Intra-Patient Evolution of HIV-2 Molecular Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Amplification and Sequence Analysis
2.3. Phylogenetic Analysis
2.4. Diversity Analysis
2.5. Statistical Analysis
3. Results
3.1. Study Cohort and Stratification as Faster and Slower Disease Progressors
3.2. Phylogenetic Analyses and Genetic Diversity of the Env V1–C3 Regions
3.3. Potential N-Linked Glycosylation Sites in the Env V1–C3 Regions
3.4. Length and Charge Variation of the Env V1–C3 Regions
3.5. Predicted Coreceptor Use
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaffar, S.; Grant, A.D.; Whitworth, J.; Smith, P.G.; Whittle, H. The natural history of HIV-1 and HIV-2 infections in adults in Africa: A literature review. Bull. World Health Organ. 2004, 82, 462–469. [Google Scholar] [PubMed]
- Esbjornsson, J.; Mansson, F.; Kvist, A.; da Silva, Z.J.; Andersson, S.; Fenyo, E.M.; Isberg, P.E.; Biague, A.J.; Lindman, J.; Palm, A.A.; et al. Long-term follow-up of HIV-2-related AIDS and mortality in Guinea-Bissau: A prospective open cohort study. Lancet HIV 2019, 6, e25–e31. [Google Scholar] [CrossRef]
- Nyamweya, S.; Hegedus, A.; Jaye, A.; Rowland-Jones, S.; Flanagan, K.L.; Macallan, D.C. Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev. Med. Virol. 2013, 23, 221–240. [Google Scholar] [CrossRef] [PubMed]
- Gueudin, M.; Damond, F.; Braun, J.; Taieb, A.; Lemee, V.; Plantier, J.C.; Chene, G.; Matheron, S.; Brun-Vezinet, F.; Simon, F. Differences in proviral DNA load between HIV-1- and HIV-2-infected patients. AIDS 2008, 22, 211–215. [Google Scholar] [CrossRef]
- Andersson, S.; Norrgren, H.; da Silva, Z.; Biague, A.; Bamba, S.; Kwok, S.; Christopherson, C.; Biberfeld, G.; Albert, J. Plasma viral load in HIV-1 and HIV-2 singly and dually infected individuals in Guinea-Bissau, West Africa: Significantly lower plasma virus set point in HIV-2 infection than in HIV-1 infection. Arch. Intern. Med. 2000, 160, 3286–3293. [Google Scholar] [CrossRef]
- Berry, N.; Ariyoshi, K.; Jaffar, S.; Sabally, S.; Corrah, T.; Tedder, R.; Whittle, H. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J. Hum. Virol. 1998, 1, 457–468. [Google Scholar]
- Popper, S.J.; Sarr, A.D.; Travers, K.U.; Gueye-Ndiaye, A.; Mboup, S.; Essex, M.E.; Kanki, P.J. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 1999, 180, 1116–1121. [Google Scholar] [CrossRef]
- MacNeil, A.; Sarr, A.D.; Sankale, J.L.; Meloni, S.T.; Mboup, S.; Kanki, P. Direct evidence of lower viral replication rates in vivo in human immunodeficiency virus type 2 (HIV-2) infection than in HIV-1 infection. J. Virol. 2007, 81, 5325–5330. [Google Scholar] [CrossRef]
- Blaak, H.; van der Ende, M.E.; Boers, P.H.; Schuitemaker, H.; Osterhaus, A.D. In vitro replication capacity of HIV-2 variants from long-term aviremic individuals. Virology 2006, 353, 144–154. [Google Scholar] [CrossRef][Green Version]
- Borrego, P.; Marcelino, J.M.; Rocha, C.; Doroana, M.; Antunes, F.; Maltez, F.; Gomes, P.; Novo, C.; Barroso, H.; Taveira, N. The role of the humoral immune response in the molecular evolution of the envelope C2, V3 and C3 regions in chronically HIV-2 infected patients. Retrovirology 2008, 5, 78. [Google Scholar] [CrossRef]
- MacNeil, A.; Sankale, J.L.; Meloni, S.T.; Sarr, A.D.; Mboup, S.; Kanki, P. Long-term intrapatient viral evolution during HIV-2 infection. J. Infect. Dis. 2007, 195, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Sankale, J.L.; de la Tour, R.S.; Renjifo, B.; Siby, T.; Mboup, S.; Marlink, R.G.; Essex, M.E.; Kanki, P.J. Intrapatient variability of the human immunodeficiency virus type 2 envelope V3 loop. AIDS Res. Hum. Retrovir. 1995, 11, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Borggren, M.; Repits, J.; Sterjovski, J.; Uchtenhagen, H.; Churchill, M.J.; Karlsson, A.; Albert, J.; Achour, A.; Gorry, P.R.; Fenyo, E.M.; et al. Increased sensitivity to broadly neutralizing antibodies of end-stage disease R5 HIV-1 correlates with evolution in Env glycosylation and charge. PLoS ONE 2011, 6, e20135. [Google Scholar] [CrossRef] [PubMed]
- Bjorndal, A.; Deng, H.; Jansson, M.; Fiore, J.R.; Colognesi, C.; Karlsson, A.; Albert, J.; Scarlatti, G.; Littman, D.R.; Fenyo, E.M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 1997, 71, 7478–7487. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.; Backstrom, E.; Scarlatti, G.; Bjorndal, A.; Matsuda, S.; Rossi, P.; Albert, J.; Wigzell, H. Length variation of glycoprotein 120 V2 region in relation to biological phenotypes and coreceptor usage of primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 2001, 17, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, A.; Parsmyr, K.; Sandstrom, E.; Fenyo, E.M.; Albert, J. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 1994, 32, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Mild, M.; Kvist, A.; Esbjornsson, J.; Karlsson, I.; Fenyo, E.M.; Medstrand, P. Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection. Infect. Genet. Evol. 2010, 10, 356–364. [Google Scholar] [CrossRef]
- Repits, J.; Sterjovski, J.; Badia-Martinez, D.; Mild, M.; Gray, L.; Churchill, M.J.; Purcell, D.F.; Karlsson, A.; Albert, J.; Fenyo, E.M.; et al. Primary HIV-1 R5 isolates from end-stage disease display enhanced viral fitness in parallel with increased gp120 net charge. Virology 2008, 379, 125–134. [Google Scholar] [CrossRef]
- Connor, R.I.; Sheridan, K.E.; Ceradini, D.; Choe, S.; Landau, N.R. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. J. Exp. Med. 1997, 185, 621–628. [Google Scholar] [CrossRef]
- Blaak, H.; Boers, P.H.; Gruters, R.A.; Schuitemaker, H.; van der Ende, M.E.; Osterhaus, A.D. CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. J. Virol. 2005, 79, 1686–1700. [Google Scholar] [CrossRef]
- Marcelino, J.M.; Borrego, P.; Nilsson, C.; Familia, C.; Barroso, H.; Maltez, F.; Doroana, M.; Antunes, F.; Quintas, A.; Taveira, N. Resistance to antibody neutralization in HIV-2 infection occurs in late stage disease and is associated with X4 tropism. AIDS 2012, 26, 2275–2284. [Google Scholar] [CrossRef] [PubMed]
- Morner, A.; Bjorndal, A.; Albert, J.; Kewalramani, V.N.; Littman, D.R.; Inoue, R.; Thorstensson, R.; Fenyo, E.M.; Bjorling, E. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 1999, 73, 2343–2349. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Brandin, E.; Vincic, E.; Jansson, M.; Blaxhult, A.; Gyllensten, K.; Moberg, L.; Brostrom, C.; Fenyo, E.M.; Albert, J. Evolution of human immunodeficiency virus type 2 coreceptor usage, autologous neutralization, envelope sequence and glycosylation. J. Gen. Virol. 2005, 86, 3385–3396. [Google Scholar] [CrossRef] [PubMed]
- Trevino, A.; Soriano, V.; Poveda, E.; Parra, P.; Cabezas, T.; Caballero, E.; Roc, L.; Rodriguez, C.; Eiros, J.M.; Lopez, M.; et al. HIV-2 viral tropism influences CD4+ T cell count regardless of viral load. J. Antimicrob. Chemother. 2014, 69, 2191–2194. [Google Scholar] [CrossRef]
- Visseaux, B.; Charpentier, C.; Rouard, C.; Fagard, C.; Glohi, D.; Tubiana, R.; Damond, F.; Brun-Vezinet, F.; Matheron, S.; Descamps, D.; et al. HIV-2 X4 tropism is associated with lower CD4+ cell count in treatment-experienced patients. AIDS 2014, 28, 2160–2162. [Google Scholar] [CrossRef]
- Bunnik, E.M.; Pisas, L.; van Nuenen, A.C.; Schuitemaker, H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol 2008, 82, 7932–7941. [Google Scholar] [CrossRef]
- Wei, X.; Decker, J.M.; Wang, S.; Hui, H.; Kappes, J.C.; Wu, X.; Salazar-Gonzalez, J.F.; Salazar, M.G.; Kilby, J.M.; Saag, M.S.; et al. Antibody neutralization and escape by HIV-1. Nature 2003, 422, 307–312. [Google Scholar] [CrossRef]
- Sagar, M.; Wu, X.; Lee, S.; Overbaugh, J. Human immunodeficiency virus type 1 V1–V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 2006, 80, 9586–9598. [Google Scholar] [CrossRef]
- Bjorling, E.; Scarlatti, G.; von Gegerfelt, A.; Albert, J.; Biberfeld, G.; Chiodi, F.; Norrby, E.; Fenyo, E.M. Autologous neutralizing antibodies prevail in HIV-2 but not in HIV-1 infection. Virology 1993, 193, 528–530. [Google Scholar] [CrossRef]
- de Silva, T.I.; Aasa-Chapman, M.; Cotten, M.; Hue, S.; Robinson, J.; Bibollet-Ruche, F.; Sarge-Njie, R.; Berry, N.; Jaye, A.; Aaby, P.; et al. Potent autologous and heterologous neutralizing antibody responses occur in HIV-2 infection across a broad range of infection outcomes. J. Virol. 2012, 86, 930–946. [Google Scholar] [CrossRef]
- Palm, A.A.; Lemey, P.; Jansson, M.; Mansson, F.; Kvist, A.; Szojka, Z.; Biague, A.; da Silva, Z.J.; Rowland-Jones, S.L.; Norrgren, H.; et al. Low Postseroconversion CD4(+) T-cell Level Is Associated with Faster Disease Progression and Higher Viral Evolutionary Rate in HIV-2 Infection. mBio 2019, 8, e01245-18. [Google Scholar] [CrossRef] [PubMed]
- Mansson, F.; Biague, A.; da Silva, Z.J.; Dias, F.; Nilsson, L.A.; Andersson, S.; Fenyo, E.M.; Norrgren, H. Prevalence and incidence of HIV-1 and HIV-2 before, during and after a civil war in an occupational cohort in Guinea-Bissau, West Africa. AIDS 2009, 23, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Norrgren, H.; Andersson, S.; Naucler, A.; Dias, F.; Johansson, I.; Biberfeld, G. HIV-1, HIV-2, HTLV-I/II and Treponema pallidum infections: Incidence, prevalence, and HIV-2-associated mortality in an occupational cohort in Guinea-Bissau. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995, 9, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.L.; Taylor, J.M.; Detels, R.; Hofmann, B.; Melmed, R.; Nishanian, P.; Giorgi, J.V. The prognostic value of cellular and serologic markers in infection with human immunodeficiency virus type 1. N. Engl. J. Med. 1990, 322, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.M.; Fahey, J.L.; Detels, R.; Giorgi, J.V. CD4 percentage, CD4 number, and CD4:CD8 ratio in HIV infection: Which to choose and how to use. J. Acquir Immune Defic. Syndr. 1989, 2, 114–124. [Google Scholar] [PubMed]
- Anglaret, X.; Diagbouga, S.; Mortier, E.; Meda, N.; Verge-Valette, V.; Sylla-Koko, F.; Cousens, S.; Laruche, G.; Ledru, E.; Bonard, D.; et al. CD4+ T-lymphocyte counts in HIV infection: Are European standards applicable to African patients? J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1997, 14, 361–367. [Google Scholar] [CrossRef]
- Barroso, H.; Borrego, P.; Bartolo, I.; Marcelino, J.M.; Familia, C.; Quintas, A.; Taveira, N. Evolutionary and structural features of the C2, V3 and C3 envelope regions underlying the differences in HIV-1 and HIV-2 biology and infection. PLoS ONE 2011, 6, e14548. [Google Scholar] [CrossRef]
- Zhang, M.; Gaschen, B.; Blay, W.; Foley, B.; Haigwood, N.; Kuiken, C.; Korber, B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 2004, 14, 1229–1246. [Google Scholar] [CrossRef]
- Visseaux, B.; Charpentier, C.; Taieb, A.; Damond, F.; Benard, A.; Larrouy, L.; Chene, G.; Brun-Vezinet, F.; Matheron, S.; Descamps, D.; et al. Concordance between HIV-2 genotypic coreceptor tropism predictions based on plasma RNA and proviral DNA. AIDS 2013, 27, 292–295. [Google Scholar] [CrossRef]
- Visseaux, B.; Hurtado-Nedelec, M.; Charpentier, C.; Collin, G.; Storto, A.; Matheron, S.; Larrouy, L.; Damond, F.; Brun-Vezinet, F.; Descamps, D.; et al. Molecular determinants of HIV-2 R5-X4 tropism in the V3 loop: Development of a new genotypic tool. J. Infect. Dis. 2012, 205, 111–120. [Google Scholar] [CrossRef]
- Doring, M.; Borrego, P.; Buch, J.; Martins, A.; Friedrich, G.; Camacho, R.J.; Eberle, J.; Kaiser, R.; Lengauer, T.; Taveira, N.; et al. A genotypic method for determining HIV-2 coreceptor usage enables epidemiological studies and clinical decision support. Retrovirology 2016, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- HIV Sequence Database. Available online: http://www.hiv.lanl.gov/ (accessed on 5 November 2012).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Zwickl, D.J. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. Ph.D. Thesis, The University of Texas at Austin, Austin, TX, USA, 2006. [Google Scholar]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Anisimova, M.; Gil, M.; Dufayard, J.F.; Dessimoz, C.; Gascuel, O. Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-based Approximation Schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef]
- Drummond, A.; Pybus, O.G.; Rambaut, A. Inference of viral evolutionary rates from molecular sequences. Adv. Parasitol. 2003, 54, 331–358. [Google Scholar]
- Firth, C.; Kitchen, A.; Shapiro, B.; Suchard, M.A.; Holmes, E.C.; Rambaut, A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol. Biol. Evol. 2010, 27, 2038–2051. [Google Scholar] [CrossRef]
- Polzer, S.; Dittmar, M.T.; Schmitz, H.; Meyer, B.; Muller, H.; Krausslich, H.G.; Schreiber, M. Loss of N-linked glycans in the V3-loop region of gp120 is correlated to an enhanced infectivity of HIV-1. Glycobiology 2001, 11, 11–19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, W.; Nie, J.; Prochnow, C.; Truong, C.; Jia, Z.; Wang, S.; Chen, X.S.; Wang, Y. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology 2013, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.R.; Lassen, K.G.; Yang, H.C.; Quinn, T.C.; Ray, S.C.; Blankson, J.N.; Siliciano, R.F. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J. Virol. 2006, 80, 4758–4770. [Google Scholar] [CrossRef]
- Shankarappa, R.; Gupta, P.; Learn, G.H., Jr.; Rodrigo, A.G.; Rinaldo, C.R., Jr.; Gorry, M.C.; Mullins, J.I.; Nara, P.L.; Ehrlich, G.D. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 1998, 241, 251–259. [Google Scholar] [CrossRef][Green Version]
- Esbjornsson, J.; Mansson, F.; Kvist, A.; Isberg, P.E.; Nowroozalizadeh, S.; Biague, A.J.; da Silva, Z.J.; Jansson, M.; Fenyo, E.M.; Norrgren, H.; et al. Inhibition of HIV-1 disease progression by contemporaneous HIV-2 infection. N. Engl. J. Med. 2012, 367, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Barroso, H.; Taveira, N. Evidence for negative selective pressure in HIV-2 evolution in vivo. Infect. Genet. Evol. 2005, 5, 239–246. [Google Scholar] [CrossRef] [PubMed]
| Individual | Sample | CD4% | CD4abs | AIDS a | ART a | Clones b | Stratification c | C2 d | V3 d | C3 d | Total nr PNGS e | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 & 3 | 206 | 238 | 241 | 248 | 272 | 278 | 289 | 296 | 300 | 310 | 343 | 357 | 365 | 366 | 367 | 368 | 369 | 371 | 371.25 | 371.75 | 375 | 389 | C2–C3 | V1–C3 | |||||||
| DL3405 | 2001-10-20 | 37 | 798 | N | N | 11 | Slower | Slower | R | 11 | 20 | |||||||||||||||||||||
| 2004-06-26 | 25 | 198 | N | N | 8 | R | 11 | 20.5 | ||||||||||||||||||||||||
| 2008-10-07 | 46 | 782 | N | N | 9 | R | 11 | 21 | ||||||||||||||||||||||||
| DL3542 | 1994-11-07 | 35 | 235 | N | N | 9 | Faster | Slower | K | 10 | 17 | |||||||||||||||||||||
| 2013-09-25 | 36 | n/a | Y | N | 8 | E | 11 | 20 | ||||||||||||||||||||||||
| DL2051 | 1996-02-12 | 33 | 253 | N | N | 7 | Faster | Slower | 11 | 16 | ||||||||||||||||||||||
| 1997-09-26 | 38 | 613 | N | N | 5 | 10 | 20 | |||||||||||||||||||||||||
| 2002-07-31 | 20 | 179 | N | N | 11 | 10 | 13 | |||||||||||||||||||||||||
| 2006-11-28 | 33 | 369 | N | N | 5 | 11 | 16 | |||||||||||||||||||||||||
| 2009-10-07 | 22 | 386 | N | N | 2 | 11 | 16 | |||||||||||||||||||||||||
| DL2876 | 1995-12-28 | 35 | 559 | N | N | 6 | Faster | Slower | 10.5 | 17 | ||||||||||||||||||||||
| 1996-12-19 | 30 | 594 | N | N | 6 | 10 | 17 | |||||||||||||||||||||||||
| 2000-08-23 | 23 | 282 | N | N | 9 | 11 | 18 | |||||||||||||||||||||||||
| DL3654 | 1998-02-04 | 25 | 592 | N | N | 7 | Slower | Slower | 11 | 17 | ||||||||||||||||||||||
| 2013-09-27 | 29 | n/a | N | N | 7 | 11 | 19 | |||||||||||||||||||||||||
| DL2533 | 1991-04-19 | n/a | n/a | N | N | 10 | Slower | Slower | 11 | 20 | ||||||||||||||||||||||
| 2001-12-04 | 29 | 515 | N | N | 7 | 11 | 18 | |||||||||||||||||||||||||
| 2002-07-24 | 25 | 300 | N | N | 4 | 10.5 | 16 | |||||||||||||||||||||||||
| 2006-11-14 | 32 | 611 | N | N | 12 | 12 | 18 | |||||||||||||||||||||||||
| DL2316 | 2001-10-21 | 23 | 305 | N | N | 10 | Faster | Faster | 10 | 18.5 | ||||||||||||||||||||||
| 2002-07-22 | 26 | 161 | Y | N | 7 | 10 | 19 | |||||||||||||||||||||||||
| DL2794 | 1993-11-10 | 11 | 116 | N | N | 10 | Slower | Faster | 10 | 18 | ||||||||||||||||||||||
| 1996-04-08 | 25 | 330 | N | N | 7 | 11 | 20 | |||||||||||||||||||||||||
| 2004-06-26 | 13 | 91 | Y | N | 5 | 10 | 18 | |||||||||||||||||||||||||
| DL3941 | 2004-12-08 | n/a | n/a | N | N | 12 | Faster | Faster | 11 | 19 | ||||||||||||||||||||||
| 2008-09-18 | 23 | 386 | N | N | 5 | 10 | 16 | |||||||||||||||||||||||||
| 2010-03-10 | 18 | 267 | N | N | 8 | 10.5 | 15 | |||||||||||||||||||||||||
| 2013-09-23 | 13 | n/a | Y | N | 6 | 10 | 14 | |||||||||||||||||||||||||
| DL2381 | 2003-03-13 | 32 | 1156 | N | N | 7 | Faster | Faster | 11 | 18 | ||||||||||||||||||||||
| 2004-06-10 | 23 | n/a | N | N | 11 | 11 | 18 | |||||||||||||||||||||||||
| 2009-09-28 | 15 | 253 | N | N | 12 | 11 | 19 | |||||||||||||||||||||||||
| DL2335 | 2002-07-16 | 21 | 192 | N | N | 9 | Faster | Faster | 10 | 17 | ||||||||||||||||||||||
| 2003-11-14 | 20 | 240 | N | N | 12 | 10 | 16 | |||||||||||||||||||||||||
| 2004-11-05 | 21 | 295 | N | N | 12 | 8 | 14 | |||||||||||||||||||||||||
| 2007-10-11 | n/a | n/a | N | N | 7 | 9 | 15 | |||||||||||||||||||||||||
| 2009-09-29 | 25 | 196 | Y | N | 2 | 10 | 17.5 | |||||||||||||||||||||||||
| DL3647 | 2001-06-25 | 25 | 318 | N | N | 9 | Faster | Faster | 9 | 17 | ||||||||||||||||||||||
| 2006-11-11 | 17 | 138 | Y | N | 5 | 8 | 20 | |||||||||||||||||||||||||
| 2010-03-19 | 6 | 60 | Y | N | 7 | 10 | 20 | |||||||||||||||||||||||||
| DL3646 | 1996-04-08 | 21 | 670 | N | N | 9 | Slower | Faster | K | 11 | 19 | |||||||||||||||||||||
| 2006-11-10 | 17 | 373 | N | N | 6 | K | 10 | 18 | ||||||||||||||||||||||||
| 2008-10-04 | 14 | 326 | N | N | 9 | K | 10 | 18 | ||||||||||||||||||||||||
| 2009-10-19 | n/a | n/a | N | N | 10 | K | 10 | 18 | ||||||||||||||||||||||||
| 2010-05-11 | 20 | 231 | N | Y | 6 | K | 10 | 18 | ||||||||||||||||||||||||
| DL3222 | 1997-02-06 | 17 | 748 | N | N | 9 | Slower | Faster | K | 11 | 18 | |||||||||||||||||||||
| 2001-07-26 | 25 | 702 | N | N | 11 | K | 10 | 16 | ||||||||||||||||||||||||
| DL3740 | 2007-10-24 | 16 | 352 | N | N | 7 | Slower | Faster | 10 | 16 | ||||||||||||||||||||||
| 2009-03-30 | 17 | 318 | N | N | 8 | 11 | 19 | |||||||||||||||||||||||||
| 2009-10-23 | n/a | n/a | N | N | 4 | 11 | 18.5 | |||||||||||||||||||||||||
| 2013-09-16 | 16 | n/a | N | Y | 9 | 10 | 17 | |||||||||||||||||||||||||
| DL2386 | 2008-10-16 | n/a | n/a | N | N | 5 | Faster | Faster | 10 | 17 | ||||||||||||||||||||||
| 2010-02-19 | 16 | 280 | N | N | 6 | 11 | 18 | |||||||||||||||||||||||||
| 2013-09-26 | 15 | n/a | N | N | 4 | 11 | 18 | |||||||||||||||||||||||||
| Percentage of clones with PNGS | 98.5 | 99.3 | 88.5 | 97.1 | 99.3 | 99.0 | 95.8 | 7.6 | 89.0 | 96.1 | 5.6 | 1.0 | 88.3 | 5.6 | 0.5 | 2.4 | 25.2 | 28.6 | 4.6 | 0.2 | 0.2 | 0.2 | ||||||||||
| Slope a | Slope b | CD4% = 35 c | CD4% = 14 c | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Property | Slower | Faster | p | All Individuals | p | Slower | Faster | p | Slower | Faster | p |
| Diversity | −2.94 × 10−4 | −4.18 × 10−4 | 0.876 | −3.59 × 10−4 | 0.364 | 0.012 | 0.013 | 0.822 | 0.019 | 0.021 | 0.822 |
| V1-C3 PNGS | 0.149 | −0.011 | 0.033 * | N/A | N/A | 18.528 | 17.440 | 0.316 | 15.389 | 17.682 | 0.049 * |
| V1-C3 PNGS d | 0.143 | −0.005 | 0.089 | N/A | N/A | 18.355 | 17.381 | 0.432 | 15.355 | 17.495 | 0.073 |
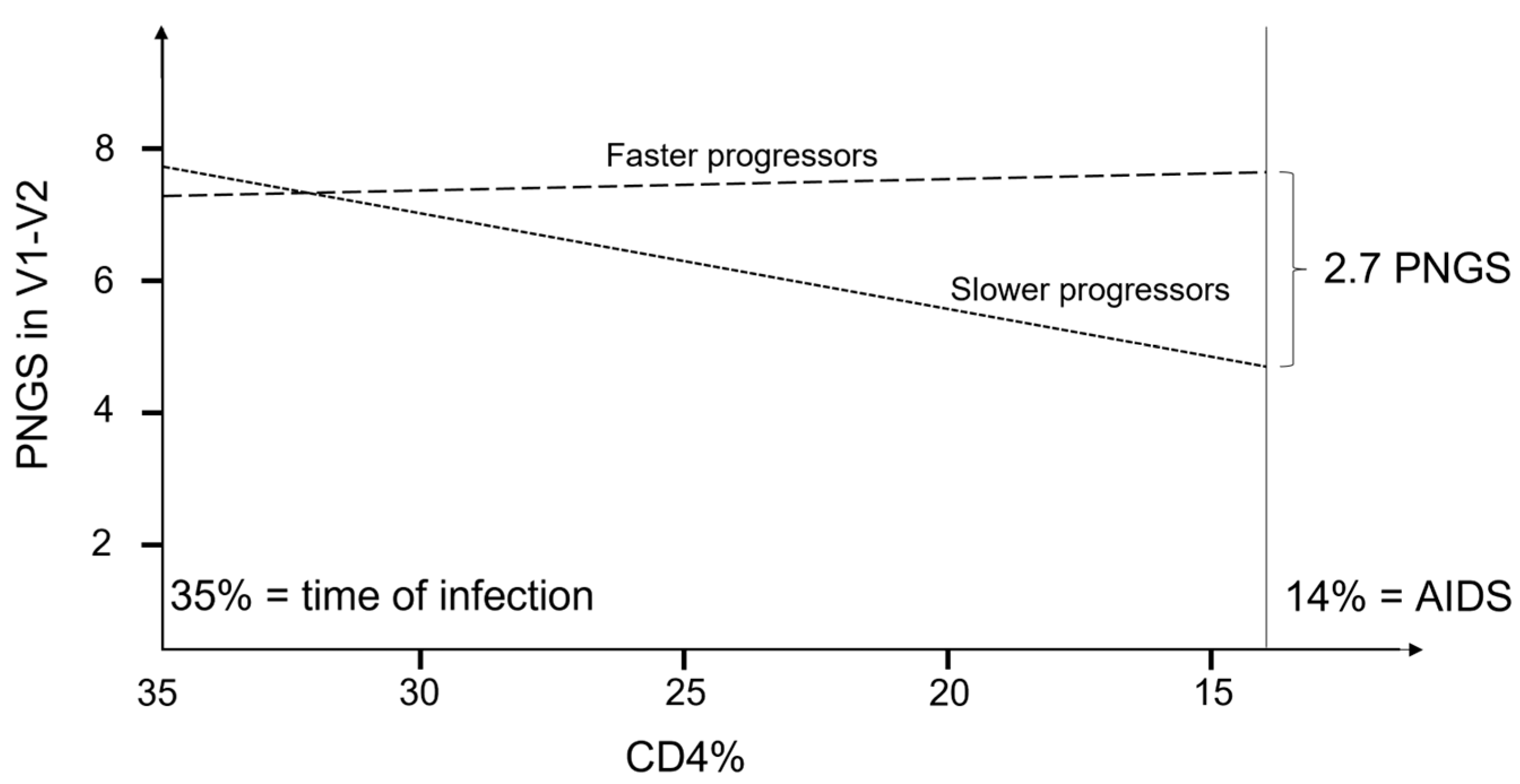
| V1-V2 PNGS | 0.141 | −0.010 | 0.026 * | N/A | N/A | 7.718 | 7.270 | 0.662 | 4.747 | 7.489 | 0.014 * |
| V1-V2 PNGS d | 0.145 | −0.002 | 0.055 | N/A | N/A | 7.604 | 7.145 | 0.666 | 4.565 | 7.179 | 0.013 * |
| C2 PNGS | −2.28 × 10−4 | 4.28 × 10−4 | 0.978 | 4.60 × 10−5 | 0.997 | 7.886 | 7.729 | 0.514 | 7.885 | 7.728 | 0.514 |
| V3 PNGS e | - | - | - | - | - | - | - | - | - | - | - |
| C3 PNGS | 0.004 | 0.003 | 0.976 | 0.004 | 0.718 | 1.921 | 1.533 | 0.121 | 1.843 | 1.455 | 0.121 |
| V1-C3 length | 0.162 | −0.206 | 0.112 | 0.007 | 0.957 | 284.916 | 284.792 | 0.965 | 284.764 | 284.640 | 0.965 |
| V1-V2 length | 0.152 | −0.202 | 0.119 | 0.003 | 0.978 | 93.995 | 93.674 | 0.897 | 93.922 | 93.602 | 0.897 |
| C2 length e | - | - | - | - | - | - | - | - | - | - | - |
| V3 length e | - | - | - | - | - | - | - | - | - | - | - |
| C3 length e | - | - | - | - | - | - | - | - | - | - | - |
| V1-C3 charge | −0.056 | −0.062 | 0.952 | −0.059 | 0.178 | 7.319 | 6.695 | 0.519 | 8.567 | 7.943 | 0.519 |
| V1-V2 charge | −0.087 | −0.050 | 0.511 | −0.068 | 0.017 * | −3.154 | −4.417 | 0.093 | −1.719 | −2.982 | 0.093 |
| C2 charge | 0.027 | 0.037 | 0.805 | 0.032 | 0.109 | 3.596 | 3.842 | 0.623 | 2.929 | 3.174 | 0.623 |
| V3 charge | −0.013 | 0.011 | 0.272 | −0.001 | 0.931 | 5.064 | 5.612 | 0.167 | 5.084 | 5.632 | 0.167 |
| C3 charge | 0.013 | −0.099 | 0.131 | −0.053 | 0.166 | 1.386 | 1.192 | 0.782 | 2.493 | 2.299 | 0.782 |
| Slope a | Slope b | ||||
|---|---|---|---|---|---|
| Property | Slower | Faster | p | All individuals | p |
| Diversity | −4.15 × 10−4 | 1.54 × 10−3 | 0.009 * | N/A | N/A |
| V1-C3 PNGS | 3.24 × 10−2 | −6.09 × 10−2 | 0.273 | −1.17 × 10−2 | 0.782 |
| V1-V2 PNGS | −6.86 × 10−3 | −3.58 × 10−2 | 0.709 | −2.00 × 10−2 | 0.602 |
| C2 PNGS | 5.97 × 10−3 | 7.54 × 10−5 | 0.801 | 3.20 × 10−3 | 0.781 |
| V3 PNGS c | - | - | - | - | - |
| C3 PNGS | 3.15 × 10−2 | −3.23 × 10−2 | 0.003 * | N/A | N/A |
| V1-C3 length | 1.42 × 10−1 | 1.25 × 10−1 | 0.945 | 1.36 × 10−1 | 0.257 |
| V1-V2 length | 1.44 × 10−1 | 9.17 × 10−2 | 0.827 | 1.23 × 10−1 | 0.295 |
| C2 length c | - | - | - | - | - |
| V3 length c | - | - | - | - | - |
| C3 length c | - | - | - | - | - |
| V1-C3 charge | −2.49 × 10−2 | 3.39 × 10−2 | 0.551 | −1.48 × 10−3 | 0.976 |
| V1-V2 charge | 1.51 × 10−2 | 1.13 × 10−1 | 0.137 | 5.39 × 10−2 | 0.109 |
| C2 charge | 5.96 × 10−4 | −1.96 × 10−2 | 0.613 | −7.92 × 10−3 | 0.685 |
| V3 charge | −7.93 × 10−3 | −3.25 × 10−2 | 0.347 | −1.83 × 10−2 | 0.158 |
| C3 charge | −8.00 × 10−4 | 3.42 × 10−2 | 0.574 | 1.51 × 10−2 | 0.624 |
| Prediction by Major Determinants a | Prediction by Geno2Pheno b | ||||||
|---|---|---|---|---|---|---|---|
| Individual | Sample | CD4 Count | CD4% | Fulfilled Criteria for CXCR4 Use | Fraction of CXCR4 Using Sequences | X4-Capable | Fraction of CXCR4 Using Sequences |
| DL2533 | 2001-12-04 | 515 | 29 | V3 charge +7 | 1/6 | No c | 0/6 |
| DL2876 | 1996-12-19 | 594 | 30 | L18S | 1/7 | Yes | 1/7 |
| DL3647 | 2006-12-11 | 138 | 17 | V3 charge +7 | 4/5 | No c | 0/5 |
| DL3740 | 2007-10-24 | 352 | 16 | V3 charge +7 | 2/7 | No c | 0/7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palm, A.A.; Esbjörnsson, J.; Kvist, A.; Månsson, F.; Biague, A.; Norrgren, H.; Jansson, M.; Medstrand, P. Intra-Patient Evolution of HIV-2 Molecular Properties. Viruses 2022, 14, 2447. https://doi.org/10.3390/v14112447
Palm AA, Esbjörnsson J, Kvist A, Månsson F, Biague A, Norrgren H, Jansson M, Medstrand P. Intra-Patient Evolution of HIV-2 Molecular Properties. Viruses. 2022; 14(11):2447. https://doi.org/10.3390/v14112447
Chicago/Turabian StylePalm, Angelica A., Joakim Esbjörnsson, Anders Kvist, Fredrik Månsson, Antonio Biague, Hans Norrgren, Marianne Jansson, and Patrik Medstrand. 2022. "Intra-Patient Evolution of HIV-2 Molecular Properties" Viruses 14, no. 11: 2447. https://doi.org/10.3390/v14112447
APA StylePalm, A. A., Esbjörnsson, J., Kvist, A., Månsson, F., Biague, A., Norrgren, H., Jansson, M., & Medstrand, P. (2022). Intra-Patient Evolution of HIV-2 Molecular Properties. Viruses, 14(11), 2447. https://doi.org/10.3390/v14112447






