Effects of the NF-κB Signaling Pathway Inhibitor BAY11-7082 in the Replication of ASFV
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus
2.2. Reagents and Antibodies
2.3. HAD Assay
2.4. Immunofluorescence Assay
2.5. RNA Isolation, cDNA Library Preparation, and Sequencing
2.6. Data Analysis of RNA-Seq
2.7. qPCR
2.8. Western Blot Analysis
2.9. Cell Viability Assay
2.10. Statistical Analysis
3. Results
3.1. Proliferation and Growth Curve of ASFV In Vitro
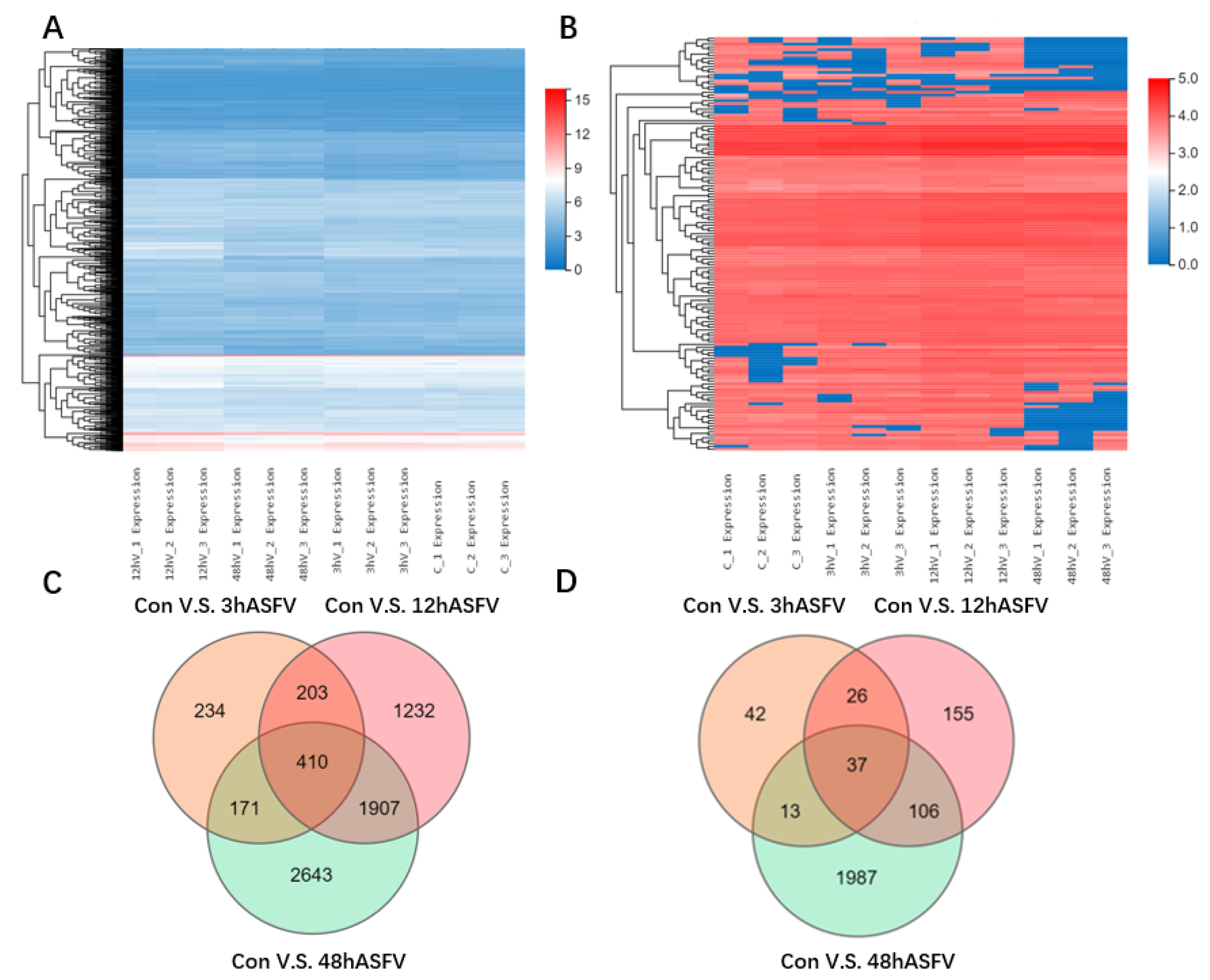
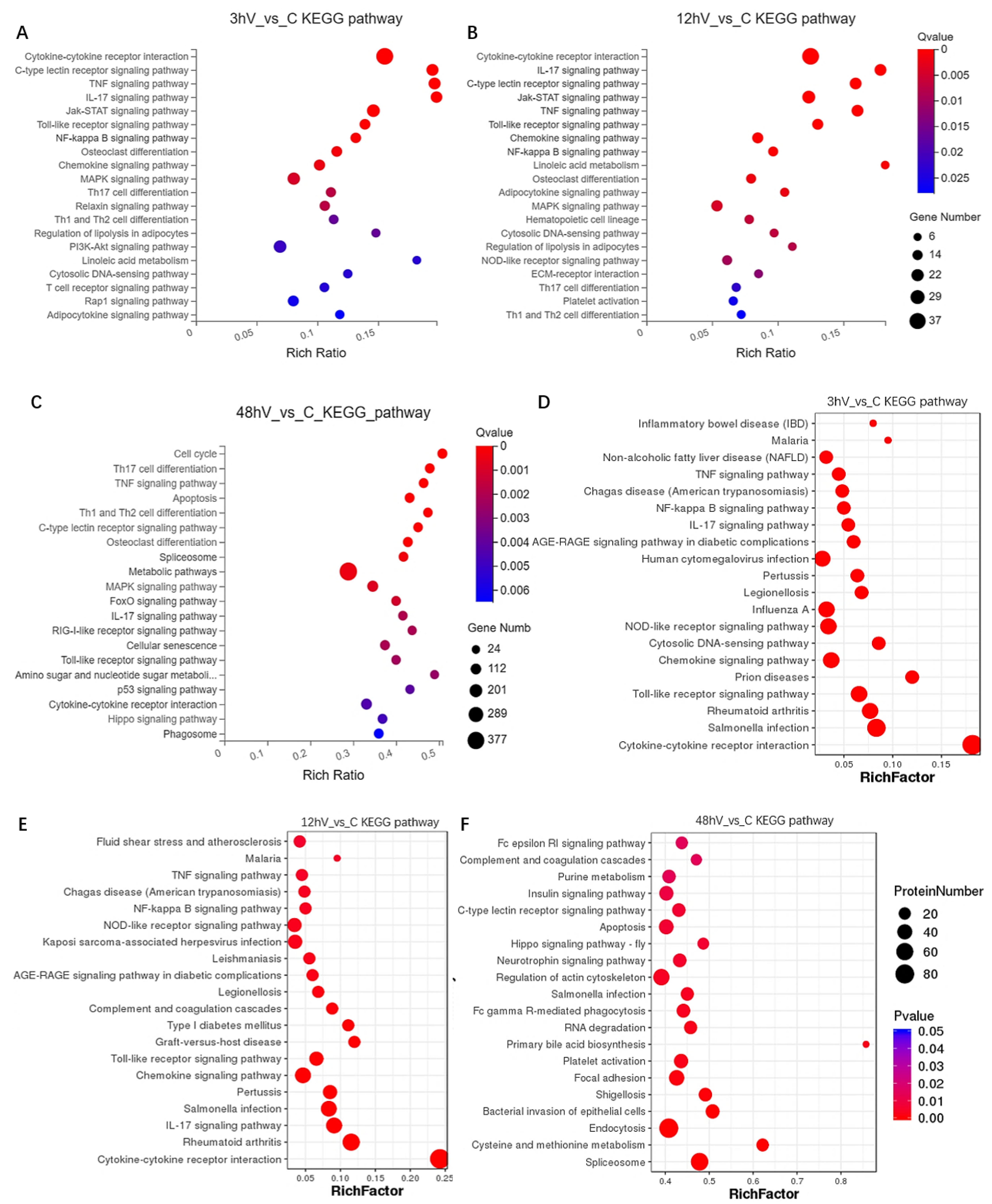
3.2. Conjoint Analysis of Transcriptomics and Proteomics
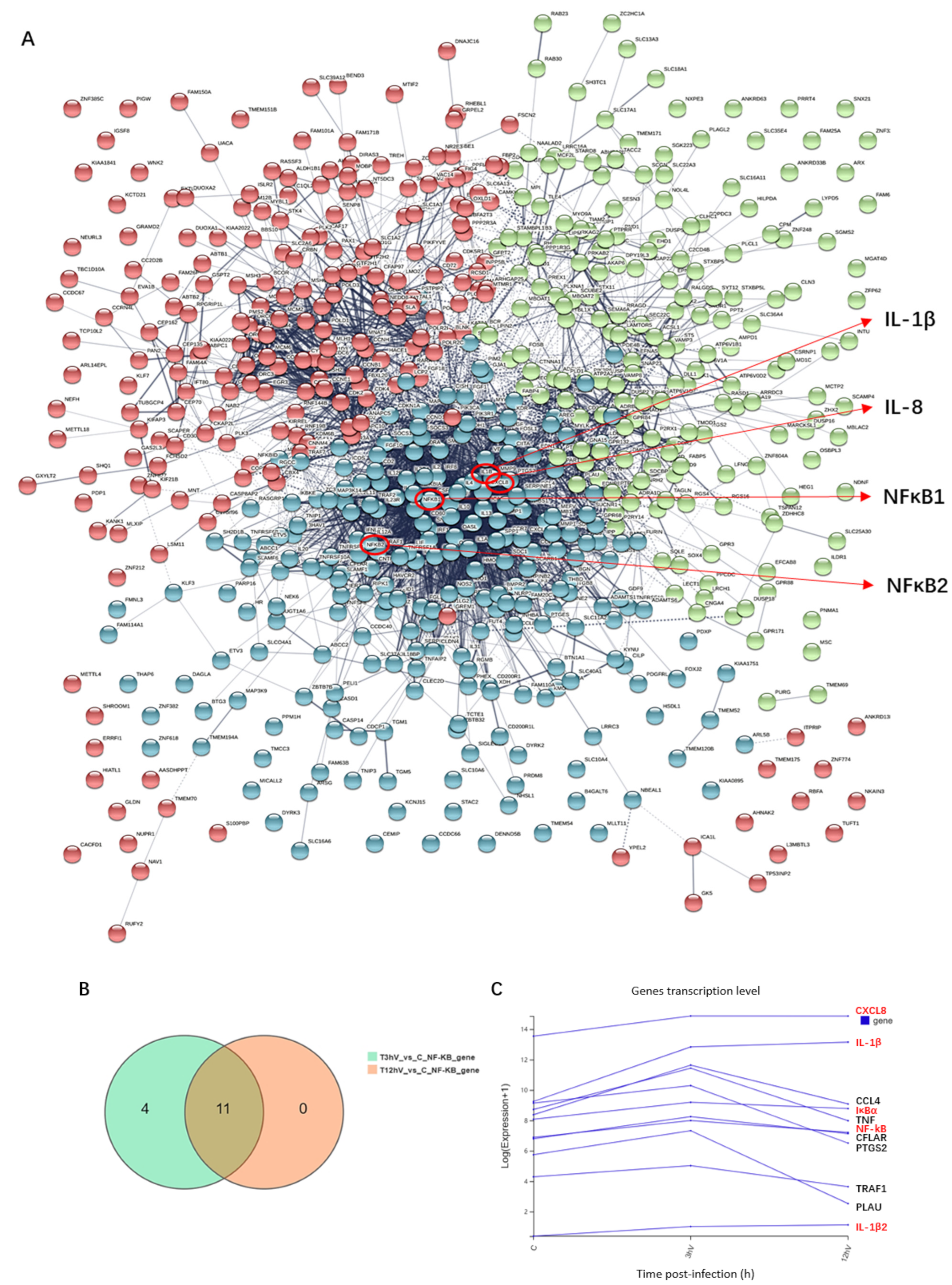
3.3. ASFV Early Infection Activates NF-κB Signaling Pathway
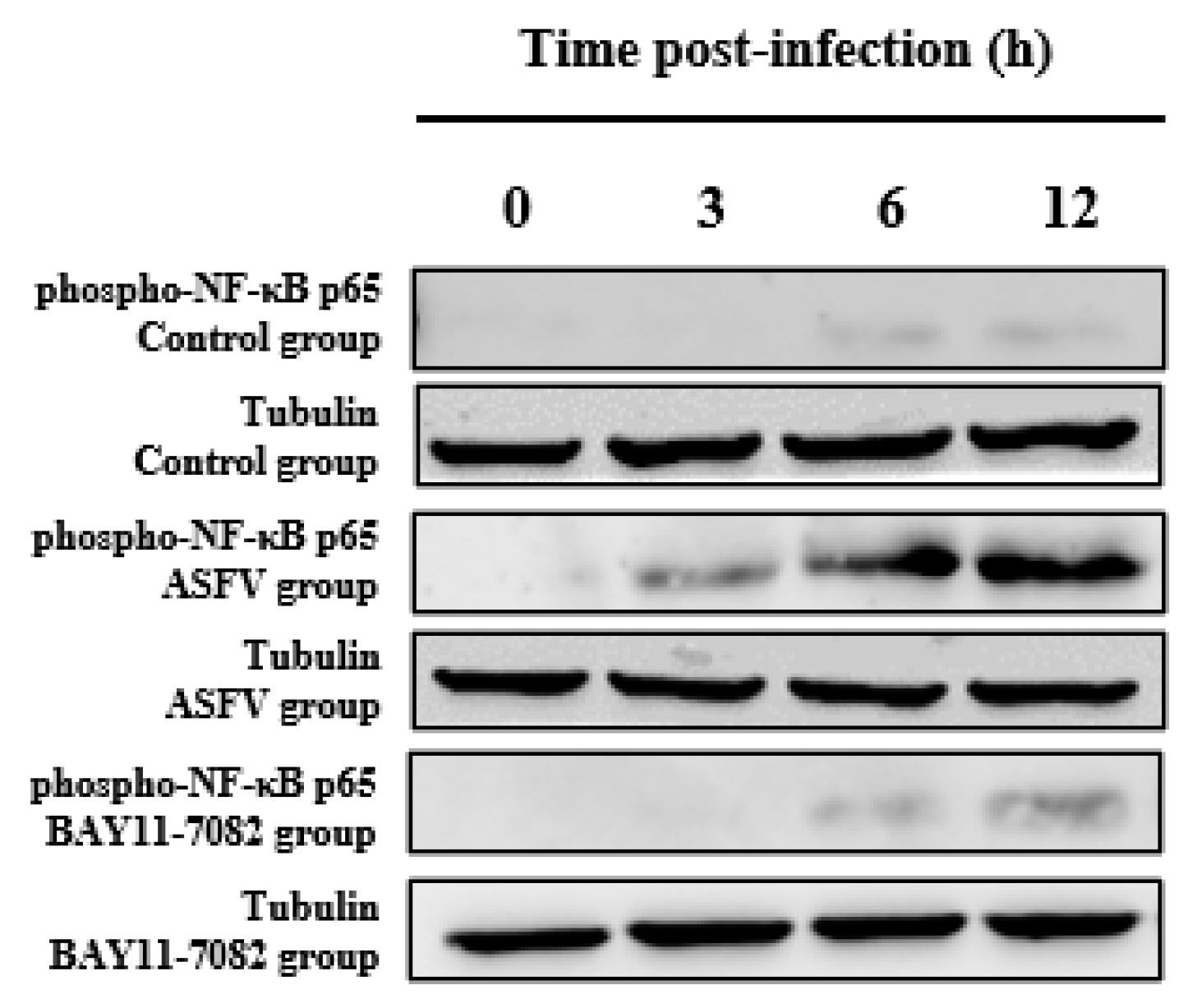
3.4. BAY11-7082 Inhibits the Overexpression of Phospho-NF-κB p65 Caused by ASFV Infection
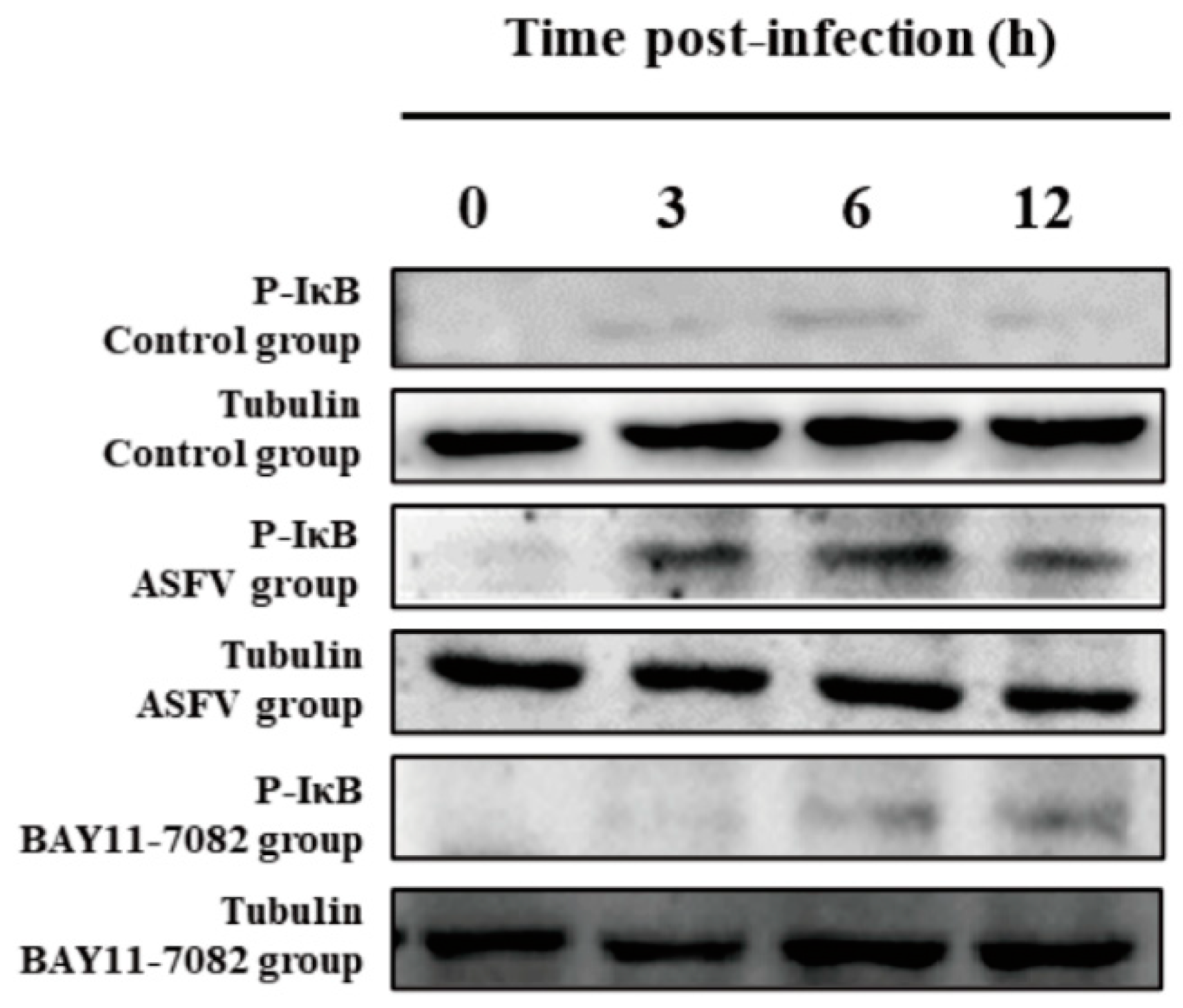
3.5. BAY11-7082 Inhibits the Overexpression of p-IκB Caused by ASFV Infection
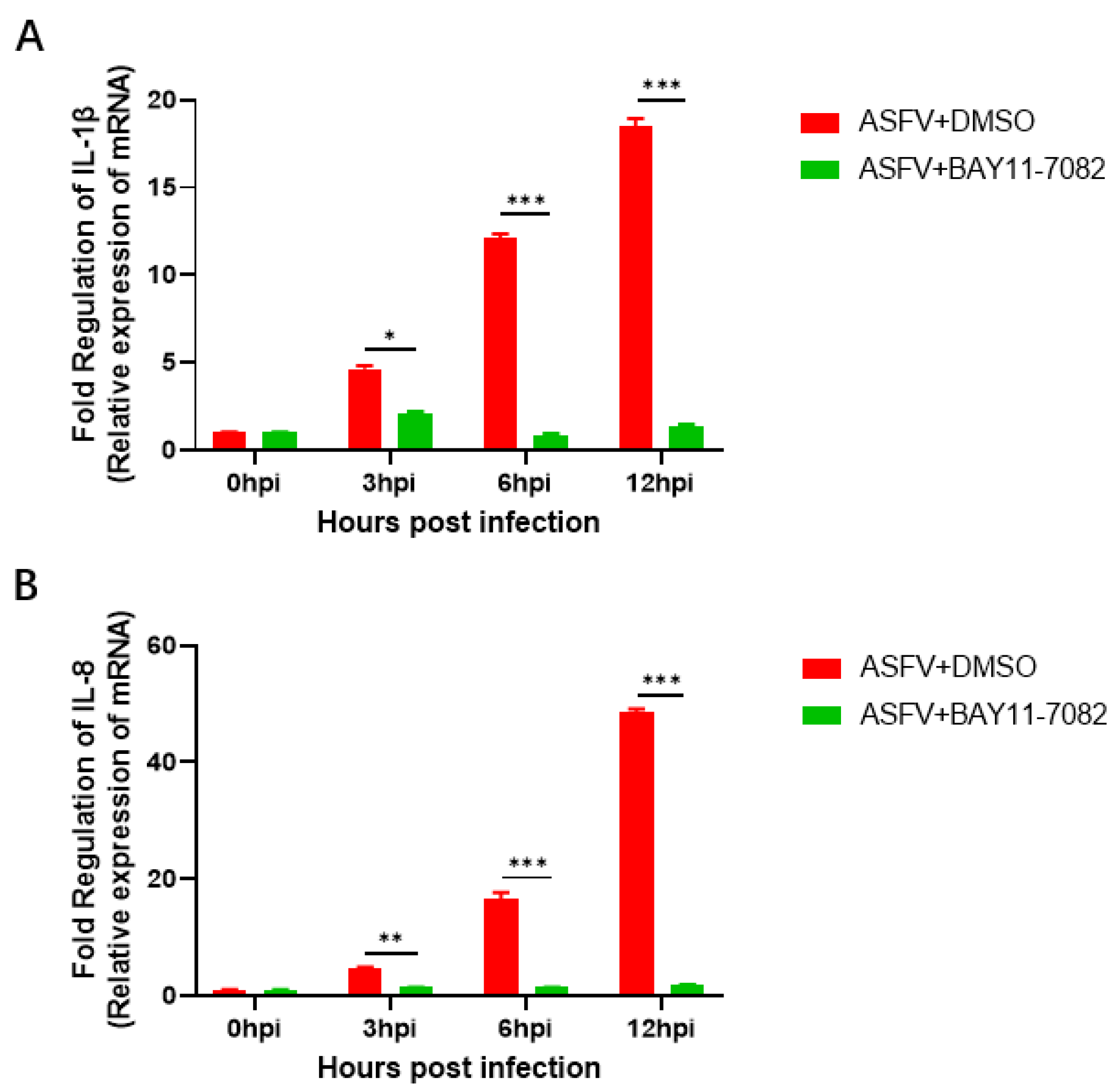
3.6. BAY11-7082 Inhibits the Overexpression of IL-1β and IL-8 Caused by ASFV Infection
3.7. BAY11-7082 Inhibits ASFV Replication
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ann, S.; Miyako, K.; Louise, L.; Francesc, A.; Thomas, B.; Christina, M.; Morten, T.M.; Thomas, P.; Anette, B.; Graham, J. Identification of African Swine Fever Virus Transcription within Peripheral Blood Mononuclear Cells of Acutely Infected Pigs. Viruses 2021, 13, 2333. [Google Scholar]
- Malogolovkin, A.; Burmakina, G.; Titov, I.; Sereda, A.; Gogin, A.; Baryshnikova, E.; Kolbasov, D. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg. Infect. Dis. 2015, 21, 312–315. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, R.; Zhang, X.; Li, F.; Wang, J.; Zhang, J.; Liu, X.; Wang, L.; Zhang, J.; Wu, X.; et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes Infect. 2019, 8, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Angel, L.M.; Jose, B.; de Patricia, L. Methods for growing and titrating African swine fever virus: Field and laboratory samples. Curr. Protoc. Cell Biol. 2011, 26, 14–26. [Google Scholar]
- Busch, F.; Haumont, C.; Penrith, M.-L.; Laddomada, A.; Dietze, K.; Globig, A.; Guberti, V.; Zani, L.; Depner, K.; Netherton, C.; et al. Evidence-Based African Swine Fever Policies: Do We Address Virus and Host Adequately? Front. Vet. Sci. 2021, 8, 637487. [Google Scholar] [PubMed]
- Sánchez-Cordón, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K.; Andrés, G. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet. J. 2018, 233, 41–48. [Google Scholar]
- Gallardo, C.; Soler, A.; Nieto, R.; Cano, C.; Pelayo, V.; Sánchez, M.A.; Pridotkas, G.; Fernandez-Pinero, J.; Briones, V.; Arias, M. Experimental Infection of Domestic Pigs with African Swine Fever Virus Lithuania 2014 Genotype II Field Isolate. Transbound. Emerg. Dis. 2017, 64, 300–304. [Google Scholar] [CrossRef]
- Rowlands, R.J.; Michaud, V.; Heath, L.; Hutchings, G.; Oura, C.; Vosloo, W.; Dwarka, R.; Onashvili, T.; Albina, E.; Dixon, L.K. African Swine Fever Virus Isolate, Georgia, 2007. Emerg. Infect. Dis. 2008, 14, 1870–1874. [Google Scholar] [CrossRef]
- de Villiers, E.P.; Gallardo, C.; Arias, M.; da Silva, M.; Upton, C.; Martin, R.; Bishop, R.P. Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology 2010, 400, 128–136. [Google Scholar] [CrossRef]
- David, L.; Huber, W.; Granovskaia, M.; Toedling, J.; Palm, C.J.; Bofkin, L.; Jones, T.; Davis, W.R.; Steinmetz, L.M. A high-resolution map of transcription in the yeast genome. Proc. Nat. Acad. Sci. USA 2006, 103, 5320–5325. [Google Scholar] [CrossRef]
- Breckpot, J.; Thienpont, B.; Gewillig, M.; Allegaert, K.; Vermeesch, J.R.; Devriendt, K. Differences in Copy Number Variation between Discordant Monozygotic Twins as a Model for Exploring Chromosomal Mosaicism in Congenital Heart Defects. Mol. Syndr. 2012, 2, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Rebollar, E.A.; Antwis, R.E.; Becker, M.H.; Belden, L.K.; Bletz, M.C.; Brucker, R.M.; Harrison, X.A.; Hughey, M.C.; Kueneman, J.G.; Loudon, H.A.; et al. Using “omics” and integrated Multi-Omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front. Microbiol. 2016, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Csh. Perspect. Biol. 2009, 1, a1651. [Google Scholar]
- Dejardin, E.; Droin, N.M.; Delhase, M.; Haas, E.; Cao, Y.; Makris, C.; Li, Z.; Karin, M.; Ware, C.F.; Green, D.R. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 2002, 17, 525–535. [Google Scholar] [CrossRef]
- Bonizzi, G.; Karin, M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004, 25, 280–288. [Google Scholar] [CrossRef]
- Beg, A.A.; Baltimore, D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science 1996, 274, 782–784. [Google Scholar] [CrossRef]
- Lavagno, L.; Gunella, G.; Bardelli, C.; Spina, S.; Fresu, L.G.; Viano, I.; Brunelleschi, S. Anti-inflammatory drugs and tumor necrosis factor-alpha production from monocytes: Role of transcription factor NF-kappa B and implication for rheumatoid arthritis therapy. Eur. J. Pharmacol. 2004, 501, 199–208. [Google Scholar] [CrossRef]
- Naik, K.K.; Thangavel, S.; Alam, A. Cytotoxicity and Anti-inflammatory Activity of Flavonoid Derivatives Targeting NF-kappaB. Recent Pat. Inflamm. Allergy Drug Discov. 2017, 10, 119–132. [Google Scholar] [CrossRef]
- Freitas, R.H.; Fraga, C.A. NF-κB-IKKβ Pathway as a Target for Drug Development: Realities, Challenges and Perspectives. Curr. Drug Targets 2018, 19, 1933–1942. [Google Scholar] [CrossRef]
- Takamatsu, H.; Denyer, M.S.; Lacasta, A.; Stirling, C.M.; Argilaguet, J.M.; Netherton, C.L.; Oura, C.A.; Martins, C.; Rodriguez, F. Cellular immunity in ASFV responses. Virus Res. 2013, 173, 110–121. [Google Scholar] [CrossRef]
- Carlson, J.; O’Donnell, V.; Alfano, M.; Salinas, L.V.; Holinka, L.G.; Krug, P.W.; Gladue, D.P.; Higgs, S.; Borca, M.V. Association of the Host Immune Response with Protection Using a Live Attenuated African Swine Fever Virus Model. Viruses 2016, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Gil, S.; Sepulveda, N.; Albina, E.; Leitao, A.; Martins, C. The low-virulent African swine fever virus (ASFV/NH/P68) induces enhanced expression and production of relevant regulatory cytokines (IFN alpha, TNF alpha and IL12p40) on porcine macrophages in comparison to the highly virulent ASFV/L60. Arch. Virol. 2008, 153, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Pintaric, M.; Gerner, W.; Saalmueller, A. Synergistic effects of IL-2, IL-12 and IL-18 on cytolytic activity, perforin expression and IFN-gamma production of porcine natural killer cells. Vet. Immunol. Immunop. 2008, 121, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J. IPC: Professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Ann. Rev. Immunol. 2005, 23, 275–306. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, G.; Graham, S.P.; Giudici, S.D.; Bonelli, P.; Pilo, G.; Anfossi, A.G.; Pittau, M.; Nicolussi, P.S.; Laddomada, A.; Oggiano, A. Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Vet. Microbiol. 2017, 198, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Han, J.; Zhang, Y.; Duan, D.; Zhang, S. Porcine circovirus type 2 induces type I interferon production via MyD88–IKKα–IRFs signaling rather than NF-κB in porcine alveolar macrophages in vitro. Res. Vet. Sci. 2016, 104, 188–194. [Google Scholar] [CrossRef]
- Sultuybek, G.K.; Soydas, T.; Yenmis, G. NF-κB as the mediator of metformin’s effect on ageing and ageing-related diseases. Clin. Exp. Pharmacol. Physiol. 2019, 46, 413–422. [Google Scholar] [CrossRef]
- Qin, J.; Cheng, X.F.; Lei, F.; Akolkar, G.; Cai, J.; Zhang, X.; Blet, A.; Xie, J.; Zhang, P.; Liu, Y.; et al. Redefining Cardiac Biomarkers in Predicting Mortality of Inpatients With COVID-19. Hypertension 2020, 76, 1104–1112. [Google Scholar] [CrossRef]
- Ye, J.; Zhu, N.; Sun, R.; Liao, W.; Fan, S.; Shi, F.; Lin, H.S.; Ying, Y. Metformin Inhibits Chemokine Expression Through the AMPK/NF-κB Signaling Pathway. J. Interferon Cytokine Res. 2018, 38, 363–369. [Google Scholar] [CrossRef]
- Zhu, Z.; Meng, G. ASFVdb: An integrative resource for genomic and proteomic analyses of African swine fever virus. J. Biol. Databases Curation 2020, 2020, a23. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tian, K. African swine fever in China. Vet. Rec. 2018, 183, 300–301. [Google Scholar] [CrossRef] [PubMed]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Cao, Y.; Jiao, P.; Yu, P.; Zhang, H.; Chen, T.; Zhou, X.; Qi, Y.; Sun, L.; Liu, D.; et al. Synergistic effect of the responses of different tissues against African swine fever virus. Transb. Emerg. Dis. 2021, 8, 1–12. [Google Scholar] [CrossRef]
- Ju, X.; Li, F.; Li, J.; Wu, C.; Xiang, G.; Zhao, X.; Nan, Y.; Zhao, D.; Ding, Q. Genome-wide transcriptomic analysis of highly virulent African swine fever virus infection reveals complex and unique virus host interaction. Vet. Microbiol. 2021, 261, 109211. [Google Scholar] [CrossRef]
- Sun, H.; Niu, Q.; Yang, J.; Zhao, Y.; Tian, Z.; Fan, J.; Zhang, Z.; Wang, Y.; Geng, S.; Zhang, Y.; et al. Transcriptome Profiling Reveals Features of Immune Response and Metabolism of Acutely Infected, Dead and Asymptomatic Infection of African Swine Fever Virus in Pigs. Front. Immunol. 2021, 12, 1–15. [Google Scholar]
- Yang, B.; Shen, C.; Zhang, D.; Zhang, T.; Shi, X.; Yang, J.; Hao, Y.; Zhao, D.; Cui, H.; Yuan, X.; et al. Mechanism of interaction between virus and host is inferred from the changes of gene expression in macrophages infected with African swine fever virus CN/GS/2018 strain. Virol. J. 2021, 18, 1–16. [Google Scholar] [CrossRef]
- Cuesta-Geijo, M.Á.; Chiappi, M.; Galindo, I.; Barrado-Gil, L.; Muñoz-Moreno, R.; Carrascosa, J.L.; Alonso, C. Cholesterol Flux Is Required for Endosomal Progression of African Swine Fever Virions during the Initial Establishment of Infection. J. Virol. 2016, 90, 1534–1543. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.Q.Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; Wang, Y.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2021, 7, 601641. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-kappaB signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Dudziak, D.; Dirmeier, U.; Hobom, G.; Riedel, A.; Schlee, M.; Staudt, L.M.; Rosenwald, A.; Behrends, U.; Bornkamm, G.W. Active NF-kappaB signalling is a prerequisite for influenza virus infection. J. Gen. Virol. 2004, 85, 2347–2356. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Kwon, H.; Chen, L.F.; Greene, W.C. Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 2007, 81, 6043–6056. [Google Scholar] [CrossRef] [PubMed]
- Sehnert, B.; Burkhardt, H.; Dübel, S.; Voll, R.E. Cell-Type Targeted NF-kappaB Inhibition for the Treatment of Inflammatory Diseases. Cells 2020, 9, 1627. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Galindo, I.; Cuesta-Geijo, M.A.; Cabezas, M.; Hernaez, B.; Muñoz-Moreno, R. African swine fever virus-cell interactions: From virus entry to cell survival. Virus Res. 2013, 173, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Rhiannon, N.S.; Gavin, C.B.; Charles, C.A.; Linda, K.D. African swine fever virus A238L inhibitor of NF-κB and of calcineurin phosphatase is imported actively into the nucleus and exported by a CRM1-mediated pathway. J. Gen. Virol. 2007, 88, 2. [Google Scholar]
- Clara, I.R.; María, L.N.; Angel, L.C.; María, L.S.; Manuel, F.; Yolanda, R. African Swine Fever Virus IAP-Like Protein Induces the Activation of Nuclear Factor Kappa B. J. Virol. 2002, 76, 3936–3942. [Google Scholar]
- Lucía, B.; del Puerto, A.; Inmaculada, G.; Miguel, Á.; Isabel, G.; Carlos, M.; Covadonga, A. African Swine Fever Virus Ubiquitin-Conjugating Enzyme Is an Immunomodulator Targeting NF-κB Activation. Viruses 2021, 13, 1160. [Google Scholar]


| Gene | Primer Sequence (5′-3′) |
|---|---|
| CADC-B646L-rPCRF | ATAGAGATACAGCTCTTCCAG |
| CADC-B646L-rPCRR | GTATGTAAGAGCTGCAGAAC |
| CADC-B646L-Probe | FAM-TATCGATAAGATTGAT-MGB |
| B646L-F | TGAAATAAAATGGAAGCCCACAGATC |
| B646L-R | ACACTGTACAACATTGCGTAAAAGC |
| GAPDH-F | GCAAAGACTGAACCCACTAATT |
| GAPDH-R | TTGCCTCTGTTGTTACTTGGAG |
| IL-1β-F | ACCTGGACCTTGGTTCTCTG |
| IL-1β-R | CATCTGCCTGATGCTCTTG |
| IL-8-F | CACTGTGAAAATTCAGAAATCATTGT |
| IL-8-R | CTTCACAAATACCTGCACAACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Q.; Yang, Y.; Feng, Y.; Quan, W.; Luo, Y.; Wang, H.; Zheng, J.; Chen, X.; Huang, Z.; Chen, X.; et al. Effects of the NF-κB Signaling Pathway Inhibitor BAY11-7082 in the Replication of ASFV. Viruses 2022, 14, 297. https://doi.org/10.3390/v14020297
Gao Q, Yang Y, Feng Y, Quan W, Luo Y, Wang H, Zheng J, Chen X, Huang Z, Chen X, et al. Effects of the NF-κB Signaling Pathway Inhibitor BAY11-7082 in the Replication of ASFV. Viruses. 2022; 14(2):297. https://doi.org/10.3390/v14020297
Chicago/Turabian StyleGao, Qi, Yunlong Yang, Yongzhi Feng, Weipeng Quan, Yizhuo Luo, Heng Wang, Jiachen Zheng, Xiongnan Chen, Zhao Huang, Xiaojun Chen, and et al. 2022. "Effects of the NF-κB Signaling Pathway Inhibitor BAY11-7082 in the Replication of ASFV" Viruses 14, no. 2: 297. https://doi.org/10.3390/v14020297
APA StyleGao, Q., Yang, Y., Feng, Y., Quan, W., Luo, Y., Wang, H., Zheng, J., Chen, X., Huang, Z., Chen, X., Xu, R., Zhang, G., & Gong, L. (2022). Effects of the NF-κB Signaling Pathway Inhibitor BAY11-7082 in the Replication of ASFV. Viruses, 14(2), 297. https://doi.org/10.3390/v14020297






