Cell Type Variability in the Incorporation of Lipids in the Dengue Virus Virion
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Large Scale Virus Preparation
2.3. Lipid Extraction
2.4. Lipid Analysis
2.5. Normalisation and Statistical Analysis
2.6. cDNA Synthesis by RT-PCR
2.7. RNA Standard Preparation
2.8. One Step qRT-PCR
2.9. Western Blot Analysis
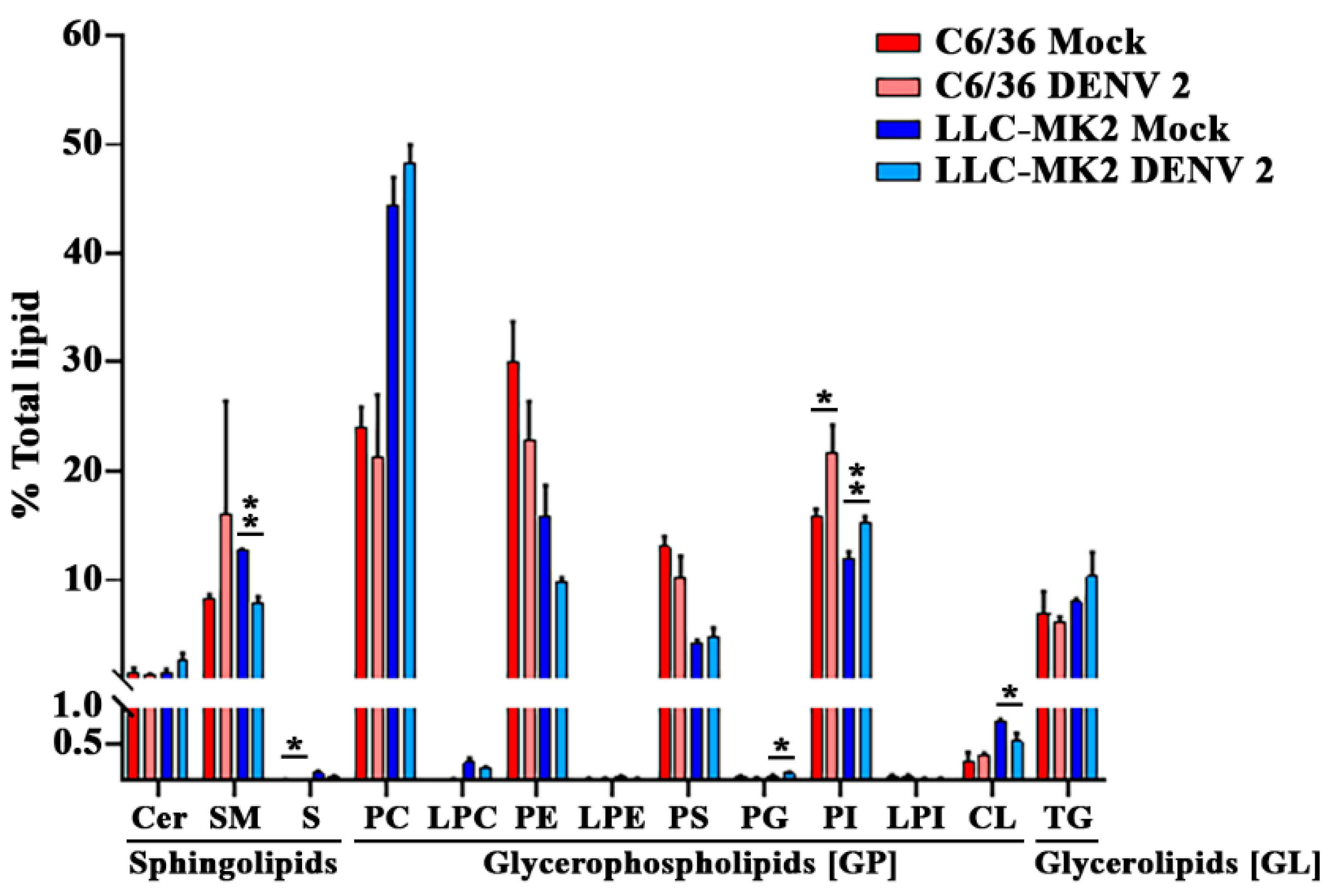
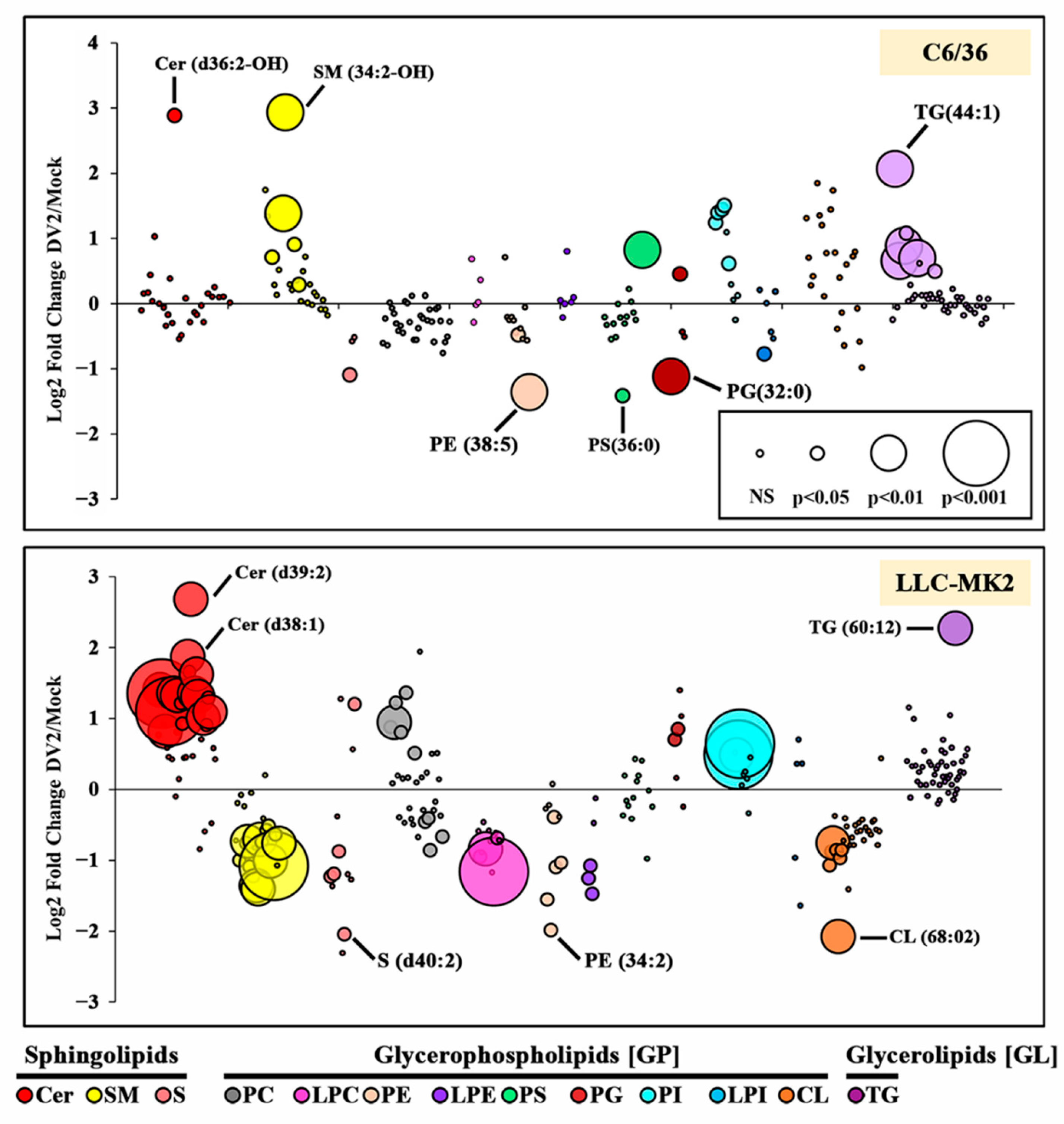
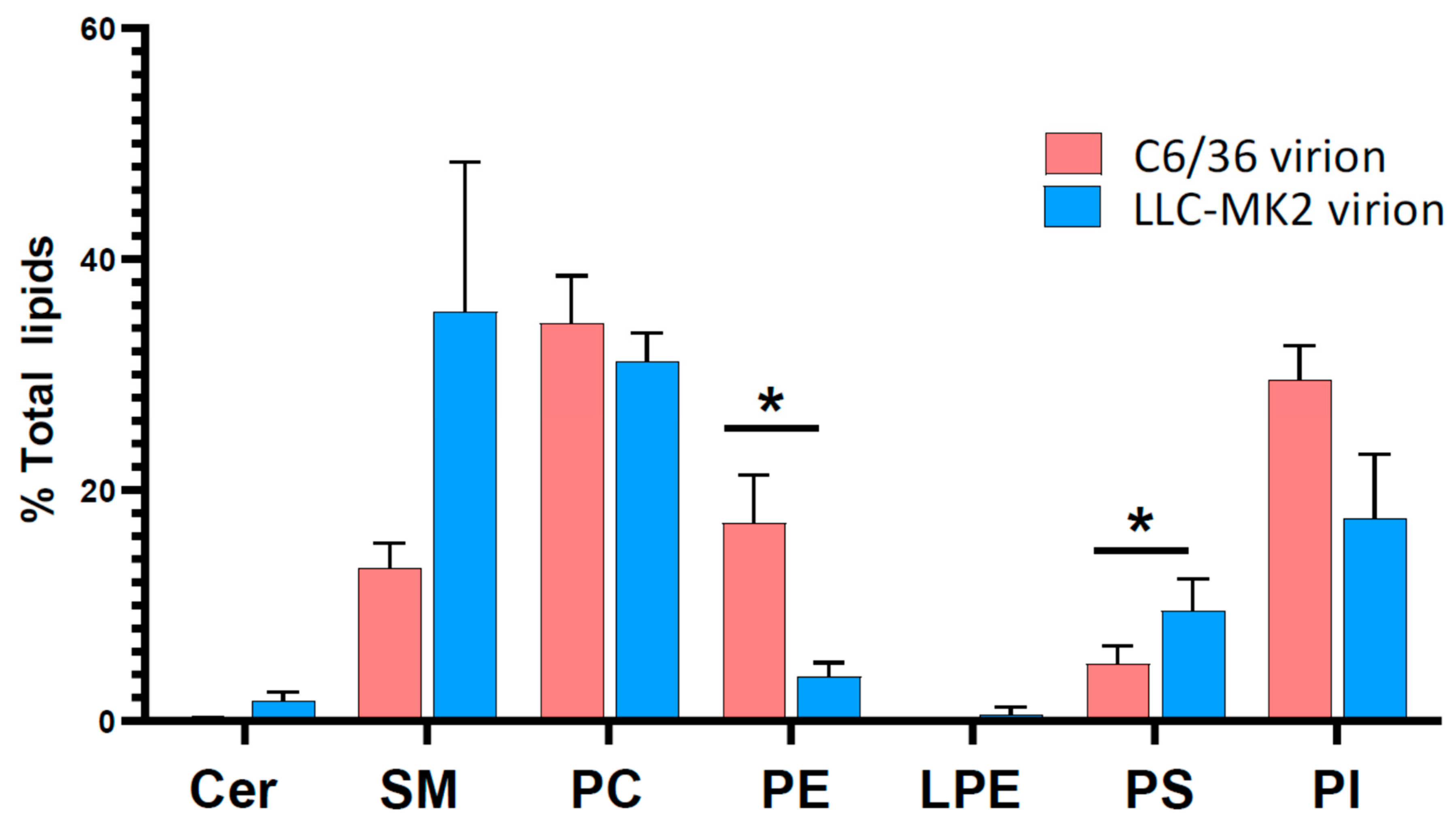
3. Results
3.1. Purification of Viral Particles
3.2. DENV Infection Alters the Cellular Lipid Profile
3.3. Lipid Profiles of DENV Virions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat Microbiol 2020, 5, 796–812. [Google Scholar] [CrossRef]
- Gould, E.A.; Solomon, T. Pathogenic flaviviruses. Lancet 2008, 371, 500–509. [Google Scholar] [CrossRef]
- Smith, D.R. Waiting in the wings: The potential of mosquito transmitted flaviviruses to emerge. Crit. Rev. Microbiol. 2017, 4, 405–422. [Google Scholar] [CrossRef]
- Roukens, A.H.; Visser, L.G. Yellow fever vaccine: Past, present and future. Expert Opin. Biol. Ther. 2008, 8, 1787–1795. [Google Scholar] [CrossRef]
- Hegde, N.R.; Gore, M.M. Japanese encephalitis vaccines: Immunogenicity, protective efficacy, effectiveness, and impact on the burden of disease. Hum. Vaccin. Immunother. 2017, 13, 1–18. [Google Scholar] [CrossRef]
- Paz-Bailey, G.; Adams, L.; Wong, J.M.; Poehling, K.A.; Chen, W.H.; McNally, V.; Atmar, R.L.; Waterman, S.H. Dengue Vaccine: Recommendations of the Advisory Committee on Immunization Practices, United States, 2021. MMWR Recomm Rep. 2021, 70, 1–16. [Google Scholar] [CrossRef]
- Ooi, E.E.; Goh, K.T.; Gubler, D.J. Dengue prevention and 35 years of vector control in Singapore. Emerg Infect. Dis 2006, 12, 887–893. [Google Scholar] [CrossRef]
- Lindenbach, B.; Thiel, H.J.; Rice, C.M. Flaviviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, O.M., Eds.; Lippincot William & Wilkins: Philadelphia, PA, USA, 2011; pp. 1101–1151. [Google Scholar]
- Lescar, J.; Soh, S.; Lee, L.T.; Vasudevan, S.G.; Kang, C.; Lim, S.P. The Dengue Virus Replication Complex: From RNA Replication to Protein-Protein Interactions to Evasion of Innate Immunity. Adv. Exp. Med. Biol. 2018, 1062, 115–129. [Google Scholar]
- Fibriansah, G.; Ibarra, K.D.; Ng, T.S.; Smith, S.A.; Tan, J.L.; Lim, X.N.; Ooi, J.S.; Kostyuchenko, V.A.; Wang, J.; de Silva, A.M.; et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 2015, 349, 88–91. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef]
- Russell, P.K.; Brandt, W.E.; Dalrymple, J.M. Chemical and Antigenic Structure of Flaviviruses; Academic Press: New York, NY, USA, 1980; pp. 503–529. [Google Scholar]
- Vial, T.; Marti, G.; Misse, D.; Pompon, J. Lipid Interactions Between Flaviviruses and Mosquito Vectors. Front. Physiol. 2021, 12, 763195. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.; Sansom, M.S. The Role of the Membrane in the Structure and Biophysical Robustness of the Dengue Virion Envelope. Structure 2016, 24, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Pfefferkorn, E.R.; Hunter, H.S. Purification and Partial Chemical Analysis of Sindbis Virus. Virology 1963, 20, 433–445. [Google Scholar] [CrossRef]
- Luukkonen, A.; Kaariainen, L.; Renkonen, O. Phospholipids of Semliki Forest virus grown in cultured mosquito cells. Biochim. Biophys Acta 1976, 450, 109–120. [Google Scholar] [CrossRef]
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim. Biophys Acta 2011, 1811, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed]
- Martin-Acebes, M.A.; Merino-Ramos, T.; Blazquez, A.B.; Casas, J.; Escribano-Romero, E.; Sobrino, F.; Saiz, J.C. The composition of West Nile virus lipid envelope unveils a role of sphingolipid metabolism in flavivirus biogenesis. J. Virol. 2014, 88, 12041–12054. [Google Scholar] [CrossRef]
- Aktepe, T.E.; Pham, H.; Mackenzie, J.M. Differential utilisation of ceramide during replication of the flaviviruses West Nile and dengue virus. Virology 2015, 484, 241–250. [Google Scholar] [CrossRef]
- Igarashi, A. Isolation of a Singh′s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 1978, 40, 531–544. [Google Scholar] [CrossRef]
- Hull, R.N.; Cherry, W.R.; Johnson, I.S. The adaption and maintenance of mammalian cells to continuous growth in tissue culture. Anat. Rec. 1956, 124, 490. [Google Scholar]
- Halstead, S.B.; Simasthien, P. Observations related to the pathogenesis of dengue hemorrhagic fever. II. Antigenic and biologic properties of dengue viruses and their association with disease response in the host. Yale J. Biol. Med. 1970, 42, 276–292. [Google Scholar]
- Hitakarun, A.; Khongwichit, S.; Wikan, N.; Roytrakul, S.; Yoksan, S.; Rajakam, S.; Davidson, A.D.; Smith, D.R. Evaluation of the antiviral activity of orlistat (tetrahydrolipstatin) against dengue virus, Japanese encephalitis virus, Zika virus and chikungunya virus. Sci. Rep. 2020, 10, 1499. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Guo, X.J.; Feng, J.N. Comparisons of Expression Levels of Heat Shock Proteins (hsp70 and hsp90) From Anaphothrips obscurus (Thysanoptera: Thripidae) in Polymorphic Adults Exposed to Different Heat Shock Treatments. J. Insect. Sci. 2018, 18, 15. [Google Scholar] [CrossRef]
- Ivanova, P.T.; Myers, D.S.; Milne, S.B.; McClaren, J.L.; Thomas, P.G.; Brown, H.A. Lipid composition of viral envelope of three strains of influenza virus - not all viruses are created equal. ACS Infect. Dis. 2015, 1, 399–452. [Google Scholar] [CrossRef]
- Liebscher, S.; Ambrose, R.L.; Aktepe, T.E.; Mikulasova, A.; Prier, J.E.; Gillespie, L.K.; Lopez-Denman, A.J.; Rupasinghe, T.W.T.; Tull, D.; McConville, M.J.; et al. Phospholipase A2 activity during the replication cycle of the flavivirus West Nile virus. PLoS Pathog. 2018, 14, e1007029. [Google Scholar] [CrossRef]
- Chotiwan, N.; Andre, B.G.; Sanchez-Vargas, I.; Islam, M.N.; Grabowski, J.M.; Hopf-Jannasch, A.; Gough, E.; Nakayasu, E.; Blair, C.D.; Belisle, J.T.; et al. Dynamic remodeling of lipids coincides with dengue virus replication in the midgut of Aedes aegypti mosquitoes. PLoS Pathog. 2018, 14, e1006853. [Google Scholar] [CrossRef] [PubMed]
- Martín-Acebes, M.A.; Blázquez, A.-B.; Jiménez de Oya, N.; Escribano-Romero, E.; Saiz, J.-C. West Nile Virus Replication Requires Fatty Acid Synthesis but Is Independent on Phosphatidylinositol-4-Phosphate Lipids. PLoS ONE 2011, 6, e24970. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.; Riley, C.; Isaac, G.; Hopf-Jannasch, A.S.; Moore, R.J.; Weitz, K.W.; Pasa-Tolic, L.; Metz, T.O.; Adamec, J.; Kuhn, R.J. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012, 8, e1002584. [Google Scholar] [CrossRef] [PubMed]
- Vial, T.; Tan, W.L.; Wong Wei Xiang, B.; Misse, D.; Deharo, E.; Marti, G.; Pompon, J. Dengue virus reduces AGPAT1 expression to alter phospholipids and enhance infection in Aedes aegypti. PLoS Pathog. 2019, 15, e1008199. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S. Mammalian lipids: Structure, synthesis and function. Essays Biochem. 2021, 65, 813–845. [Google Scholar] [CrossRef]
- Luukkonen, A.; Brummer-Korvenkontio, M.; Renkonen, O. Lipids of cultured mosquito cells (Aedes albopictus). Comparison with cultured mammalian fibroblasts (BHK 21 cells). Biochim. Biophys. Acta. 1973, 326, 256–261. [Google Scholar] [CrossRef]
- Jenkin, H.M.; McMeans, E.; Anderson, L.E.; Yang, T.K. Comparison of phospholipid composition of Aedes aegypti and Aedes albopictus cells obtained from logarithmic and stationary phases of growth. Lipids 1975, 10, 686–694. [Google Scholar] [CrossRef]
- Tongluan, N.; Ramphan, S.; Wintachai, P.; Jaresitthikunchai, J.; Khongwichit, S.; Wikan, N.; Rajakam, S.; Yoksan, S.; Wongsiriroj, N.; Roytrakul, S.; et al. Involvement of fatty acid synthase in dengue virus infection. Virol. J. 2017, 14, 28. [Google Scholar] [CrossRef]
- Vial, T.; Tan, W.L.; Deharo, E.; Misse, D.; Marti, G.; Pompon, J. Mosquito metabolomics reveal that dengue virus replication requires phospholipid reconfiguration via the remodeling cycle. Proc. Natl. Acad. Sci. USA 2020, 117, 27627–27636. [Google Scholar] [CrossRef]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Junjhon, J.; Pennington, J.G.; Edwards, T.J.; Perera, R.; Lanman, J.; Kuhn, R.J. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 2014, 88, 4687–4697. [Google Scholar] [CrossRef]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Amara, A. Flavivirus entry receptors: An update. Viruses 2013, 6, 69–88. [Google Scholar] [CrossRef]
- Rashid, M.U.; Coombs, K.M. Serum-reduced media impacts on cell viability and protein expression in human lung epithelial cells. J. Cell Physiol. 2019, 234, 7718–7724. [Google Scholar] [CrossRef]
- White, E.Z.; Pennant, N.M.; Carter, J.R.; Hawsawi, O.; Odero-Marah, V.; Hinton, C.V. Serum deprivation initiates adaptation and survival to oxidative stress in prostate cancer cells. Sci. Rep. 2020, 10, 12505. [Google Scholar] [CrossRef]
- Zanluca, C.; Mazzarotto, G.A.; Bordignon, J.; Duarte Dos Santos, C.N. Development, characterization and application of monoclonal antibodies against Brazilian Dengue virus isolates. PLoS ONE 2014, 9, e110620. [Google Scholar] [CrossRef]

| Cell Type Detected * | ||||
|---|---|---|---|---|
| Lipid Class | Full Name | C6/36 and LLC-MK2 | C6/36 Only | LLC-MK2 Only |
| Cer | Ceramide | 23 | 4 | 8 |
| SM | Sphingomyelin | 23 | 2 | 4 |
| S | Sulfoglycosphingolipids | 6 | - | 9 |
| PC | Phosphatidylcholine | 31 | - | - |
| LPC | Lyso-PC | 5 | 1 | 9 |
| PE | Phosphatidylethanolamine | 11 | 1 | - |
| LPE | Lyso-PE | 5 | 1 | 1 |
| PS | Phosphatidylserine | 13 | 2 | 3 |
| PG | Phosphatidylglycerol | 3 | 2 | 3 |
| PI | Phosphatidylinositol | 10 | - | - |
| LPI | Lyso-PI | 5 | 2 | 1 |
| CL | Cardiolipin | 23 | - | 9 |
| TG | Triglyceride | 42 | - | 6 |
| Total | 200 | 15 | 53 | |
| C6/36: DENV 2 Infected/Mock | LLC-MK2: DENV 2 Infected/Mock | |||
|---|---|---|---|---|
| Lipid Class | Upregulated | Downregulated | Upregulated | Downregulated |
| Cer | (d36:2-OH) | (d32:1), (d33:1), (d34:0), (d35:1), (d36:1), (d36:2), (d37:0-OH), (d37:1), (d38:1), (d39:1), (d39:2), (d40:1), (d40:2), (d41:2), (d42:2), (d43:1), (d43:1-OH), (d43:2), | ||
| SM | (33:1), (34:2), (34:2-OH), (36:2), (38:0) | (32:1), (34:0-OH), (34:1), (34:2), (35:2), (36:1), (36:2), (37:1), (38:1), (40:1), (40:2), (41:1), (42:1), (42:2), (44:2) | ||
| S | (d34-OH:1) | (d44-OH:2) | (d32:1), (d34:1), (d36-OH:2), (d40:2) | |
| PC | (30:0), (31:0), (32:0), (33:0), (34:0), (35:0), | (36:3), (37:2), (37:3), (40:3) | ||
| LPC | (16:1), (18:1), (20:0), (20:4) | |||
| PE | (36:1) *, (38:5) * | (32:1), (34:2), (36:1) *, (36:2), (38:5) * | ||
| LPE | (16:0), (18:0), (18:1) | |||
| PS | (40:4) | (36:0) | ||
| PG | (36:2) | (32:0) | (32:0), (36:1) | |
| PI | (34:1), (34:2) *, (35:2) *, (36:1) *, (36:3) | (34:2) *, (35:2) *, (36:1) * | ||
| LPI | (18:0) | |||
| CL | (66:04), (68:02), (68:03), (67:03), (68:04), (66:02), (66:03) | |||
| TG | (44:1), (45:1), (46:1), (46:2), (48:2), (50:3) | (60:12) | ||
| Total | 19 | 6 | 31 | 41 |
| C6/36 | LLC-MK2 | |||
|---|---|---|---|---|
| Lipid Class | Mock | DENV 2 | Mock | DENV 2 |
| Cer | (d40:0) | (d37:2) | (d33:1-OH) | |
| (d34:0-OH) | ||||
| (d36:0) | ||||
| (30:1) | (30:0) | |||
| SM | (33:0) | (38:0) | ||
| (42:0) | ||||
| (d32:1) | (d34:1) | (d33:1) | (d38:0) | |
| S | (d33:1) | (d42:1) | (d36:1) | |
| (d36:1) | (d42:2) | (d41:1) | ||
| (22:6) | (14:0) | |||
| LPC | (17:1) | |||
| (18:2) | ||||
| PE | (36:3) | |||
| LPE | (18:4) | |||
| (34:3) | ||||
| PS | (38:1) | |||
| (38:5) | ||||
| PG | (38:2) | |||
| PI | (35:0) | (35:0) | ||
| LPI | (17:0) | (20:5) | (22:6) | |
| CL | (74:10) | (66:02) | (74:11) | |
| (75:05) | ||||
| TG | (45:2) | (56:1) | (62:12) | |
| Total | 7 | 11 | 17 | 7 |
| C6/36 Lipid Species | ||||||||
| Cer | (d34:1) * | CL | (66:03) | PC | (30:0) | (34:3) | PE | (36:2) |
| (d36:1) | (66:04) | (31:0) | (34:4) * | |||||
| (d36:2) | (68:03) | (32:1) * | (35:1) | |||||
| (d38:1) | (68:04) | (32:2) * | (36:3) * | |||||
| (d39:2) | (72:08) | (33:0) | (37:1) | |||||
| (d40:1) | (33:2) | (38:1) | ||||||
| (d42:2) | (34:0) | (38:5) * | ||||||
| (34:1) * | (38:6) * | |||||||
| (34.2) * | ||||||||
| (32:0) | PI | (34:1) * | PS | (32:1) | SM | (32:1) | ||
| (34:1) * | (35:2) | (33:1) | (33:1) | |||||
| (36:2) | (36:1) * | (34:1) | (34:2) * | |||||
| PG | (36:3) | (36:2) | (38:1) | (34:2-OH) | ||||
| (36:4) | (36:3)* | (38:5) | (36:0) | |||||
| (38:1) | (38:4)* | (38:6) | (36:2) * | |||||
| (38:2) | (40:4) | (38:1) | ||||||
| (39:1) | ||||||||
| (40:2) | ||||||||
| LLC-MK2 Lipid Species | ||||||||
| Cer | (d40:2) * | CL | (68:03) | PC | (30:0) * | (32:2) | S | (d42:2) |
| (72:07) | (30:1) | (33:0) | ||||||
| (31:0) | ||||||||
| (32:0) * | ||||||||
| (32:1) * | ||||||||
| PG | (36:1) | PI | (34:1) * | PS | (36:1) * | |||
| (36:2) | (35:2) | (36:2) | ||||||
| (36:1) * | (38:1) | |||||||
| (36:4) | (40:2) | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hitakarun, A.; Williamson, M.K.; Yimpring, N.; Sornjai, W.; Wikan, N.; Arthur, C.J.; Pompon, J.; Davidson, A.D.; Smith, D.R. Cell Type Variability in the Incorporation of Lipids in the Dengue Virus Virion. Viruses 2022, 14, 2566. https://doi.org/10.3390/v14112566
Hitakarun A, Williamson MK, Yimpring N, Sornjai W, Wikan N, Arthur CJ, Pompon J, Davidson AD, Smith DR. Cell Type Variability in the Incorporation of Lipids in the Dengue Virus Virion. Viruses. 2022; 14(11):2566. https://doi.org/10.3390/v14112566
Chicago/Turabian StyleHitakarun, Atitaya, Maia Kavanagh Williamson, Nathamon Yimpring, Wannapa Sornjai, Nitwara Wikan, Christopher J. Arthur, Julien Pompon, Andrew D. Davidson, and Duncan R. Smith. 2022. "Cell Type Variability in the Incorporation of Lipids in the Dengue Virus Virion" Viruses 14, no. 11: 2566. https://doi.org/10.3390/v14112566
APA StyleHitakarun, A., Williamson, M. K., Yimpring, N., Sornjai, W., Wikan, N., Arthur, C. J., Pompon, J., Davidson, A. D., & Smith, D. R. (2022). Cell Type Variability in the Incorporation of Lipids in the Dengue Virus Virion. Viruses, 14(11), 2566. https://doi.org/10.3390/v14112566





