of CA-CA interactions contributing to formation of immature Gag lattice are shown on the right. The two-fold, three-fold, and six-fold interaction interfaces are denoted by pink bar, triangle, and hexagon, respectively. Six-fold interactions involving the MHR and six-helix bundle (formed by CA-CTD and SP1 regions) form Gag hexamer subunits, with interactions at the two-fold and two-fold interfaces binding adjacent hexamer rings to one another. In the immature lattice, MA trimers form above the two-fold CA interaction interface, with an MA domain presumably contributed from each of three adjacent hexamer rings.
4. Enhancement of Gag Multimerization by NC-RNA Interaction
While the NC domain of HIV-1 Gag promotes selective packaging of the genomic RNA (gRNA) through its specific interaction with the psi element (reviewed in [76,77,78]), the RNA binding capacity of the NC domain also promotes efficient assembly of the immature lattice. The presence of nucleic acids promotes efficient in vitro assembly of Gag [79,80,81,82,83]. In cells, deletion or substitution of basic arginine and lysine residues within NC, which reduces NC-RNA binding, disrupts Gag–Gag interaction and particle assembly/release [84,85]. Together, these findings suggest that non-specific RNA binding by NC basic residues contribute to Gag multimerization and efficient particle assembly. Multiple studies have demonstrated that replacement of NC with heterologous domains mediating protein–protein interaction allows for efficient particle production regardless of the ability to bind RNA [86,87,88,89,90], indicating that the specific NC-RNA interaction is not required for efficient Gag multimerization. In the absence of packageable gRNA, full length HIV-1 Gag encapsidates cellular RNA species non-selectively [91], suggesting that non-specific RNA binding can also play a role in enhancing Gag assembly in the cellular environment.
It is likely that NC-RNA interactions enhance Gag multimerization through scaffolding of Gag assembly, with nucleic acids bridging multiple adjacent NC domains, facilitating inter-CA interactions. This effect has been observed in in vitro assembly reactions with short oligonucleotides (10–15 bases) promoting Gag assembly [81]. A similar effect is seen with RSV Gag, where short oligonucleotides (16 bases) promote assembly through dimerization of the Gag polyprotein [92,93]. In the context of Gag assembly in cells, single particle tracking PALM (photoactivated localization microscopy) and coarse grained computational modeling data suggest that longer gRNA serves as a scaffolding for Gag assembly, regulating the rate of Gag protein incorporation into the immature lattice as well as the recruitment and tethering of Gag assembly precursors to the membrane at the site of viral assembly [94,95]. Additionally, it has been observed that even extremely short oligonucleotides, which cannot bind multiple NC domains, may nucleate Gag assembly through the formation of nucleic acid-triggered NC-dimers, which enhance Gag dimerization more efficiently than CA-CA interactions [81,96].
In addition to these general mechanisms mediated by any RNA, specific interaction of Gag with packageable viral gRNA also contribute to efficient Gag multimerization. In cells, both Gag-viral gRNA interaction in the cytoplasm and trafficking of the gRNA to the plasma membrane are dependent on interactions with two zinc finger motifs, which play partially redundant roles [97]. Particle production is enhanced by the presence of packageable viral RNA in a manner reliant on specific recognition of packageable RNA by the Gag NC domain [98]. In vitro experiments demonstrate that Gag assembly is more efficient in the presence of HIV RNA containing clusters of unpaired G residues within the nucleocapsid interaction domain of the psi sequence (identified in [99]) compared to RNA constructs lacking these clusters [100]. Additionally, if assembly is seeded with complexes capable of high affinity NC-psi interaction, addition of bulk Gag that lacks specific high affinity interaction is sufficient for efficient growth of the immature lattice [101]. This suggests that high affinity NC-psi interaction is only required for the formation of the initial Gag complex but is not necessary for lattice growth. Additionally, NC domains in the context of full length Gag show greater affinity for psi containing RNA, forming “early complexes” in association with 5′ leader sequence which are dependent on CA dimerization [102]. These data are consistent with a recently proposed model where specific NC binding to the viral genomic packaging element psi promotes nucleation of the initial Gag assembly complex (reviewed in [77]). Interestingly, while many NC binding sites have been identified within the HIV-1 genomic leader sequence, a subset of non-electrostatic high affinity binding sites exist at a GGAG sequence that is associated with a three-way junction within the dimerized leader [103,104]. Binding of NC at these locations triggers re-modeling of the RNA structure of the leader, which is essential for specific packaging of genomic RNA [103]. Based on these data, Ding et al. suggest a mechanism where initial NC binding to these high affinity sites stimulates recruitment of additional Gag molecules to the gRNA, nucleating Gag assembly in association with packageable genome.
5. Formation of the Gag Lattice at the Plasma Membrane and Potential Contribution of Cytosolic Gag-RNA Complexes
A previous chemical crosslinking study suggested that in the cytoplasm of the cell, Gag exists primarily as a monomer or a low order multimer [56]. In this study, much of the Gag population progresses to higher order multimerization only when it reaches the plasma membrane. This is supported by FRET experiments which demonstrate that the majority of Gag-Gag oligomerization occurs at the plasma membrane, although some FRET signal is found in the cytosol [105,106,107]. Both VLP assembly studies and image-based analysis of Gag assembly in cells by FRET or transmission electron microscopy demonstrate that MA-membrane binding and NC-RNA interaction play functionally redundant roles promoting Gag oligomerization [49,107,108]. In agreement with these results, a coarse-grained simulation model suggests that both RNA and membranes function as scaffolds promoting Gag assembly [95].
Using a photoconvertible labeling system paired with TIRF excitation, Ivanchenko et al. showed that Gag clusters that nucleate sites of viral assembly are primarily recruited from the cytoplasm [109]. Consistent with this result, quantitative measurements of Gag plasma membrane association using a combination of dual-color, z-scan florescence fluctuation spectroscopy with epifluorescence and TIRF microscopy showed that HIV-1 Gag does not bind membrane until cytosolic concentration is high. This, in turn, suggests that Gag does not bind the membrane as a monomer and, hence, is unlikely to be recruited laterally as monomers into sites of viral assembly [110].
Cytosolic complexes containing Gag proteins have been identified [111,112,113], which have been proposed to function as assembly intermediates involving both Gag and viral gRNA upstream of the establishment of the Gag lattice on the plasma membrane [114]. Gag has been shown to traffic progressively through these complexes by pulse chase labeling approaches [112,115] as well as by analysis of Gag mutants defective in different steps of multimerization [114,116]. In this proposed assembly pathway, Gag first associates with the genomic RNA in the ~80S complex, which is associated with RNA granule proteins DDX6 and ABCE1 [111]. Of note, a recent work has suggested that these smaller cytosolic complexes (up to ~80S) likely consist primarily of monomeric or dimeric Gag associated with ribosomes or other RNA-containing cellular complexes and might not represent assembly intermediates containing cytosolic Gag multimers [117].
Although the identity of the “~80S intermediate” requires further refinement, this complex may play a role in Gag assembly as the cytosolic site of Gag-gRNA binding and may even nucleate lattice formation upon binding to the plasma membrane. Consistent with this model, a fluorescence fluctuation imaging study observed Gag oligomerization in the cytosol, supporting the possibility that formation of cytosolic viral gRNA-associated Gag clusters precedes immature lattice assembly [118]. Additionally, the presence of packageable gRNA dimers has been shown to enhance viral assembly [98,119]. Early experiments utilizing TIRF imaging of HIV-1 assembly sites suggest that gRNA arrives to the plasma membrane as a pre-assembled dimer complex [120], while later TIRF experiments utilizing two-color imaging observed gRNA dimerization primarily at the plasma membrane [121]. However, a 3D super-resolution microscopy using a two-color strategy revealed that dimerization of gRNA occurs in the cytosol, and Gag facilitates this process [122]. Although the spatiotemporal details of gRNA dimerization during HIV-1 assembly are still unclear, it is conceivable that gRNA-associated Gag clusters at the plasma membrane “seed” assembly of the immature Gag lattice [77,100,119,123] (Figure 3A). Consistent with this possibility, recently published work demonstrated that mutations disrupting Gag dimerization (CA W184A, W185A) or membrane binding (MA 1GA) introduced in Gag molecules bound to viral RNA impair viral RNA packaging even when these mutant Gag molecules can coassemble into particles formed by assembly competent but RNA-binding-incompetent Gag that is coexpressed [124]. Because the initial Gag-gRNA complex likely plays a key role in the establishment of the immature lattice at the site of assembly and ensures efficient viral RNA packaging, more in-depth study of when this complex is established and how it is incorporated specifically into the assembly site at the plasma membrane is needed.
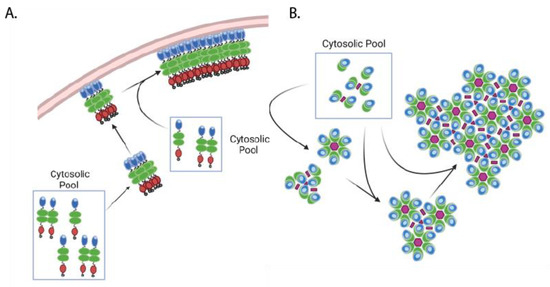
Figure 3.
Immature lattice establishment and assembly progression. Side view (A) and top view (B) of lattice formation and growth. Initial Gag complexes arrive at the plasma membrane from the cytosol, potentially in complex with genomic RNA. Higher order multimerization occurs at the plasma membrane, with monomeric and dimeric Gag (from the cytosolic pool) added to the growing lattice edges. Gag domains are shown in the same color scheme as in Figure 1 and Figure 2. Pink polygons depict different CA-CA interfaces as described in Figure 2.
Single particle tracking data indicate that after the nucleation of assembly on the membrane, Gag is primarily added to the growing immature lattice as lower order multimers such as dimer [94] (Figure 3B) [94]. In agreement with these findings, in silico modeling of lattice growth and measurement of membrane-bound lattice dynamics by atomic force microscopy demonstrate that the immature Gag lattice grows through the addition of monomer and dimer subunits and not by the addition of completed hexamer rings [125,126]. Together, these results suggest a model for HIV-1 particle assembly where the arrival of a Gag complex, which likely contains viral gRNA, from the cytosol to the plasma membrane nucleates Gag lattice formation, which is followed by its growth driven by the recruitment of monomeric or dimeric Gag from the cytosolic pool.
6. Organization of MA in Membrane-Bound Gag Multimer
While the MA-MA interaction is generally not thought to contribute to the overall oligomerization of Gag or formation of the Gag lattice, the MA domain also multimerizes into trimers within the immature Gag lattice. MA domains have been observed to form trimers both in crystal structures and in solution [15,127,128]. Studies utilizing transmission electron microscopy analysis of membrane-bound protein crystals demonstrate that upon binding to membranes containing phosphatidylserine (PS) MA organizes into hexamers [22]; however, on PI(4,5)P2-containing membranes MA domains from both purified MA and MA-CA proteins organize as trimers [36]. In agreement with these two-dimensional crystal data, recent cryo-electron tomography showed that MA within the Gag lattice of immature particles organizes in a similar hexamer of trimers arrangement [129]. Interestingly, in the context of either membrane-bound MACA protein [36] or the immature full length Gag lattice [129], the MA trimer is situated above the three-fold CA interface, suggesting that each MA domain within the trimer is contributed by a Gag molecule from three adjacent hexamer rings (Figure 2). The trimerization of the MA domain within the immature lattice is thought to play functional roles in the incorporation of the envelope glycoprotein into the assembling viral particle [130,131] and in the exposure of myristate [128], which is discussed later in more detail.
7. Genetic Evidence for Multimerization-Mediated Enhancement of Gag Membrane Binding
Oligomerization-competent Gag precursor shows greater affinity for model membranes than mature Gag proteins that correspond to domains of the precursor (including MA) [19]. In the context of full-length Gag protein, mutations disrupting Gag multimerization, such as whole deletion of the MHR or point mutations of invariant residues within this region (CA residues 153–172), cause a reduction in Gag binding to membranes in vitro and in cell homogenates [132,133]. Similarly, mutations and/or truncations disrupting the CA dimer interface (CA W184A, M185A) [134,135], the helix contributing to the CA-SP1 six-helix bundle (CA L232A and SP1 M4A) [136,137], or NC-RNA-mediated multimerization (such as NC deletion) [138,139,140] disrupt the strong membrane binding of Gag. The proportion of Gag bound to the plasma membrane is correlated with its cellular concentration, with Gag at low concentrations remaining primarily cytosolic while higher concentrations, which presumably allow for greater multimerization, drive Gag localization primarily to the plasma membrane [110,141]. Together, these results strongly suggest that Gag–Gag interactions facilitating multimerization play a role in modulating Gag association with the membrane.
8. Mechanisms of Multimerization-Dependent Enhancement of Membrane Binding
In agreement with the genetic evidence of multimerization-dependent enhancement of membrane binding, in vitro membrane binding assays demonstrate that myristoylated MA fused to CA show a greater affinity for acidic lipid-containing membranes than myr-MA alone [22]. Additionally, dimerization-competent HIV-1 CA-CTD enhances membrane binding of MA-CTD fusion proteins relative to dimerization-deficient MA-CTD [142]. Similarly, HIV-1 MA protein chimeras containing inducible dimerization (FKBP) or constitutive hexamerization (Ccmk4) domains show enhanced binding to membranes containing PS, PS and cholesterol, or PS and PI(4,5)P2 [42]. Multiple mechanisms may contribute to the multimerization-dependent enhancement of Gag membrane binding including increased membrane binding avidity of the Gag multimer, multimerization-triggered exposure of myristate, Gag assembly mediated remodeling of the local plasma membrane microenvironment, and multimerization-triggered displacement of tRNA from the MA HBR.
8.1. Avidity
In early studies demonstrating that mutations disrupting NC-RNA-mediated multimerization also reduce HIV-1 Gag membrane binding [138,139], the authors proposed a model in which complexes of multimerized MA have a higher avidity for membranes due to clustered membrane binding domains. Although it lacks a myristate moiety, RSV MA has a patch of basic amino acids, which mediates membrane binding through interactions with acidic phospholipid head groups [143,144]. Relative to non-multimerizing constructs, which bind membrane poorly, dimerization-competent RSV proteins or chimeras containing RSV MA fused with strongly dimerizing HIV-1 CA-CTD (Q192A) show increased membrane binding [145]. A similar effect is seen when the membrane binding of RSV chimeras containing inducible dimerization (FKBP) or constitutive hexamerization (Ccmk4) domains is tested. In these experiments, dimerized or hexamerized MA demonstrates stronger binding to multiple membrane types [42]. Although myristate plays an important role in HIV-1 MA membrane binding, forced dimerization of non-myristoylated HIV-1 MA increases its affinity for binding membranes compared with monomeric MA that is either myristoylated or non-myristoylated [142]. Together, these experimental data support the model described earlier in which multimerization increases the avidity of Gag membrane binding through increased electrostatic interactions of clustered basic patches with acidic phospholipids.
8.2. Myristoyl Switch
A myristoyl switch model has been proposed to explain the differential binding of MA and full length Pr55Gag to membranes where, in the context of the isolated MA domain, myristate is sequestered within the MA globular head, but in the context of Pr55Gag, it is exposed to facilitate membrane binding [5,11,26,146,147,148,149]. More specifically, myristate exposure depends on the multimerization state of the MA domain; the myristate moiety is primarily sequestered within the MA hydrophobic pocket of monomeric MA but primarily exposed when MA is trimerized [128]. These studies agree with a model where Pr55Gag multimerization mediated by CA and NC domains stabilizes the MA-trimer interactions, leading to the exposure of myristate and priming MA for stable membrane binding.
8.3. Membrane Domain Reorganization
It has been recognized for some time that the lipid composition of the envelope of HIV-1 is different from the host cell plasma membrane from which it was derived. Compared with the lipid composition of the producer cell plasma membrane, HIV-1 particles are particularly enriched in specific lipids including PI(4,5)P2 and cholesterol [150,151,152]. While the profile of enriched membrane components was not identical between these studies, they supported the possibility that Gag membrane binding and viral assembly occur at specific microdomains of the plasma membrane, which would likely be compositionally distinct from the rest of the host cell plasma membrane [153,154,155,156]. While these studies are consistent with preferential Gag binding at potentially pre-existing PI(4,5)P2-enriched microdomains, more recent work has suggested that the multimerization of the Gag protein can re-model the membrane composition at the nascent site of assembly, thereby potentially driving the de novo formation of membrane micro-environments enriched in membrane components promoting further recruitment of Gag protein. Gag assembly has been shown to remodel the local membrane environment by driving the fusion of pre-existing microdomains, including lipid rafts and tetraspanin-enriched microdomains [157,158]. In addition to the remodeling of existing membrane domains, a study using an in vitro model membrane system [159] showed that Gag multimerization can drive the formation of membrane nanoclusters, which are enriched in both PI(4,5)P2 and cholesterol and display reduced mobility compared to free membrane lipids. A similar observation has been reported in cells, where Gag multimerization specifically traps PI(4,5)P2 and cholesterol within the site of assembly, while allowing for the free diffusion of non-enriched membrane components [160]. Recent in vitro membrane binding and fluorescent lipid quenching experiments further demonstrate that the Gag mutants, which lack an intact PI(4,5)P2 binding site (MA K29E, K31E) or efficient multimerization (CA W184A, M185A) fail to cluster PI(4,5)P2 [161]. This study also demonstrated that in contrast to the cellular PI(4,5)P2-specific membrane binding domain PH, which preferentially binds membranes containing freely diffusing PI(4,5)P2, myristoylated HIV-1 MA preferentially binds to membranes containing clustered PI(4,5)P2. Together, these results suggest a model in which Gag oligomerization drives the formation of its own micro-domain enriched in PI(4,5)P2 and cholesterol. Given that both PI(4,5)P2 (described above) and cholesterol [162,163] promote Gag membrane binding, the Gag multimerization-mediated enrichment of these membrane components in the local environment likely accelerates the recruitment of additional Gag molecules to the growing site of assembly.
8.4. Dissociation of tRNA
It is still unclear whether or how Gag oligomerization contributes to RNA-mediated regulation of Gag membrane binding. In the presence of tRNA, full length Gag retains the ability to bind membranes containing PI(4,5)P2 [34]. Purified MA, which lacks the capacity to multimerize through CA and NC, is not able to overcome tRNA-mediated inhibition of membrane binding even in the presence of PI(4,5)P2-containing membranes [52]. A recent co-crystal of MA-tRNALys3 complex suggests that tRNA could freely interact with MA domains in a trimer orientation but would be sterically prohibited from binding to the HBR of MA involved in higher-order oligomers [54]. Therefore, it is conceivable that Gag oligomerization, driven by CA and NC domains downstream of MA, promotes membrane binding in part by promoting the steric displacement of tRNA from MA HBR, priming some MA domains within a Gag complex for membrane binding.
9. Summary and Open Questions
Specific localization of HIV-1 Gag to the plasma membrane during viral assembly is primarily driven by specific binding to PI(4,5)P2 at the plasma membrane, paired with inhibition of non-PI(4,5)P2-containing membrane binding by tRNA. During assembly, Gag oligomerization is driven by inter-protein interactions between adjacent CA domains as well as scaffolding via NC-RNA and MA-membrane interactions. Gag multimerizes primarily on the plasma membrane, and lattice growth occurs through the addition of monomeric or dimeric Gag to the lattice edges from the cytosolic pool. In addition to its primary role in lattice assembly, Gag multimerization through CA-CA and NC-RNA interactions also promotes membrane binding via the MA domain. Multimerization-mediated enhancement presumably likely occurs through a combination of the following mechanisms: increase in membrane binding domain avidity, myristate exposure, remodeling of the local membrane microenvironment, and steric displacement of tRNA from the MA HBR.
There are still many remaining questions about mechanisms by which multimerization enhances the membrane binding of Gag and the implications of this effect for its trafficking and assembly. As mentioned above, it is unclear if Gag multimerization impacts RNA binding to the MA HBR. Whether tRNA is indeed removed upon Gag multimerization and whether the presence of PI(4,5)P2 in the membrane also affects the dependence of Gag membrane binding on multimerization need to be tested. If tRNA is displaced from MA-HBR upon Gag multimerization, this mechanism likely contributes to the temporal/spatial regulation of Gag membrane binding and the growth of the Gag lattice in two major ways: (1) enhancing efficient incorporation of Gag into active assembly sites by preventing non-productive membrane binding of monomeric or dimeric Gag and (2) priming cytosolic Gag complexes for membrane binding by reducing tRNA imposed inhibition. Related to the latter, evidence suggests that Gag-gRNA complexes seed lattice and particle assembly more efficiently than Gag multimer lacking gRNA. How do the characteristics of Gag multimers change when they contain gRNA? Do Gag multimers containing gRNA bind more efficiently to the plasma membrane than Gag multimers not associated with gRNA, which in turn would ensure gRNA packaging? Do gRNA-associated Gag clusters bound to the plasma membrane recruit cytosolic Gag more efficiently than those without gRNA? The enrichment of membrane components such as PI(4,5)P2 and cholesterol is observed at sites of Gag multimerization. How does this membrane remodeling contribute to viral assembly? Does the enrichment of PI(4,5)P2 or cholesterol promote recruitment of Gag preferentially to the site where Gag multimer pre-exists, thereby facilitating growth of the Gag lattice? Does the remodeling of the local membrane environment at the site of assembly regulate the incorporation of host-derived co-factors into the assembling virus particle? Addressing these questions will undoubtedly advance our understanding of the process of Gag membrane binding and multimerization and will contribute to our understanding of the mechanisms promoting the productive assembly of HIV-1 particles specifically at the plasma membrane of infected cells.
Author Contributions
C.S. and A.O., writing—original draft, review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by NIH grants R37 AI 071727 (to A.O.), T32 AI 007528 (to C.S.), and F31 AI 134287 (to C.S.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the past and present members of the Ono lab for fruitful discussions on the topics discussed in this review. The figures in this review were created using BioRender.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Göttlinger, H.G.; Sodroski, J.G.; Haseltine, W.A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 1989, 86, 5781–5785. [Google Scholar] [CrossRef] [PubMed]
- Bryant, M.; Ratner, L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 1990, 87, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Spearman, P.; Wang, J.J.; Vander Heyden, N.; Ratner, L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 1994, 68, 3232–3242. [Google Scholar] [CrossRef]
- Zhou, W.; Resh, M.D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 1996, 70, 8540–8548. [Google Scholar] [CrossRef]
- Ono, A.; Ablan, S.D.; Lockett, S.J.; Nagashima, K.; Freed, E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA 2004, 101, 14889–14894. [Google Scholar] [CrossRef]
- Ono, A.; Orenstein, J.M.; Freed, E.O. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 2000, 74, 2855–2866. [Google Scholar] [CrossRef]
- Freed, E.O.; Orenstein, J.M.; Buckler-White, A.J.; Martin, M.A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 1994, 68, 5311–5320. [Google Scholar] [CrossRef]
- Zhou, W.; Parent, L.J.; Wills, J.W.; Resh, M.D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 1994, 68, 2556–2569. [Google Scholar] [CrossRef]
- Yuan, X.; Yu, X.; Lee, T.H.; Essex, M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 1993, 67, 6387–6394. [Google Scholar] [CrossRef]
- Ono, A.; Freed, E.O. Binding of human immunodeficiency virus type 1 Gag to membrane: Role of the matrix amino terminus. J. Virol. 1999, 73, 4136–4144. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Huang, M.; Freed, E.O. Characterization of human immunodeficiency virus type 1 matrix revertants: Effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 1997, 71, 4409–4418. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Fatty acylation of proteins: The long and the short of it. Prog. Lipid Res. 2016, 63, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Schultz, A.M.; Henderson, L.E.; Oroszlan, S. Fatty acylation of proteins. Annu. Rev. Cell Biol. 1988, 4, 611–647. [Google Scholar] [CrossRef]
- Hill, C.P.; Worthylake, D.; Bancroft, D.P.; Christensen, A.M.; Sundquist, W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 1996, 93, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
- Massiah, M.A.; Worthylake, D.; Christensen, A.M.; Sundquist, W.I.; Hill, C.P.; Summers, M.F. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: Evidence for conformational changes during viral assembly. Protein Sci. 1996, 5, 2391–2398. [Google Scholar] [CrossRef]
- Massiah, M.A.; Starich, M.R.; Paschall, C.; Summers, M.F.; Christensen, A.M.; Sundquist, W.I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 1994, 244, 198–223. [Google Scholar] [CrossRef]
- Matthews, S.; Barlow, P.; Clark, N.; Kingsman, S.; Kingsman, A.; Campbell, I. Refined solution structure of p17, the HIV matrix protein. Biochem. Soc. Trans. 1995, 23, 725–729. [Google Scholar] [CrossRef]
- Ehrlich, L.S.; Fong, S.; Scarlata, S.; Zybarth, G.; Carter, C. Partitioning of HIV-1 Gag and Gag-related proteins to membranes. Biochemistry 1996, 35, 3933–3943. [Google Scholar] [CrossRef]
- Scarlata, S.; Ehrlich, L.S.; Carter, C.A. Membrane-induced alterations in HIV-1 Gag and matrix protein-protein interactions. J. Mol. Biol. 1998, 277, 161–169. [Google Scholar] [CrossRef]
- Murray, P.S.; Li, Z.; Wang, J.; Tang, C.L.; Honig, B.; Murray, D. Retroviral matrix domains share electrostatic homology: Models for membrane binding function throughout the viral life cycle. Structure 2005, 13, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Alfadhli, A.; Huseby, D.; Kapit, E.; Colman, D.; Barklis, E. Human immunodeficiency virus type 1 matrix protein assembles on membranes as a hexamer. J. Virol. 2007, 81, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Neil, S.J.; Bess, C.; Johnson, M.C.; Virgen, C.A.; Simon, S.M.; Bieniasz, P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006, 4, e435. [Google Scholar] [CrossRef]
- Finzi, A.; Orthwein, A.; Mercier, J.; Cohen, E.A. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 2007, 81, 7476–7490. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chukkapalli, V.; Hogue, I.B.; Boyko, V.; Hu, W.S.; Ono, A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J. Virol. 2008, 82, 2405–2417. [Google Scholar] [CrossRef]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef] [PubMed]
- Shkriabai, N.; Datta, S.A.; Zhao, Z.; Hess, S.; Rein, A.; Kvaratskhelia, M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 2006, 45, 4077–4083. [Google Scholar] [CrossRef] [PubMed]
- Anraku, K.; Fukuda, R.; Takamune, N.; Misumi, S.; Okamoto, Y.; Otsuka, M.; Fujita, M. Highly sensitive analysis of the interaction between HIV-1 Gag and phosphoinositide derivatives based on surface plasmon resonance. Biochemistry 2010, 49, 5109–5116. [Google Scholar] [CrossRef]
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 2008, 454, 236–240. [Google Scholar] [CrossRef]
- Gerber, P.P.; Cabrini, M.; Jancic, C.; Paoletti, L.; Banchio, C.; von Bilderling, C.; Sigaut, L.; Pietrasanta, L.I.; Duette, G.; Freed, E.O.; et al. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 2015, 209, 435–452. [Google Scholar] [CrossRef]
- Gonzales, B.; de Rocquigny, H.; Beziau, A.; Durand, S.; Burlaud-Gaillard, J.; Lefebvre, A.; Krull, S.; Emond, P.; Brand, D.; Piver, E. Type I Phosphatidylinositol-4-Phosphate 5-Kinases α and γ Play a Key Role in Targeting HIV-1 Pr55. J. Virol. 2020, 94, e00189-20. [Google Scholar] [CrossRef] [PubMed]
- Mücksch, F.; Laketa, V.; Müller, B.; Schultz, C.; Kräusslich, H.G. Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife 2017, 6, e25287. [Google Scholar] [CrossRef]
- Inlora, J.; Chukkapalli, V.; Derse, D.; Ono, A. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J. Virol. 2011, 85, 3802–3810. [Google Scholar] [CrossRef] [PubMed]
- Chukkapalli, V.; Oh, S.J.; Ono, A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA 2010, 107, 1600–1605. [Google Scholar] [CrossRef] [PubMed]
- Alfadhli, A.; Still, A.; Barklis, E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 2009, 83, 12196–12203. [Google Scholar] [CrossRef]
- Alfadhli, A.; Barklis, R.L.; Barklis, E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology 2009, 387, 466–472. [Google Scholar] [CrossRef]
- Inlora, J.; Collins, D.R.; Trubin, M.E.; Chung, J.Y.; Ono, A. Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA. mBio 2014, 5, e02202. [Google Scholar] [CrossRef]
- Barros, M.; Heinrich, F.; Datta, S.A.K.; Rein, A.; Karageorgos, I.; Nanda, H.; Lösche, M. Membrane Binding of HIV-1 Matrix Protein: Dependence on Bilayer Composition and Protein Lipidation. J. Virol. 2016, 90, 4544–4555. [Google Scholar] [CrossRef]
- Carlson, L.A.; Bai, Y.; Keane, S.C.; Doudna, J.A.; Hurley, J.H. Reconstitution of selective HIV-1 RNA packaging in vitro by membrane-bound Gag assemblies. Elife 2016, 5, e14663. [Google Scholar] [CrossRef]
- Keller, H.; Kräusslich, H.G.; Schwille, P. Multimerizable HIV Gag derivative binds to the liquid-disordered phase in model membranes. Cell Microbiol. 2013, 15, 237–247. [Google Scholar] [CrossRef]
- Tran, R.J.; Lalonde, M.S.; Sly, K.L.; Conboy, J.C. Mechanistic Investigation of HIV-1 Gag Association with Lipid Membranes. J. Phys. Chem. B 2019, 123, 4673–4687. [Google Scholar] [CrossRef]
- Dick, R.A.; Kamynina, E.; Vogt, V.M. Effect of multimerization on membrane association of Rous sarcoma virus and HIV-1 matrix domain proteins. J. Virol. 2013, 87, 13598–13608. [Google Scholar] [CrossRef] [PubMed]
- Mercredi, P.Y.; Bucca, N.; Loeliger, B.; Gaines, C.R.; Mehta, M.; Bhargava, P.; Tedbury, P.R.; Charlier, L.; Floquet, N.; Muriaux, D.; et al. Structural and molecular determinants of membrane binding by the HIV-1 matrix protein. J. Mol. Biol. 2016, 428, 1637–1655. [Google Scholar] [CrossRef] [PubMed]
- Purohit, P.; Dupont, S.; Stevenson, M.; Green, M.R. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 2001, 7, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Lochrie, M.A.; Waugh, S.; Pratt, D.G.; Clever, J.; Parslow, T.G.; Polisky, B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997, 25, 2902–2910. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Chang, Y.F.; Wang, S.M.; Tseng, Y.T.; Huang, K.J.; Wang, C.T. HIV-1 matrix protein repositioning in nucleocapsid region fails to confer virus-like particle assembly. Virology 2008, 378, 97–104. [Google Scholar] [CrossRef]
- Hearps, A.C.; Wagstaff, K.M.; Piller, S.C.; Jans, D.A. The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry 2008, 47, 2199–2210. [Google Scholar] [CrossRef]
- Cimarelli, A.; Luban, J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1999, 73, 5388–5401. [Google Scholar] [CrossRef]
- Ott, D.E.; Coren, L.V.; Gagliardi, T.D. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 2005, 79, 13839–13847. [Google Scholar] [CrossRef]
- Ramalingam, D.; Duclair, S.; Datta, S.A.; Ellington, A.; Rein, A.; Prasad, V.R. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011, 85, 305–314. [Google Scholar] [CrossRef]
- Alfadhli, A.; McNett, H.; Tsagli, S.; Bächinger, H.P.; Peyton, D.H.; Barklis, E. HIV-1 matrix protein binding to RNA. J. Mol. Biol. 2011, 410, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Gaines, C.R.; Tkacik, E.; Rivera-Oven, A.; Somani, P.; Achimovich, A.; Alabi, T.; Zhu, A.; Getachew, N.; Yang, A.L.; McDonough, M.; et al. HIV-1 Matrix Protein Interactions with tRNA: Implications for Membrane Targeting. J. Mol. Biol. 2018, 430, 2113–2127. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, D.; Olety, B.; Ono, A. Relationships between MA-RNA Binding in Cells and Suppression of HIV-1 Gag Mislocalization to Intracellular Membranes. J. Virol. 2019, 93, e00756-19. [Google Scholar] [CrossRef] [PubMed]
- Bou-Nader, C.; Muecksch, F.; Brown, J.B.; Gordon, J.M.; York, A.; Peng, C.; Ghirlando, R.; Summers, M.F.; Bieniasz, P.D.; Zhang, J. HIV-1 matrix-tRNA complex structure reveals basis for host control of Gag localization. Cell Host Microbe 2021, 29, 1421–1436. [Google Scholar] [CrossRef]
- Chukkapalli, V.; Inlora, J.; Todd, G.C.; Ono, A. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J. Virol. 2013, 87, 7155–7159. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Bieniasz, P.D. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010, 6, e1001200. [Google Scholar] [CrossRef]
- Holmes, M.; Zhang, F.; Bieniasz, P.D. Single-Cell and Single-Cycle Analysis of HIV-1 Replication. PLoS Pathog. 2015, 11, e1004961. [Google Scholar] [CrossRef]
- Mammano, F.; Ohagen, A.; Höglund, S.; Göttlinger, H.G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 1994, 68, 4927–4936. [Google Scholar] [CrossRef]
- Strambio-de-Castillia, C.; Hunter, E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 1992, 66, 7021–7032. [Google Scholar] [CrossRef]
- Gamble, T.R.; Yoo, S.; Vajdos, F.F.; von Schwedler, U.K.; Worthylake, D.K.; Wang, H.; McCutcheon, J.P.; Sundquist, W.I.; Hill, C.P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 1997, 278, 849–853. [Google Scholar] [CrossRef]
- Borsetti, A.; Ohagen, A.; Göttlinger, H.G. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 1998, 72, 9313–9317. [Google Scholar] [CrossRef]
- Schur, F.K.; Hagen, W.J.; Rumlová, M.; Ruml, T.; Müller, B.; Kräusslich, H.G.; Briggs, J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature 2015, 517, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.M.; Zadrozny, K.K.; Chrustowicz, J.; Purdy, M.D.; Yeager, M.; Ganser-Pornillos, B.K.; Pornillos, O. Crystal structure of an HIV assembly and maturation switch. Elife 2016, 5, e17063. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.; Obr, M.; Hagen, W.J.; Wan, W.; Jakobi, A.J.; Kirkpatrick, J.M.; Sachse, C.; Kräusslich, H.G.; Briggs, J.A. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016, 353, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Accola, M.A.; Höglund, S.; Göttlinger, H.G. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 1998, 72, 2072–2078. [Google Scholar] [CrossRef]
- Gross, I.; Hohenberg, H.; Wilk, T.; Wiegers, K.; Grättinger, M.; Müller, B.; Fuller, S.; Kräusslich, H.G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000, 19, 103–113. [Google Scholar] [CrossRef]
- Datta, S.A.; Temeselew, L.G.; Crist, R.M.; Soheilian, F.; Kamata, A.; Mirro, J.; Harvin, D.; Nagashima, K.; Cachau, R.E.; Rein, A. On the role of the SP1 domain in HIV-1 particle assembly: A molecular switch? J. Virol. 2011, 85, 4111–4121. [Google Scholar] [CrossRef]
- Mendonça, L.; Sun, D.; Ning, J.; Liu, J.; Kotecha, A.; Olek, M.; Frosio, T.; Fu, X.; Himes, B.A.; Kleinpeter, A.B.; et al. CryoET structures of immature HIV Gag reveal six-helix bundle. Commun. Biol. 2021, 4, 481. [Google Scholar] [CrossRef]
- Bharat, T.A.; Castillo Menendez, L.R.; Hagen, W.J.; Lux, V.; Igonet, S.; Schorb, M.; Schur, F.K.; Kräusslich, H.G.; Briggs, J.A. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc. Natl. Acad. Sci. USA 2014, 111, 8233–8238. [Google Scholar] [CrossRef]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef]
- Dostálková, A.; Kaufman, F.; Křížová, I.; Vokatá, B.; Ruml, T.; Rumlová, M. Quantification of the effects of IP6 and other small polyanions on immature HIV-1 particle assembly and core Stability. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Mallery, D.L.; Kleinpeter, A.B.; Renner, N.; Faysal, K.M.R.; Novikova, M.; Kiss, L.; Wilson, M.S.C.; Ahsan, B.; Ke, Z.; Briggs, J.A.G.; et al. A stable immature lattice packages IP. Sci. Adv. 2021, 7, eabe4716. [Google Scholar] [CrossRef] [PubMed]
- von Schwedler, U.K.; Stray, K.M.; Garrus, J.E.; Sundquist, W.I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 2003, 77, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.H.; Chang, Y.F.; Wang, C.T. Mutations in the alpha-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid protein disrupt Gag multimerization and markedly impair virus particle production. J. Biomed. Sci. 2006, 13, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Novikova, M.; Adams, L.J.; Fontana, J.; Gres, A.T.; Balasubramaniam, M.; Winkler, D.C.; Kudchodkar, S.B.; Soheilian, F.; Sarafianos, S.G.; Steven, A.C.; et al. Identification of a structural element in HIV-1 Gag required for virus particle assembly and maturation. mBio 2018, 9, e01567-18. [Google Scholar] [CrossRef] [PubMed]
- Comas-Garcia, M.; Davis, S.R.; Rein, A. On the selective packaging of genomic RNA by HIV-1. Viruses 2016, 8, 246. [Google Scholar] [CrossRef]
- Rein, A. RNA packaging in HIV. Trends Microbiol. 2019, 27, 715–723. [Google Scholar] [CrossRef]
- Olson, E.D.; Musier-Forsyth, K. Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly. Semin. Cell Dev. Biol. 2019, 86, 129–139. [Google Scholar] [CrossRef]
- Gross, I.; Hohenberg, H.; Kräusslich, H.G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 1997, 249, 592–600. [Google Scholar] [CrossRef]
- Campbell, S.; Vogt, V.M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995, 69, 6487–6497. [Google Scholar] [CrossRef]
- Campbell, S.; Rein, A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999, 73, 2270–2279. [Google Scholar] [CrossRef] [PubMed]
- Burniston, M.T.; Cimarelli, A.; Colgan, J.; Curtis, S.P.; Luban, J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 1999, 73, 8527–8540. [Google Scholar] [CrossRef] [PubMed]
- El Meshri, S.E.; Dujardin, D.; Godet, J.; Richert, L.; Boudier, C.; Darlix, J.L.; Didier, P.; Mély, Y.; de Rocquigny, H. Role of the nucleocapsid domain in HIV-1 Gag oligomerization and trafficking to the plasma membrane: A fluorescence lifetime imaging microscopy investigation. J. Mol. Biol. 2015, 427, 1480–1494. [Google Scholar] [CrossRef]
- Cimarelli, A.; Luban, J. Human immunodeficiency virus type 1 virion density is not determined by nucleocapsid basic residues. J. Virol. 2000, 74, 6734–6740. [Google Scholar] [CrossRef] [PubMed]
- Dawson, L.; Yu, X.F. The role of nucleocapsid of HIV-1 in virus assembly. Virology 1998, 251, 141–157. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Y.; Qian, H.; Love, Z.; Barklis, E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J. Virol. 1998, 72, 1782–1789. [Google Scholar] [CrossRef]
- Johnson, M.C.; Scobie, H.M.; Ma, Y.M.; Vogt, V.M. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 2002, 76, 11177–11185. [Google Scholar] [CrossRef]
- Crist, R.M.; Datta, S.A.; Stephen, A.G.; Soheilian, F.; Mirro, J.; Fisher, R.J.; Nagashima, K.; Rein, A. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: Implications for retrovirus assembly. J. Virol. 2009, 83, 2216–2225. [Google Scholar] [CrossRef]
- Alfadhli, A.; Dhenub, T.C.; Still, A.; Barklis, E. Analysis of human immunodeficiency virus type 1 Gag dimerization-induced assembly. J. Virol. 2005, 79, 14498–14506. [Google Scholar] [CrossRef]
- Accola, M.A.; Strack, B.; Göttlinger, H.G. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 2000, 74, 5395–5402. [Google Scholar] [CrossRef]
- Rulli, S.J.; Hibbert, C.S.; Mirro, J.; Pederson, T.; Biswal, S.; Rein, A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007, 81, 6623–6631. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.M.; Vogt, V.M. Nucleic acid binding-induc.ced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 2004, 78, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.M.; Vogt, V.M. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 2002, 76, 5452–5462. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qu, N.; Tan, J.; Rushdi, M.N.; Krueger, C.J.; Chen, A.K. Roles of Gag-RNA interactions in HIV-1 virus assembly deciphered by single-molecule localization microscopy. Proc. Natl. Acad. Sci. USA 2018, 115, 6721–6726. [Google Scholar] [CrossRef]
- Pak, A.J.; Grime, J.M.A.; Sengupta, P.; Chen, A.K.; Durumeric, A.E.P.; Srivastava, A.; Yeager, M.; Briggs, J.A.G.; Lippincott-Schwartz, J.; Voth, G.A. Immature HIV-1 lattice assembly dynamics are regulated by scaffolding from nucleic acid and the plasma membrane. Proc. Natl. Acad. Sci. USA 2017, 114, E10056–E10065. [Google Scholar] [CrossRef]
- Zhao, H.; Datta, S.A.K.; Kim, S.H.; To, S.C.; Chaturvedi, S.K.; Rein, A.; Schuck, P. Nucleic acid-induced dimerization of HIV-1 Gag protein. J. Biol. Chem. 2019, 294, 16480–16493. [Google Scholar] [CrossRef]
- Boutant, E.; Bonzi, J.; Anton, H.; Nasim, M.B.; Cathagne, R.; Réal, E.; Dujardin, D.; Carl, P.; Didier, P.; Paillart, J.C.; et al. Zinc fingers in HIV-1 Gag precursor are not equivalent for gRNA recruitment at the plasma membrane. Biophys. J. 2020, 119, 419–433. [Google Scholar] [CrossRef]
- Dilley, K.A.; Nikolaitchik, O.A.; Galli, A.; Burdick, R.C.; Levine, L.; Li, K.; Rein, A.; Pathak, V.K.; Hu, W.S. Interactions between HIV-1 Gag and viral RNA genome enhance virion assembly. J. Virol. 2017, 91, e02319-16. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008, 6, e96. [Google Scholar] [CrossRef]
- Comas-Garcia, M.; Kroupa, T.; Datta, S.A.; Harvin, D.P.; Hu, W.S.; Rein, A. Efficient support of virus-like particle assembly by the HIV-1 packaging signal. Elife 2018, 7, e38438. [Google Scholar] [CrossRef]
- Kucharska, I.; Ding, P.; Zadrozny, K.K.; Dick, R.A.; Summers, M.F.; Ganser-Pornillos, B.K.; Pornillos, O. Biochemical reconstitution of HIV-1 assembly and maturation. J. Virol. 2020, 94, e01844-19. [Google Scholar] [CrossRef] [PubMed]
- Roldan, A.; Russell, R.S.; Marchand, B.; Götte, M.; Liang, C.; Wainberg, M.A. In vitro identification and characterization of an early complex linking HIV-1 genomic RNA recoognition and Pr55Gag multimerization. J. Biol. Chem. 2004, 279, 39886–39894. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Kharytonchyk, S.; Waller, A.; Mbaekwe, U.; Basappa, S.; Kuo, N.; Frank, H.M.; Quasney, C.; Kidane, A.; Swanson, C.; et al. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. USA 2020, 117, 17737–17746. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Hübner, W.; Chen, P.; Del Portillo, A.; Liu, Y.; Gordon, R.E.; Chen, B.K. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 2007, 81, 12596–12607. [Google Scholar] [CrossRef] [PubMed]
- Derdowski, A.; Ding, L.; Spearman, P. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 2004, 78, 1230–1242. [Google Scholar] [CrossRef] [PubMed]
- Hogue, I.B.; Hoppe, A.; Ono, A. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: Relative contributions of the CA and NC domains and membrane binding. J. Virol. 2009, 83, 7322–7336. [Google Scholar] [CrossRef]
- O’Carroll, I.P.; Crist, R.M.; Mirro, J.; Harvin, D.; Soheilian, F.; Kamata, A.; Nagashima, K.; Rein, A. Functional redundancy in HIV-1 viral particle assembly. J. Virol. 2012, 86, 12991–12996. [Google Scholar] [CrossRef]
- Ivanchenko, S.; Godinez, W.J.; Lampe, M.; Kräusslich, H.; Eils, R.; Rohr, K.; Bräuchle, C.; Müller, B.; Lamb, D.C. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009, 5, e1000652. [Google Scholar] [CrossRef]
- Fogarty, K.H.; Berk, S.; Grigsby, I.F.; Chen, Y.; Mansky, L.M.; Mueller, J.D. Interrelationship between cytoplasmic retroviral Gag concentration and Gag-membrane association. J. Mol. Biol. 2014, 426, 1611–1624. [Google Scholar] [CrossRef]
- Barajas, B.C.; Tanaka, M.; Robinson, B.A.; Phuong, D.J.; Chutiraka, K.; Reed, J.C.; Lingappa, J.R. Identifying the assembly intermediate in which Gag first associates with unspliced HIV-1 RNA suggests a novel model for HIV-1 RNA packaging. PLoS Pathog. 2018, 14, e1006977. [Google Scholar] [CrossRef] [PubMed]
- Lingappa, J.R.; Hill, R.L.; Wong, M.L.; Hegde, R.S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 1997, 136, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Yu, X.F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 1998, 243, 78–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robinson, B.A.; Reed, J.C.; Geary, C.D.; Swain, J.V.; Lingappa, J.R. A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells. J. Virol. 2014, 88, 5718–5741. [Google Scholar] [CrossRef] [PubMed]
- Dooher, J.E.; Schneider, B.L.; Reed, J.C.; Lingappa, J.R. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic 2007, 8, 195–211. [Google Scholar] [CrossRef]
- Tanaka, M.; Robinson, B.A.; Chutiraka, K.; Geary, C.D.; Reed, J.C.; Lingappa, J.R. Mutations of conserved residues in the major homology region arrest assembling HIV-1 Gag as a membrane-targeted intermediate containing genomic RNA and cellular proteins. J. Virol. 2016, 90, 1944–1963. [Google Scholar] [CrossRef]
- Deng, Y.; Hammond, J.A.; Pauszek, R.; Ozog, S.; Chai, I.; Rabuck-Gibbons, J.; Lamichhane, R.; Henderson, S.C.; Millar, D.P.; Torbett, B.E.; et al. Discrimination between functional and non-functional cellular gag complexes involved in HIV-1 assembly. J. Mol. Biol. 2021, 433, 166842. [Google Scholar] [CrossRef]
- Hendrix, J.; Baumgärtel, V.; Schrimpf, W.; Ivanchenko, S.; Digman, M.A.; Gratton, E.; Kräusslich, H.G.; Müller, B.; Lamb, D.C. Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J. Cell Biol. 2015, 210, 629–646. [Google Scholar] [CrossRef]
- Floderer, C.; Masson, J.B.; Boilley, E.; Georgeault, S.; Merida, P.; El Beheiry, M.; Dahan, M.; Roingeard, P.; Sibarita, J.B.; Favard, C.; et al. Single molecule localisation microscopy reveals how HIV-1 Gag proteins sense membrane virus assembly sites in living host CD4 T cells. Sci. Rep. 2018, 8, 16283. [Google Scholar] [CrossRef]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA 2009, 106, 19114–19119. [Google Scholar] [CrossRef]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Clerté, C.; Chamontin, C.; Basyuk, E.; Lainé, S.; Hottin, J.; Bertrand, E.; Margeat, E.; Mougel, M. Imaging HIV-1 RNA dimerization in cells by multicolor super-resolution and fluctuation microscopies. Nucleic Acids Res. 2016, 44, 7922–7934. [Google Scholar] [CrossRef] [PubMed]
- Nikolaitchik, O.A.; Dilley, K.A.; Fu, W.; Gorelick, R.J.; Tai, S.H.; Soheilian, F.; Ptak, R.G.; Nagashima, K.; Pathak, V.K.; Hu, W.S. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013, 9, e1003249. [Google Scholar] [CrossRef] [PubMed]
- Duchon, A.; Santos, S.; Chen, J.; Brown, M.; Nikolaitchik, O.A.; Tai, S.; Chao, J.A.; Freed, E.O.; Pathak, V.K.; Hu, W.S. Plasma membrane anchoring and gag: Gag multimerization on viral RNA are critical properties of HIV-1 Gag required to mediate efficient genome packaging. mBio 2021, 12, e0325421. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Pak, A.J.; Morado, D.R.; Voth, G.A.; Briggs, J.A.G. Immature HIV-1 assembles from Gag dimers leaving partial hexamers at lattice edges as potential substrates for proteolytic maturation. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Valbuena, A.; Maity, S.; Mateu, M.G.; Roos, W.H. Visualization of single molecules building a viral capsid protein lattice through stochastic pathways. ACS Nano 2020, 14, 8724–8734. [Google Scholar] [CrossRef]
- Morikawa, Y.; Zhang, W.H.; Hockley, D.J.; Nermut, M.V.; Jones, I.M. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J. Virol. 1998, 72, 7659–7663. [Google Scholar] [CrossRef]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 2004, 101, 517–522. [Google Scholar] [CrossRef]
- Qu, K.; Ke, Z.; Zila, V.; Anders-Össwein, M.; Glass, B.; Mücksch, F.; Müller, R.; Schultz, C.; Müller, B.; Kräusslich, H.G.; et al. Maturation of the matrix and viral membrane of HIV-1. Science 2021, 373, 700–704. [Google Scholar] [CrossRef]
- Tedbury, P.R.; Novikova, M.; Alfadhli, A.; Hikichi, Y.; Kagiampakis, I.; KewalRamani, V.N.; Barklis, E.; Freed, E.O. HIV-1 matrix trimerization-impaired mutants are rescued by matrix substitutions that enhance envelope glycoprotein incorporation. J. Virol. 2019, 94, e01526-19. [Google Scholar] [CrossRef]
- Tedbury, P.R.; Freed, E.O. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol. 2014, 22, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Ebbets-Reed, D.; Scarlata, S.; Carter, C.A. The major homology region of the HIV-1 gag precursor influences membrane affinity. Biochemistry 1996, 35, 14268–14275. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.F.; Wang, S.M.; Huang, K.J.; Wang, C.T. Mutations in capsid major homology region affect assembly and membrane affinity of HIV-1 Gag. J. Mol. Biol. 2007, 370, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Waheed, A.A.; Joshi, A.; Freed, E.O. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: Use of a novel Gag multimerization assay. J. Virol. 2005, 79, 14131–14140. [Google Scholar] [CrossRef]
- Joshi, A.; Nagashima, K.; Freed, E.O. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation. J. Virol. 2006, 80, 7939–7951. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Hu, J.; Russell, R.S.; Roldan, A.; Kleiman, L.; Wainberg, M.A. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J. Virol. 2002, 76, 11729–11737. [Google Scholar] [CrossRef]
- Guo, X.; Roldan, A.; Hu, J.; Wainberg, M.A.; Liang, C. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J. Virol. 2005, 79, 1803–1812. [Google Scholar] [CrossRef][Green Version]
- Platt, E.J.; Haffar, O.K. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: Requirement for a C-terminal domain. Proc. Natl. Acad. Sci. USA 1994, 91, 4594–4598. [Google Scholar] [CrossRef]
- Sandefur, S.; Varthakavi, V.; Spearman, P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 1998, 72, 2723–2732. [Google Scholar] [CrossRef]
- Ono, A.; Demirov, D.; Freed, E.O. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 2000, 74, 5142–5150. [Google Scholar] [CrossRef]
- Perez-Caballero, D.; Hatziioannou, T.; Martin-Serrano, J.; Bieniasz, P.D. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on gag precursor-membrane interactions. J. Virol. 2004, 78, 9560–9563. [Google Scholar] [CrossRef] [PubMed]
- Dalton, A.K.; Ako-Adjei, D.; Murray, P.S.; Murray, D.; Vogt, V.M. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 2007, 81, 6434–6445. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, J.M.; Fushman, D.; Cahill, S.M.; Zhou, W.; Wolven, A.; Wilson, C.B.; Nelle, T.D.; Resh, M.D.; Wills, J.; Cowburn, D. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J. Mol. Biol. 1998, 279, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Callahan, E.M.; Wills, J.W. Repositioning basic residues in the M domain of the Rous sarcoma virus gag protein. J. Virol. 2000, 74, 11222–11229. [Google Scholar] [CrossRef] [PubMed]
- Dalton, A.K.; Murray, P.S.; Murray, D.; Vogt, V.M. Biochemical characterization of rous sarcoma virus MA protein interaction with membranes. J. Virol. 2005, 79, 6227–6238. [Google Scholar] [CrossRef][Green Version]
- Spearman, P.; Horton, R.; Ratner, L.; Kuli-Zade, I.I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 1997, 71, 6582–6592. [Google Scholar] [CrossRef]
- Paillart, J.C.; Göttlinger, H.G. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of gag membrane targeting. J. Virol. 1999, 73, 2604–2612. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. A myristoyl switch regulates membrane binding of HIV-1 Gag. Proc. Natl. Acad. Sci. USA 2004, 101, 417–418. [Google Scholar] [CrossRef]
- Saad, J.S.; Loeliger, E.; Luncsford, P.; Liriano, M.; Tai, J.; Kim, A.; Miller, J.; Joshi, A.; Freed, E.O.; Summers, M.F. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 2007, 366, 574–585. [Google Scholar] [CrossRef]
- Chan, R.; Uchil, P.D.; Jin, J.; Shui, G.; Ott, D.E.; Mothes, W.; Wenk, M.R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008, 82, 11228–11238. [Google Scholar] [CrossRef]
- Mücksch, F.; Citir, M.; Lüchtenborg, C.; Glass, B.; Traynor-Kaplan, A.; Schultz, C.; Brügger, B.; Kräusslich, H.G. Quantification of phosphoinositides reveals strong enrichment of PIP. Sci. Rep. 2019, 9, 17661. [Google Scholar] [CrossRef] [PubMed]
- Lorizate, M.; Sachsenheimer, T.; Glass, B.; Habermann, A.; Gerl, M.J.; Kräusslich, H.G.; Brügger, B. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell Microbiol. 2013, 15, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Freed, E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 2001, 98, 13925–13930. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.; Hildreth, J.E. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000, 74, 3264–3272. [Google Scholar] [CrossRef] [PubMed]
- Lindwasser, O.W.; Resh, M.D. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 2001, 75, 7913–7924. [Google Scholar] [CrossRef] [PubMed]
- Holm, K.; Weclewicz, K.; Hewson, R.; Suomalainen, M. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55(gag) associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J. Virol. 2003, 77, 4805–4817. [Google Scholar] [CrossRef]
- Hogue, I.B.; Grover, J.R.; Soheilian, F.; Nagashima, K.; Ono, A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 2011, 85, 9749–9766. [Google Scholar] [CrossRef]
- Krementsov, D.N.; Rassam, P.; Margeat, E.; Roy, N.H.; Schneider-Schaulies, J.; Milhiet, P.E.; Thali, M. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 2010, 11, 1401–1414. [Google Scholar] [CrossRef]
- Yandrapalli, N.; Lubart, Q.; Tanwar, H.S.; Picart, C.; Mak, J.; Muriaux, D.; Favard, C. Self assembly of HIV-1 Gag protein on lipid membranes generates PI(4,5)P. Sci. Rep. 2016, 6, 39332. [Google Scholar] [CrossRef]
- Favard, C.; Chojnacki, J.; Merida, P.; Yandrapalli, N.; Mak, J.; Eggeling, C.; Muriaux, D. HIV-1 Gag specifically restricts PI(4,5)P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly. Sci. Adv. 2019, 5, eaaw8651. [Google Scholar] [CrossRef]
- Wen, Y.; Feigenson, G.W.; Vogt, V.M.; Dick, R.A. Mechanisms of PI(4,5)P2 Enrichment in HIV-1 Viral Membranes. J. Mol. Biol. 2020, 432, 5343–5364. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Waheed, A.A.; Freed, E.O. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology 2007, 360, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.A.; Goh, S.L.; Feigenson, G.W.; Vogt, V.M. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. USA 2012, 109, 18761–18766. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).