Network Analysis for Uncovering the Relationship between Host Response and Clinical Factors to Virus Pathogen: Lessons from SARS-CoV-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Characteristics of Analysed Data
2.3. Patient Similarity Network (PSN)
2.4. Statistics
3. Results
3.1. Univariate Analysis of Obtained Data
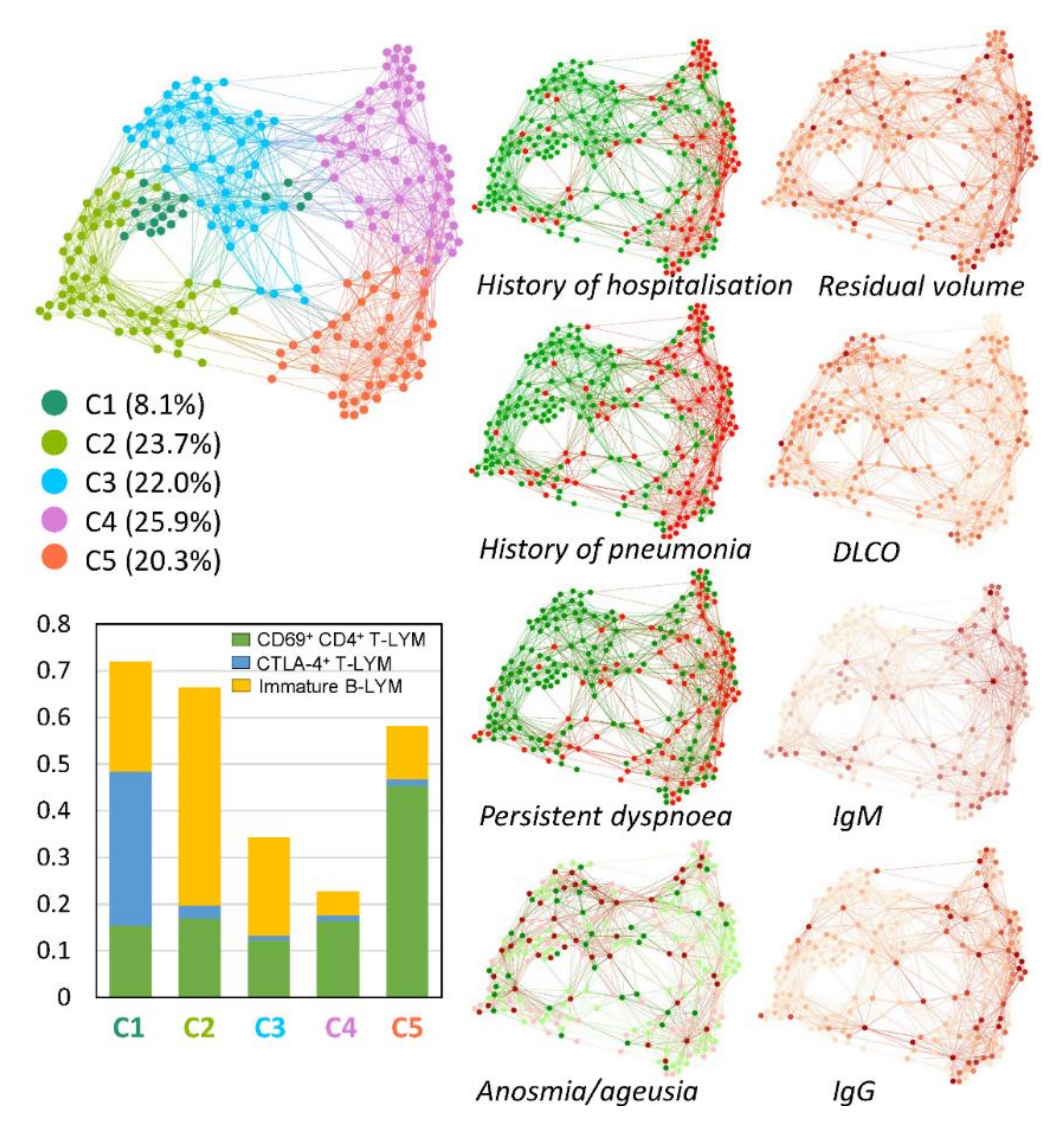
3.2. Multivariate Patient Similarity Network Analysis (PSN)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Wiech, M.; Chroscicki, P.; Swatler, J.; Stepnik, D.; De Biasi, S.; Hampel, M.; Brewinska-Olchowik, M.; Maliszewska, A.; Sklinda, K.; Durlik, M.; et al. Remodeling of T Cell Dynamics During Long COVID Is Dependent on Severity of SARS-CoV-2 Infection. Front. Immunol. 2022, 13, 886431. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtsev, I.V.; Arsentieva, N.A.; Korobova, Z.R.; Isakov, D.V.; Rubinstein, A.A.; Batsunov, O.K.; Khamitova, I.V.; Kuznetsova, R.N.; Savin, T.V.; Akisheva, T.V.; et al. Heterogenous CD8+ T Cell Maturation and ‘Polarization’ in Acute and Convalescent COVID-19 Patients. Viruses 2022, 14, 1906. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Shen, X.R.; Geng, R.; Xie, N.; Han, J.F.; Zhang, Q.M.; Shi, Z.L.; Zhou, P. A 1-year longitudinal study on COVID-19 convalescents reveals persistence of anti-SARS-CoV-2 humoral and cellular immunity. Emerg. Microbes. Infect. 2022, 11, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Rives, B.T.; Rosales, Y.Z.; Valdés, M.M.; Balbuena, H.R.; Téllez, G.M.; Pérez, J.R.; Padrón, L.C.M.; Pelier, C.R.; Lugo, F.S.; Zayas, A.V.; et al. Assessment of changes in immune status linked to COVID-19 convalescent and its clinical severity in patients and uninfected exposed relatives. Immunobiology 2022, 227, 152216. [Google Scholar] [CrossRef]
- Mishra, A.; Harichandrakumar, K.T.; Binu, V.S.; Satheesh, S.; Nair, N.S. Multivariate approach in analyzing medical data with correlated multiple outcomes: An exploration using ACCORD trial data. Clin. Epidemiol. Glob. Health 2021, 11, 100785. [Google Scholar] [CrossRef]
- Borsboom, D.; Deserno, M.K.; Rhemtulla, M.; Epskamp, S.; Fried, E.I.; McNally, R.J.; Robinaugh, D.J.; Perugini, M.; Dalege, J.; Constantini, G.; et al. Network analysis of multivariate data in psychological science. Nat. Rev. Methods Primers 2021, 1, 58. [Google Scholar] [CrossRef]
- Pérez-Segura, V.; Caro-Carretero, R.; Rua, A. Multivariate Analysis of Risk Factors of the COVID-19 Pandemic in the Community of Madrid, Spain. Int. J. Environ. Res. Public Health 2021, 18, 9227. [Google Scholar] [CrossRef]
- Li, A.Y.; Hannah, T.C.; Durbin, J.R.; Dreher, N.; McAuley, F.M.; Marayati, N.F.; Spiera, Z.; Ali, M.; Gometz, A.; Kostman, J.T.; et al. Multivariate Analysis of Black Race and Environmental Temperature on COVID-19 in the US. Am. J. Med. Sci. 2020, 360, 348–356. [Google Scholar] [CrossRef]
- Yeater, K.M.; Duke, S.E.; Riedell, W.E. Multivariate analysis: Greater insights into complex systems. Agron. J. 2015, 107, 799–810. [Google Scholar] [CrossRef]
- Everitt, B.S. Multivariate analysis: The need for data, and other problems. Br. J. Psychiatry 1975, 126, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.; Bader, G.D. Patient Similarity Networks for Precision Medicine. J. Mol. Biol. 2018, 430 Pt A, 2924–2938. [Google Scholar] [CrossRef]
- Wang, C.; Lue, W.; Kaalia, R.; Kumar, P.; Rajapakse, J.C. Network-based integration of multi-omics data for clinical outcome prediction in neuroblastoma. Sci. Rep. 2022, 12, 15425. [Google Scholar] [CrossRef] [PubMed]
- Trajerova, M.; Kriegova, E.; Mikulkova, Z.; Savara, J.; Kudelka, M.; Gallo, J. Knee osteoarthritis phenotypes based on synovial fluid immune cells correlate with clinical outcome trajectories. Osteoarthr. Cartil. 2022. Advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Gallo, J.; Kriegova, E.; Kudelka, M.; Lostak, J.; Radvansky, M. Gender Differences in Contribution of Smoking, Low Physical Activity, and High BMI to Increased Risk of Early Reoperation After TKA. J. Arthroplast. 2020, 35, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Petrackova, A.; Horak, P.; Radvansky, M.; Fillerova, R.; Smotkova Kraiczova, V.; Kudelka, M.; Mrazek, F.; Skacelova, M.; Smrzova, A.; Kriegova, E. Revealed heterogeneity in rheumatoid arthritis based on multivariate innate signature analysis. Clin. Exp. Rheumatol. 2020, 38, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Ochodkova, E.; Zehnalova, S.; Kudelka, M. Graph Construction Based on Local Representativeness. In Computing and Combinatorics: 23rd International Conference; Cao, Y., Chen, J., Eds.; COCOON: Hong Kong, China, 2017; pp. 654–665. ISBN 978-3-319-62389-4. [Google Scholar]
- Klempt, P.; Brzoň, O.; Kašný, M.; Kvapilová, K.; Hubáček, P.; Briksi, A.; Bezdíček, M.; Koudeláková, V.; Lengerová, M.; Hajdúch, M.; et al. Distribution of SARS-CoV-2 Lineages in the Czech Republic, Analysis of Data from the First Year of the Pandemic. Microorganisms 2021, 9, 1671. [Google Scholar] [CrossRef]
- Mikulkova, Z.; Manukyan, G.; Turcsanyi, P.; Kudelka, M.; Urbanova, R.; Savara, J.; Ochodkova, E.; Brychtova, Y.; Molinsky, J.; Simkovic, M.; et al. Deciphering the complex circulating immune cell microenvironment in chronic lymphocytic leukaemia using patient similarity networks. Sci. Rep. 2021, 11, 322. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 10, P10008. [Google Scholar] [CrossRef]
- Parimbelli, E.; Marini, S.; Sacchi, L.; Bellazzi, R. Patient similarity for precision medicine: A systematic review. J. Biomed. Inform. 2018, 83, 87–96. [Google Scholar] [CrossRef]
- Turcsanyi, P.; Kriegova, E.; Kudelka, M.; Radvansky, M.; Kruzova, L.; Urbanova, R.; Schneiderova, P.; Urbankova, H.; Papajik, T. Improving risk-stratification of patients with chronic lymphocytic leukemia using multivariate patient similarity networks. Leuk. Res. 2019, 79, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Parrot, T.; Gorin, J.B.; Ponzetta, A.; Maleki, K.T.; Kammann, T.; Emgård, J.; Perez-Potti, A.; Sekine, T.; Rivera-Ballesteros, O.; Karolinska COVID-19 Study Group; et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020, 5, eabe1670. [Google Scholar] [CrossRef] [PubMed]
- Shuwa, H.A.; Shaw, T.N.; Knight, S.B.; Wemyss, K.; McClure, F.A.; Pearmain, L.; Prise, I.; Jagger, C.; Morgan, D.J.; Khan, S.; et al. Alterations in T and B cell function persist in convalescent COVID-19 patients. Med 2021, 2, 720–735.e4. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Hernández, V.A.; Torres-Ruíz, J.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Páez-Franco, J.C.; Meza-Sánchez, D.E.; Juárez-Vega, G.; Pérez-Fragoso, A.; Ortiz-Navarrete, V.; Ponce-de-León, A.; et al. B Cell Subsets as Severity-Associated Signatures in COVID-19 Patients. Front. Immunol. 2020, 11, 611004. [Google Scholar] [CrossRef]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef]
- Aghbash, P.S.; Eslami, N.; Shamekh, A.; Entezari-Maleki, T.; Baghi, H.B. SARS-CoV-2 infection: The role of PD-1/PD-L1 and CTLA-4 axis. Life Sci. 2021, 270, 119124. [Google Scholar] [CrossRef]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef]
- Sorokowski, P.; Karwowski, M.; Misiak, M.; Marczak, M.K.; Dziekan, M.; Hummel, T.; Sorokowska, A. Sex Differences in Human Olfaction: A Meta-Analysis. Front. Psychol. 2019, 10, 242. [Google Scholar] [CrossRef]
- Bilinska, K.; Jakubowska, P.; Von Bartheld, C.S.; Butowt, R. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chem. Neurosci. 2020, 11, 1555–1562. [Google Scholar] [CrossRef]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Newell, K.L.; Clemmer, D.C.; Cox, J.B.; Kayode, Y.I.; Zoccoli-Rodriguez, V.; Taylor, H.E.; Endy, T.P.; Wilmore, J.R.; Winslow, G.M. Switched and unswitched memory B cells detected during SARS-CoV-2 convalescence correlate with limited symptom duration. PLoS ONE 2021, 16, e0244855. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.; Hui, S.; Isserlin, R.; Shah, M.A.; Kaka, H.; Bader, G.D. netDx: Interpretable patient classification using integrated patient similarity networks. Mol. Syst. Biol. 2019, 15, e8497. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; Vallières, F.; Bentall, R.P.; Shevlin, M.; McBride, O.; Hartman, T.K.; McKay, R.; Bennett, K.; Mason, L.; Gibson-Miller, J.; et al. Psychological characteristics associated with COVID-19 vaccine hesitancy and resistance in Ireland and the United Kingdom. Nat. Commun. 2021, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Our World in Data. Coronavirus (COVID-19) Vaccinations. Available online: https://ourworldindata.org/covid-vaccinations?country=OWID_WRL (accessed on 7 October 2022).
- World Health Organisation. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 7 October 2022).
- Dzieciolowska, S.; Hamel, D.; Gadio, S.; Dionne, M.; Gagnon, D.; Robitaille, L.; Cook, E.; Caron, I.; Talib, A.; Parkes, L.; et al. COVID-19 vaccine acceptance, hesitancy, and refusal among Canadian healthcare workers: A multicenter survey. Am. J. Infect. Control. 2021, 49, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A.; Walley, K.R.; Kalil, A.C.; Fowler, R. The Potential for Increasing Risk of Consent Refusal in COVID-19 Trials: Considering Underlying Reasons and Responses. Ann. Am. Thorac. Soc. 2022, 19, 1446–1447. [Google Scholar] [CrossRef] [PubMed]
| Factors | No of the Patients (%) | |
|---|---|---|
| Gender (men/women) | 124/126 | |
| Age, median years (min-max) | 55 (19–87) | |
| Serum IgG, median (min-max) (AU/mL) | 118 (6.21–390) | |
| Serum IgM, median (min-max) (AU/mL) | 5.65 (0.06–111) | |
| Comorbidities | ||
| Pulmonary arterial embolisation | 10 (4%) | |
| Diabetes mellitus | 31 (12%) | |
| Ischemic heart disease | 12 (5%) | |
| Medical history related to COVID-19 | ||
| Hospitalisation | 82 (33%) | |
| Pneumonia | 111 (44%) | |
| Anosmia/ageusia | 80 (32%) | |
| Pulmonary interstitial changes | 29 (12%) | |
| Systemic glucocorticoid therapy | 23 (9%) | |
| Persistent dyspnoea | 89 (36%) | |
| Persistent cough | 61 (24%) | |
| Pulmonary Function Tests | Measured±SD | Percentage of Predicted Values±SD |
| VC (l) | 3.91 ± 1.10 | 101.3 ± 16.81 |
| FVC (l) | 3.88 ± 1.10 | 104.19 ± 17.28 |
| FEV1 (l) | 3.12 ± 0.89 | 101.89 ± 17.15 |
| FEV1/VC | - | 80.06 ± 6.37 |
| PEF (l/min) | 7.30 ± 1.99 | 96.88 ± 19.33 |
| TLC (l) | 6.33 ± 1.34 | 104.77 ± 15.95 |
| DLCO (l/s) | 7.43 ± 2.33 | 80.55 ± 17.21 |
| KCO (l/s) | 1.33 ± 0.25 | 88.19 ± 15.37 |
| (a) Pneumonia | (b) Hospitalisation | |||||
|---|---|---|---|---|---|---|
| Distribution of Immune Cells [%] | No | Yes | p-Value | No | Yes | p-Value |
| Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | |||
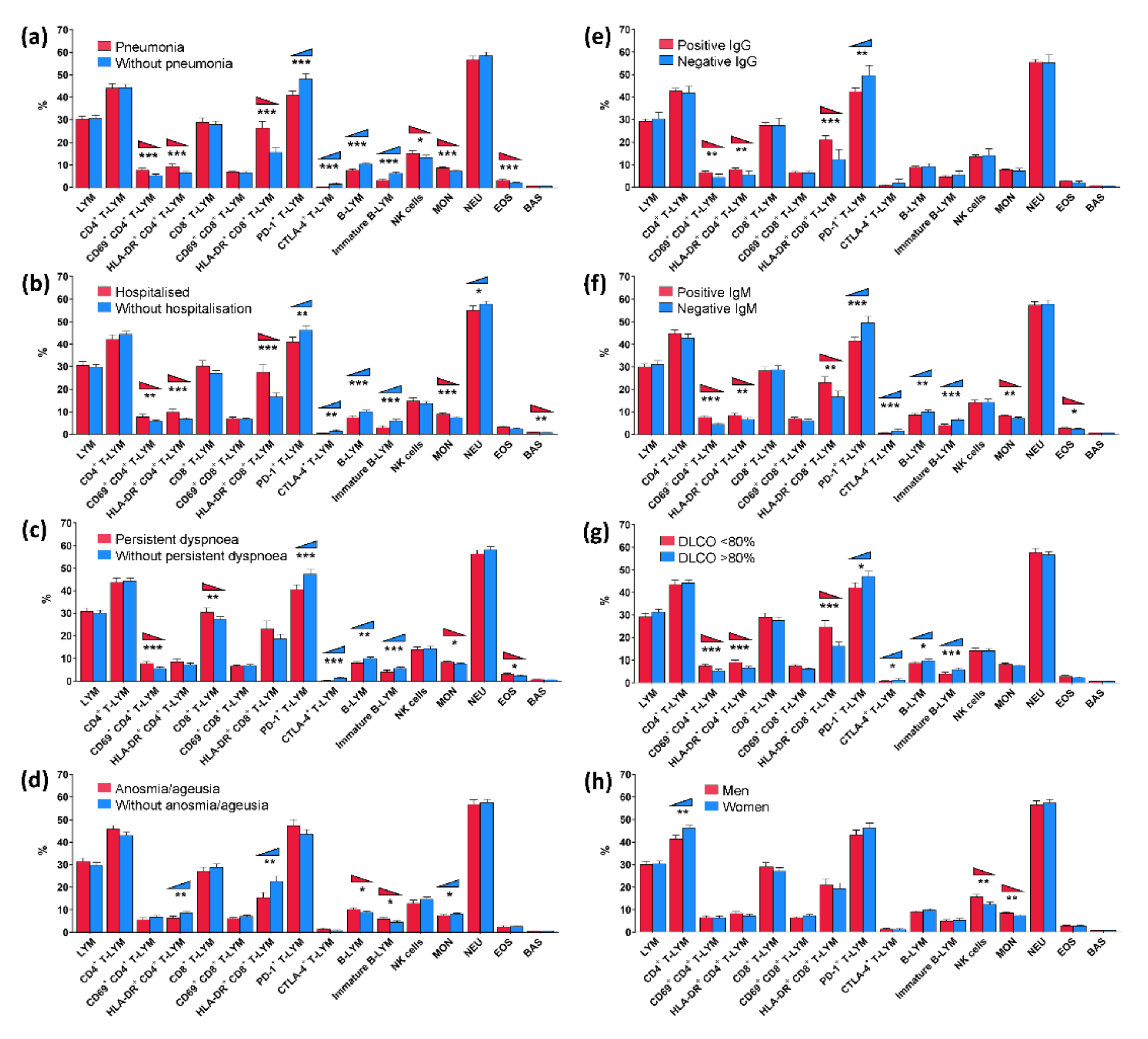
| Lymphocytes (LYM) | 30.8 (29.4–32.3) | 30.3 (28.9–31.8) | 0.856 | 30.4 (29.2–31.6) | 31.1 (29.3–33.0) | 0.534 |
| CD4+ T-LYM | 44.4 (43.0–45.9) | 44.2 (42.3–46.1) | 0.746 | 45.1 (43.7–46.4) | 42.8 (40.6–44.9) | 0.127 |
| CD69+ CD4+ T-LYM | 5.38 (4.66–6.11) | 7.77 (6.85–8.70) | <0.001 | 5.77 (5.12–6.42) | 7.75 (6.57–8.93) | 0.004 |
| HLA-DR+ CD4+ T-LYM | 6.58 (5.87–7.29) | 9.39 (8.27–10.5) | <0.001 | 6.78 (6.15–7.41) | 9.89 (8.49–11.3) | <0.001 |
| CD8+ T-LYM | 28.2 (26.6–29.7) | 29.1 (27.1–31.1) | 0.638 | 27.5 (26.2–28.9) | 30.8 (28.3–33.3) | 0.051 |
| CD69+ CD8+ T-LYM | 6.63 (6.00–7.26) | 7.00 (6.44–7.57) | 0.140 | 6.68 (6.13–7.23) | 7.01 (6.34–7.69) | 0.221 |
| HLA-DR+ CD8+ T-LYM | 15.8 (13.9–17.7) | 26.6 (23.5–29.6) | <0.001 | 16.8 (15.0–18.6) | 28.0 (24.4–31.7) | <0.001 |
| PD-1+ T-LYM | 48.3 (46.0–50.6) | 41.1 (39.2–43.0) | <0.001 | 46.9 (44.8–49.0) | 41.6 (39.4–43.8) | 0.008 |
| CTLA-4+ T-LYM | 1.56 (0.95–2.17) | 0.29 (0.20–0.39) | 0.001 | 1.35 (0.83–1.87) | 0.33 (0.19–0.46) | 0.007 |
| B-LYM | 10.5 (9.77– 11.2) | 7.72 (7.05–8.40) | <0.001 | 10.2 (9.51–10.8) | 7.50 (6.71–8.28) | <0.001 |
| Immature B-LYM | 6.39 (5.69–7.09) | 3.20 (2.45–3.94) | <0.001 | 6.03 (5.38–6.68) | 2.91 (2.06–3.76) | <0.001 |
| NK cells | 13.4 (12.3–14.5) | 15.2 (13.9–16.5) | 0.036 | 13.8 (12.8–14.8) | 15.0 (13.4–16.5) | 0.221 |
| Monocytes (MON) | 7.34 (6.96–7.73) | 8.80 (8.30–9.30) | <0.001 | 7.49 (7.14–7.84) | 8.97 (8.37–9.58) | <0.001 |
| Neutrophils (NEU) | 58.6 (57.0–60.2) | 56.8 (55.1–58.5) | 0.148 | 58.8 (57.4–60.2) | 55.9 (53.7–58.1) | 0.042 |
| Eosinophils (EOS) | 2.19 (1.91–2.47) | 3.21 (2.67–3.75) | 0.001 | 2.42 (2.13–2.71) | 3.07 (2.43–3.72) | 0.082 |
| Basophils (BAS) | 0.69 (0.64–0.74) | 0.75 (0.68–0.82) | 0.124 | 0.68 (0.63–0.73) | 0.79 (0.71–0.86) | 0.005 |
| (c) Persistent Dyspnoea | (d) Anosmia/Ageusia | |||||
| Distribution of Immune Cells | No | Yes | p-Value | No | Yes | p-Value |
| [%] | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | ||
| Lymphocytes (LYM) | 30.4 (29.0–31.7) | 31.1 (29.5–32.7) | 0.589 | 30.1 (28.9–31.4) | 31.7 (30.0–33.4) | 0.295 |
| CD4+ T-LYM | 44.5 (43.1–46.0) | 44.0 (42.0–45.9) | 0.908 | 43.4 (41.9–44.9) | 46.1 (44.5–47.8) | 0.077 |
| CD69+ CD4+ T-LYM | 5.57 (4.84–6.31) | 7.88 (6.96–8.81) | <0.001 | 6.78 (6.03–7.52) | 5.65 (4.70–6.60) | 0.093 |
| HLA-DR+ CD4+ T-LYM | 7.34 (6.61–8.07) | 8.57 (7.33–9.80) | 0.120 | 8.51 (7.67–9.35) | 6.30 (5.41–7.19) | 0.005 |
| CD8+ T-LYM | 27.4 (25.8–28.9) | 30.7 (28.7–32.7) | 0.010 | 29.2 (27.6–30.7) | 27.4 (25.5–29.2) | 0.286 |
| CD69+ CD8+ T-LYM | 6.89 (6.29–7.49) | 6.60 (6.03–7.18) | 0.908 | 7.12 (6.54–7.70) | 6.12 (5.55–6.69) | 0.093 |
| HLA-DR+ CD8+ T-LYM | 18.8 (16.8–20.8) | 23.3 (19.7–26.8) | 0.097 | 22.8 (20.5–25.2) | 15.5 (13.2–17.8) | 0.004 |
| PD-1+ T-LYM | 47.7 (45.5–49.8) | 40.8 (38.8–42.9) | <0.001 | 44.0 (42.1–45.9) | 47.7 (44.8–50.5) | 0.077 |
| CTLA-4+ T-LYM | 1.39 (0.84–1.94) | 0.36 (0.21–0.50) | <0.001 | 0.85 (0.44–1.25) | 1.37 (0.65–2.08) | 0.173 |
| B-LYM | 10.0 (9.31–10.7) | 8.06 (7.32–8.80) | 0.002 | 8.88 (8.23–9.54) | 10.1 (9.29–11.0) | 0.032 |
| Immature B-LYM | 5.63 (4.96–6.30) | 3.95 (3.03–4.87) | <0.001 | 4.62 (3.97–5.27) | 5.85 (4.85–6.84) | 0.035 |
| NK cells | 14.4 (13.3–15.4) | 13.9 (12.5–15.2) | 0.615 | 14.8 (13.7–15.8) | 13.0 (11.6–14.4) | 0.077 |
| Monocytes (MON) | 7.72 (7.31–8.12) | 8.41 (7.90–8.93) | 0.034 | 8.23 (7.84–8.61) | 7.44 (6.90–7.99) | 0.037 |
| Neutrophils (NEU) | 58.5 (57.0–60.1) | 56.6 (54.9–58.4) | 0.141 | 58.1 (56.6–59.6) | 57.4 (55.4–59.4) | 0.711 |
| Eosinophils (EOS) | 2.41 (2.07–2.75) | 3.01 (2.50–3.53) | 0.034 | 2.68 (2.34–3.02) | 2.52 (1.98–3.06) | 0.286 |
| Basophils (BAS) | 0.69 (0.64–0.74) | 0.76 (0.70–0.83) | 0.069 | 0.72 (0.67–0.77) | 0.72 (0.64–0.79) | 0.628 |
| (e) Positive Serum IgG | (f) Positive Serum IgM | |||||
| Distribution of Immune Cells | No | Yes | p-Value | No | Yes | p-Value |
| [%] | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | ||
| Lymphocytes (LYM) | 31.6 (28.6–34.6) | 30.4 (29.3–31.5) | 0.814 | 31.2 (29.5–32.9) | 30.1 (28.8–31.5) | 0.850 |
| CD4+ T-LYM | 43.6 (40.5–46.7) | 44.4 (43.1–45.7) | 0.831 | 43.0 (41.2–44.8) | 45.0 (43.5–46.6) | 0.084 |
| CD69+ CD4+ T-LYM | 4.54 (2.90–6.18) | 6.82 (6.18–7.46) | 0.002 | 4.50 (3.69–5.31) | 7.70 (6.91–8.48) | <0.001 |
| HLA-DR+ CD4+ T-LYM | 5.88 (4.15–7.61) | 8.23 (7.52–8.94) | 0.002 | 6.74 (5.83–7.65) | 8.55 (7.65–9.45) | 0.009 |
| CD8+ T-LYM | 28.5 (25.1–32.0) | 28.5 (27.2–29.9) | 0.983 | 28.8 (26.9–30.7) | 28.4 (26.8–30.1) | 0.662 |
| CD69+ CD8+ T-LYM | 6.61 (5.51–7.71) | 6.86 (6.38–7.35) | 0.983 | 6.26 (5.65–6.88) | 7.15 (6.54–7.75) | 0.072 |
| HLA-DR+ CD8+ T-LYM | 12.9 (8.42–17.5) | 22.0 (20.0–24.0) | <0.001 | 16.7 (14.1–19.3) | 23.2 (20.7–25.6) | 0.001 |
| PD-1+ T-LYM | 51.7 (47.1–56.4) | 44.2 (42.5–46.0) | 0.005 | 49.8 (46.9–52.7) | 41.8 (40.0–43.6) | <0.001 |
| CTLA-4+ T-LYM | 1.85 (-0.08–3.78) | 0.87 (0.55–1.18) | 0.798 | 1.64 (0.77–2.50) | 0.59 (0.31–0.86) | <0.001 |
| B-LYM | 9.55 (8.10–11.0) | 9.26 (8.67–9.84) | 0.937 | 10.1 (9.27–11.0) | 8.76 (8.07–9.44) | 0.009 |
| Immature B-LYM | 5.84 (4.11–7.56) | 4.83 (4.24–5.43) | 0.273 | 6.57 (5.55–7.60) | 3.99 (3.38–4.60) | <0.001 |
| NK cells | 14.7 (11.7–17.8) | 14.1 (13.2–15.0) | 0.983 | 14.3 (12.8–15.8) | 14.2 (13.1–15.3) | 0.999 |
| Monocytes (MON) | 7.61 (6.45–8.77) | 8.08 (7.74–8.41) | 0.273 | 7.29 (6.76–7.81) | 8.43 (8.02–8.84) | 0.001 |
| Neutrophils (NEU) | 57.7 (54.1–61.2) | 57.9 (56.6–59.2) | 0.983 | 58.1 (56.2–60.0) | 57.8 (56.2–59.4) | 0.662 |
| Eosinophils (EOS) | 2.05 (1.34–2.77) | 2.74 (2.42–3.06) | 0.067 | 2.34 (1.80–2.88) | 2.82 (2.48–3.17) | 0.010 |
| Basophils (BAS) | 0.63 (0.55–0.71) | 0.72 (0.68–0.77) | 0.451 | 0.73 (0.66–0.79) | 0.71 (0.65–0.76) | 0.666 |
| (g) DLCO <80% | (g) Gender | |||||
| Distribution of Immune Cells | No | Yes | p-Value | Male | Female | p-Value |
| [%] | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | ||
| Lymphocytes (LYM) | 31.6 (30.3–32.9) | 29.6 (27.9–31.2) | 0.284 | 30.5 (29.0–32.0) | 30.8 (29.4–32.2) | 0.692 |
| CD4+ T-LYM | 44.6 (43.1–46.1) | 44.1 (42.2–46.0) | 0.952 | 42.1 (40.5–43.6) | 46.8 (45.2–48.4) | 0.002 |
| CD69+ CD4+ T-LYM | 5.46 (4.69–6.23) | 7.27 (6.41–8.13) | <0.001 | 6.46 (5.65–7.27) | 6.35 (5.47–7.22) | 0.783 |
| HLA-DR+ CD4+ T-LYM | 6.59 (5.86–7.32) | 9.04 (7.97–10.1) | <0.001 | 8.30 (7.33–9.26) | 7.22 (6.37–8.07) | 0.218 |
| CD8+ T-LYM | 27.8 (26.3–29.4) | 29.2 (27.2–31.1) | 0.697 | 29.5 (27.6–31.3) | 27.6 (26.1–29.1) | 0.594 |
| CD69+ CD8+ T-LYM | 6.23 (5.79–6.67) | 7.40 (6.63–8.18) | 0.078 | 6.36 (5.85–6.87) | 7.25 (6.54–7.97) | 0.186 |
| HLA-DR+ CD8+ T-LYM | 16.3 (14.3–18.2) | 24.8 (21.8–27.8) | <0.001 | 21.2 (18.6–23.9) | 19.5 (17.0–22.0) | 0.692 |
| PD-1+ T-LYM | 47.6 (45.2–49.9) | 42.9 (40.7–45.0) | 0.018 | 43.8 (41.6–45.9) | 46.8 (44.5–49.2) | 0.186 |
| CTLA-4+ T-LYM | 1.32 (0.72–1.93) | 0.71 (0.33–1.08) | 0.039 | 1.06 (0.52–1.59) | 0.98 (0.49–1.46) | 0.770 |
| B-LYM | 9.90 (9.25–10.5) | 8.75 (7.91–9.60) | 0.012 | 8.88 (8.16–9.59) | 9.7 (8.98–10.5) | 0.285 |
| Immature B-LYM | 5.93 (5.15–6.70) | 4.13 (3.38–4.88) | <0.001 | 4.81 (4.08–5.54) | 5.26 (4.43–6.08) | 0.692 |
| NK cells | 14.2 (13.0–15.3) | 14.2 (12.9–15.5) | 0.839 | 15.8 (14.5–17.0) | 12.5 (11.4–13.6) | 0.002 |
| Monocytes (MON) | 7.66 (7.25–8.07) | 8.28 (7.78–8.79) | 0.194 | 8.51 (8.03–8.99) | 7.38 (6.99–7.76) | 0.007 |
| Neutrophils (NEU) | 57.3 (55.9–58.8) | 58.5 (56.5–60.4) | 0.790 | 57.5 (55.7–59.2) | 58.3 (56.7–59.8) | 0.783 |
| Eosinophils (EOS) | 2.38 (2.06–2.71) | 2.81 (2.32–3.29) | 0.346 | 2.64 (2.28–3.00) | 2.61 (2.16–3.06) | 0.654 |
| Basophils (BAS) | 0.70 (0.64–0.76) | 0.74 (0.68–0.81) | 0.219 | 0.71 (0.65–0.77) | 0.72 (0.66–0.78) | 0.783 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sova, M.; Kudelka, M.; Raska, M.; Mizera, J.; Mikulkova, Z.; Trajerova, M.; Ochodkova, E.; Genzor, S.; Jakubec, P.; Borikova, A.; et al. Network Analysis for Uncovering the Relationship between Host Response and Clinical Factors to Virus Pathogen: Lessons from SARS-CoV-2. Viruses 2022, 14, 2422. https://doi.org/10.3390/v14112422
Sova M, Kudelka M, Raska M, Mizera J, Mikulkova Z, Trajerova M, Ochodkova E, Genzor S, Jakubec P, Borikova A, et al. Network Analysis for Uncovering the Relationship between Host Response and Clinical Factors to Virus Pathogen: Lessons from SARS-CoV-2. Viruses. 2022; 14(11):2422. https://doi.org/10.3390/v14112422
Chicago/Turabian StyleSova, Milan, Milos Kudelka, Milan Raska, Jan Mizera, Zuzana Mikulkova, Marketa Trajerova, Eliska Ochodkova, Samuel Genzor, Petr Jakubec, Alena Borikova, and et al. 2022. "Network Analysis for Uncovering the Relationship between Host Response and Clinical Factors to Virus Pathogen: Lessons from SARS-CoV-2" Viruses 14, no. 11: 2422. https://doi.org/10.3390/v14112422
APA StyleSova, M., Kudelka, M., Raska, M., Mizera, J., Mikulkova, Z., Trajerova, M., Ochodkova, E., Genzor, S., Jakubec, P., Borikova, A., Stepanek, L., Kosztyu, P., & Kriegova, E. (2022). Network Analysis for Uncovering the Relationship between Host Response and Clinical Factors to Virus Pathogen: Lessons from SARS-CoV-2. Viruses, 14(11), 2422. https://doi.org/10.3390/v14112422





