A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9
Abstract
1. Introduction
2. Materials and Methods
2.1. Production and Purification of GP4
2.2. Cryo-EM Imaging and Image Processing
2.3. Atomic Model Building and Refinement
3. Results
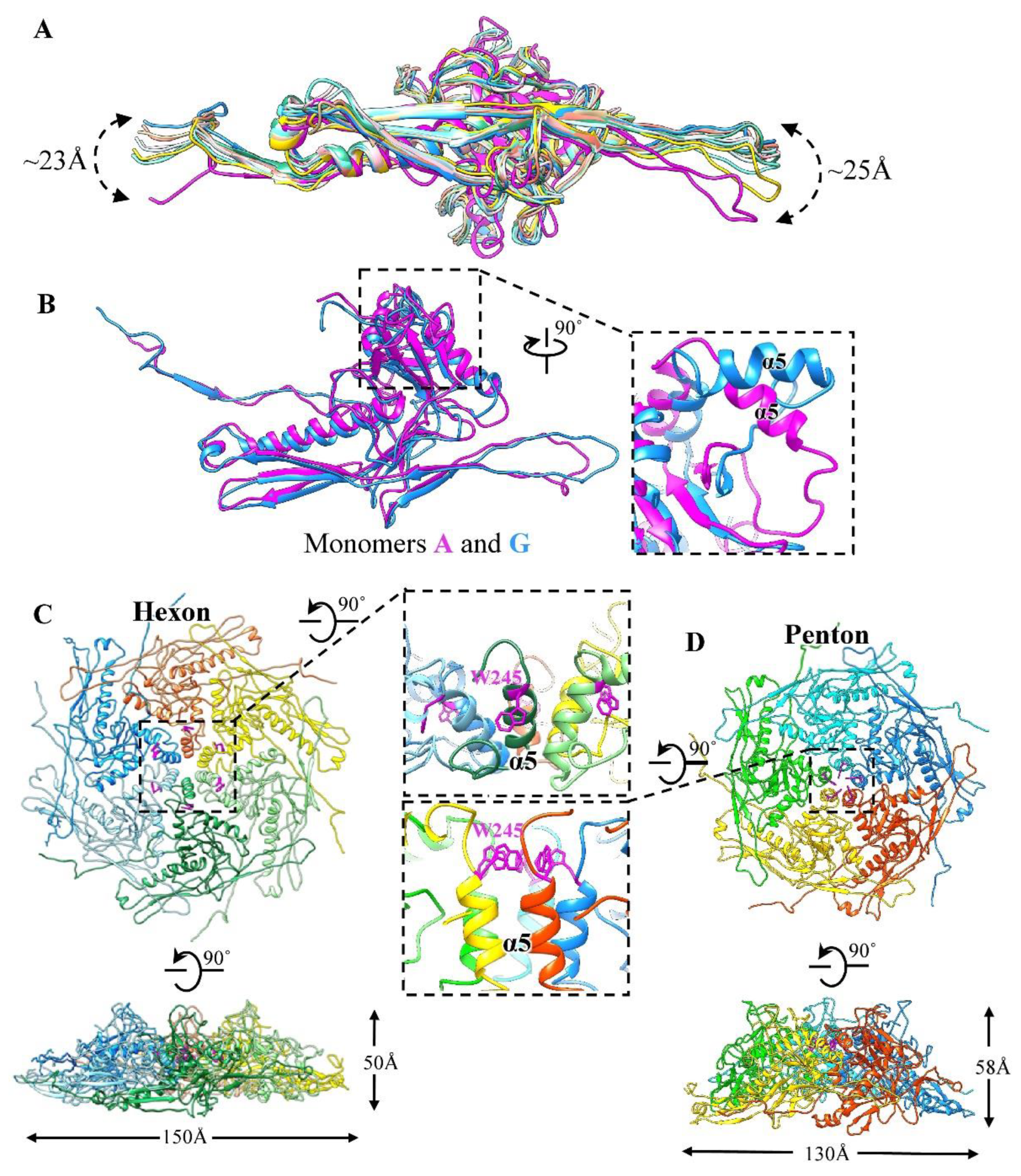
3.1. Overall Structure of the GP4 Head
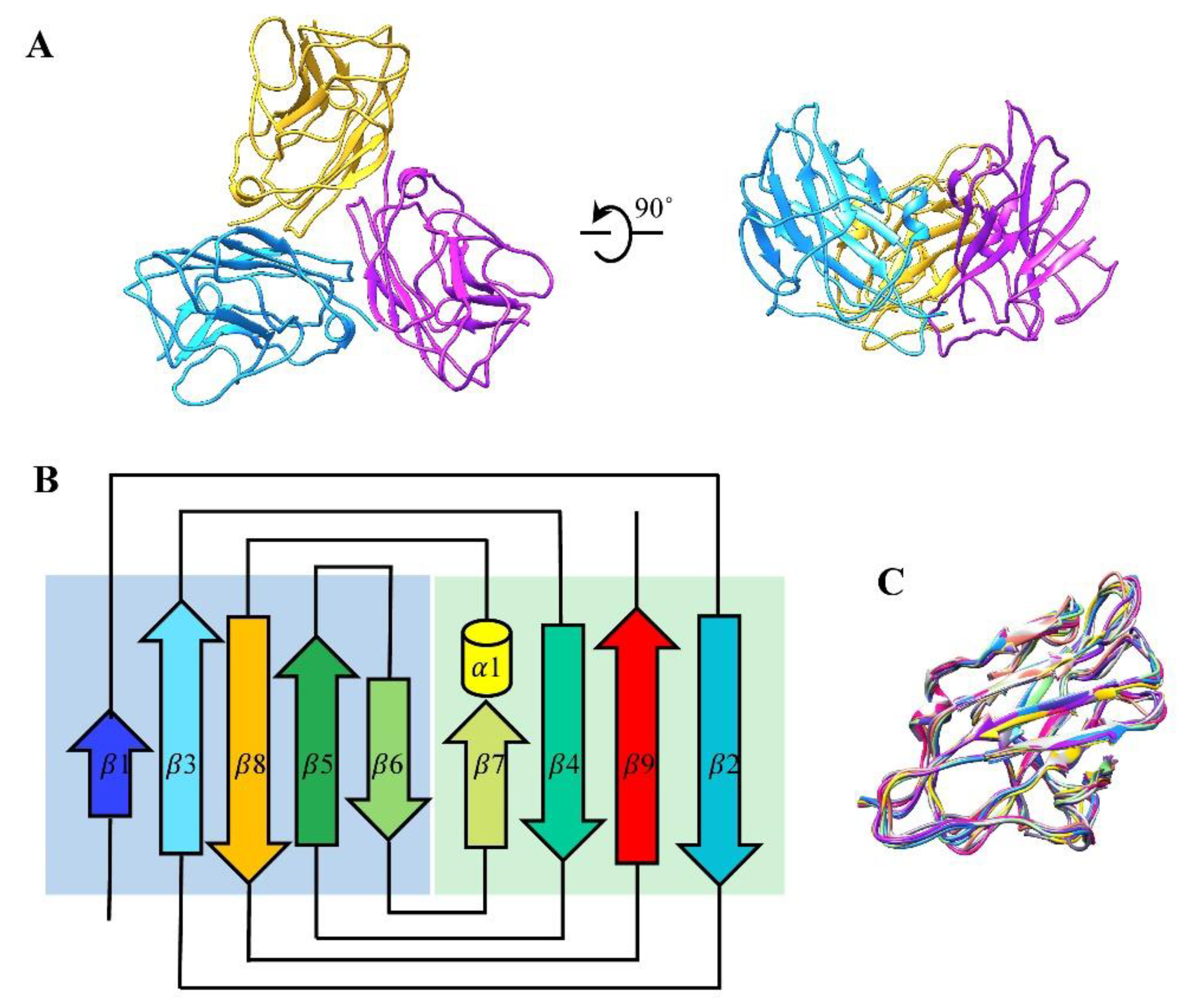
3.2. Structure of the Major Capsid Protein
3.3. Structure of Cement Proteins
3.4. Interactions among Capsid Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Veesler, D.; Cambillau, C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 2011, 75, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Fokine, A.; Rossmann, M.G. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 2014, 4, e28281. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.H.; Chiou, J. Protein chainmail variants in dsDNA viruses. AIMS Biophys. 2015, 2, 200–218. [Google Scholar] [CrossRef] [PubMed]
- Suhanovsky, M.M.; Teschke, C.M. Nature’s favorite building block: Deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 2015, 479–480, 487–497. [Google Scholar] [CrossRef]
- Aksyuk, A.A.; Rossmann, M.G. Bacteriophage assembly. Viruses 2011, 3, 172–203. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, Z.H. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 2018, 6, eaao7298. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, J.; Zhu, D.; Wang, N.; Gao, Q.; Chen, W.; Tang, H.; Wang, J.; Zhang, X.; Liu, H.; et al. Cryo-EM structure of a herpesvirus capsid at 3.1 Å. Science 2018, 6, eaao7283. [Google Scholar] [CrossRef]
- Dai, X.; Gong, D.; Xiao, Y.; Wu, T.T.; Sun, R.; Zhou, Z.H. CryoEM and mutagenesis reveal that the smallest capsid protein cements and stabilizes Kaposi’s sarcoma-associated herpesvirus capsid. Proc. Natl. Acad. Sci. USA 2015, 112, E649–E656. [Google Scholar] [CrossRef]
- Dedeo, C.L.; Teschke, C.M.; Alexandrescu, A.T. Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins. Viruses 2020, 12, 1163. [Google Scholar] [CrossRef]
- Dai, X.; Gong, D.; Lim, H.; Jih, J.; Wu, T.T.; Sun, R.; Zhou, Z.H. Structure and mutagenesis reveal essential capsid protein interactions for KSHV replication. Nature 2018, 553, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 2000, 289, 2129–2133. [Google Scholar] [CrossRef] [PubMed]
- Mannige, R.V.; Brooks, C.L., 3rd. Periodic table of virus capsids: Implications for natural selection and design. PLoS ONE 2010, 5, e9423. [Google Scholar] [CrossRef]
- Luque, A.; Benler, S.; Lee, D.Y.; Brown, C.; White, S. The Missing Tailed Phages: Prediction of Small Capsid Candidates. Microorganisms 2020, 8, 1944. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, D.; Gui, M.; Xiang, Y. Structural assembly of the tailed bacteriophage varphi29. Nat Commun. 2019, 10, 2366. [Google Scholar] [CrossRef]
- Gonzalez, B.; Monroe, L.; Li, K.; Yan, R.; Wright, E.; Walter, T.; Kihara, D.; Weintraub, S.T.; Thomas, J.A.; Serwer, P.; et al. Phage G Structure at 6.1A Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus. J. Mol. Biol. 2020, 432, 4139–4153. [Google Scholar] [CrossRef]
- Guo, F.; Liu, Z.; Fang, P.A.; Zhang, Q.; Wright, E.T.; Wu, W.; Zhang, C.; Vago, F.; Ren, Y.; Jakana, J.; et al. Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions. Proc. Natl. Acad. Sci. USA 2014, 111, E4606–E4614. [Google Scholar] [CrossRef]
- Zhao, H.; Li, K.; Lynn, A.Y.; Aron, K.E.; Yu, G.; Jiang, W.; Tang, L. Structure of a headful DNA-packaging bacterial virus at 2.9 A resolution by electron cryo-microscopy. Proc. Natl. Acad. Sci. USA 2017, 114, 3601–3606. [Google Scholar] [CrossRef]
- Baker, M.L.; Hryc, C.F.; Zhang, Q.; Wu, W.; Jakana, J.; Haase-Pettingell, C.; Afonine, P.V.; Adams, P.D.; King, J.A.; Jiang, W.; et al. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proc. Natl. Acad. Sci. USA 2013, 110, 12301–12306. [Google Scholar] [CrossRef]
- Zhang, J.T.; Yang, F.; Du, K.; Li, W.F.; Chen, Y.; Jiang, Y.L.; Li, Q.; Zhou, C.Z. Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1. Structure 2022, 30, 240–251.e244. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, H.; Jin, L.; Czornyj, E.; Hodes, A.; Hui, W.H.; Nieh, A.W.; Miller, J.F.; Zhou, Z.H. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution. Elife 2013, 2, e01299. [Google Scholar] [CrossRef] [PubMed]
- Tripler, T.N.; Kaplan, A.R.; Alexandrescu, A.T.; Teschke, C.M. Conservation and Divergence of the I-Domain Inserted into the Ubiquitous HK97 Coat Protein Fold in P22-Like Bacteriophages. J. Virol. 2019, 93, e00007–e00019. [Google Scholar] [CrossRef] [PubMed]
- Parent, K.N.; Tang, J.; Cardone, G.; Gilcrease, E.B.; Janssen, M.E.; Olson, N.H.; Casjens, S.R.; Baker, T.S. Three-dimensional reconstructions of the bacteriophage CUS-3 virion reveal a conserved coat protein I-domain but a distinct tailspike receptor-binding domain. Virology 2014, 464–465, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Yang, F.; Zhang, J.T.; Sun, H.; Chen, Y.; Yu, R.C.; Chen, Z.P.; Jiang, Y.L.; Han, S.J.; Xu, X.; et al. Capsid Structure of Anabaena Cyanophage A-1(L). J. Virol. 2021, 95, e0135621. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Jiang, Y.L.; Yang, F.; Zhang, J.T.; Li, W.F.; Zhou, K.; Ju, J.; Chen, Y.; Zhou, C.Z. Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1. Structure 2019, 27, 1508–1516.e1503. [Google Scholar] [CrossRef]
- Lander, G.C.; Evilevitch, A.; Jeembaeva, M.; Potter, C.S.; Carragher, B.; Johnson, J.E. Bacteriophage lambda stabilization by auxiliary protein gpD: Timing, location, and mechanism of attachment determined by cryo-EM. Structure 2008, 16, 1399–1406. [Google Scholar] [CrossRef]
- Stone, N.P.; Demo, G.; Agnello, E.; Kelch, B.A. Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure. Nat. Commun. 2019, 10, 4471. [Google Scholar] [CrossRef]
- Wang, Z.; Hardies, S.C.; Fokine, A.; Klose, T.; Jiang, W.; Cho, B.C.; Rossmann, M.G. Structure of the Marine Siphovirus TW1: Evolution of Capsid-Stabilizing Proteins and Tail Spikes. Structure 2018, 26, 238–248.e233. [Google Scholar] [CrossRef]
- Doore, S.M.; Schrad, J.R.; Dean, W.F.; Dover, J.A.; Parent, K.N. Shigella Phages Isolated during a Dysentery Outbreak Reveal Uncommon Structures and Broad Species Diversity. J. Virol. 2018, 92, e02117-17. [Google Scholar] [CrossRef]
- Gipson, P.; Baker, M.L.; Raytcheva, D.; Haase-Pettingell, C.; Piret, J.; King, J.A.; Chiu, W. Protruding knob-like proteins violate local symmetries in an icosahedral marine virus. Nat. Commun. 2014, 5, 4278. [Google Scholar] [CrossRef][Green Version]
- Johnson, M.C.; Sena-Velez, M.; Washburn, B.K.; Platt, G.N.; Lu, S.; Brewer, T.E.; Lynn, J.S.; Stroupe, M.E.; Jones, K.M. Structure, proteome and genome of Sinorhizobium meliloti phage PhiM5: A virus with LUZ24-like morphology and a highly mosaic genome. J. Struct. Biol. 2017, 200, 343–359. [Google Scholar] [CrossRef]
- Novacek, J.; Siborova, M.; Benesik, M.; Pantucek, R.; Doskar, J.; Plevka, P. Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc. Natl. Acad. Sci. USA 2016, 113, 9351–9356. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, L.; Zhang, Z.; Fokine, A.; Padilla-Sanchez, V.; Hanein, D.; Jiang, W.; Rossmann, M.G.; Rao, V.B. Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-A resolution and its relevance to the assembly of icosahedral viruses. Proc. Natl. Acad. Sci. USA 2017, 114, E8184–E8193. [Google Scholar] [CrossRef]
- Rao, V.B.; Black, L.W. Structure and assembly of bacteriophage T4 head. Virol. J. 2010, 7, 356. [Google Scholar] [CrossRef]
- Johnson, M.C.; Tatum, K.B.; Lynn, J.S.; Brewer, T.E.; Lu, S.; Washburn, B.K.; Stroupe, M.E.; Jones, K.M.; Hutt-Fletcher, L. Sinorhizobium meliloti Phage ΦM9 Defines a New Group of T4 Superfamily Phages with Unusual Genomic Features but a Common T=16 Capsid. J. Virol. 2015, 89, 10945–10958. [Google Scholar] [CrossRef]
- Hernando-Perez, M.; Lambert, S.; Nakatani-Webster, E.; Catalano, C.E.; de Pablo, P.J. Cementing proteins provide extra mechanical stabilization to viral cages. Nat. Commun. 2014, 5, 4520. [Google Scholar] [CrossRef]
- Gilcrease, E.B.; Winn-Stapley, D.A.; Hewitt, F.C.; Joss, L.; Casjens, S.R. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J. Bacteriol. 2005, 187, 2050–2057. [Google Scholar] [CrossRef]
- Huet, A.; Duda, R.L.; Boulanger, P.; Conway, J.F. Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 21037–21046. [Google Scholar] [CrossRef]
- Wang, R.; Cong, Y.; Mi, Z.; Fan, H.; Shi, T.; Liu, H.; Tong, Y. Characterization and complete genome sequence analysis of phage GP4, a novel lytic Bcep22-like podovirus. Arch. Virol. 2019, 164, 2339–2343. [Google Scholar] [CrossRef]
- Trotereau, A.; Boyer, C.; Bornard, I.; Pecheur, M.J.B.; Schouler, C.; Torres-Barcelo, C. High genomic diversity of novel phages infecting the plant pathogen Ralstonia solanacearum, isolated in Mauritius and Reunion islands. Sci. Rep. 2021, 11, 5382. [Google Scholar] [CrossRef]
- Grisales-Vargas, C.D.; Ramírez-Cuartas, C.A.; Pérez-Jaramillo, J.E. The First Complete Genome Resource of a Ralstonia solanacearum Phage UAM5 from Colombia. Mol. Plant-Microbe Interact. MPMI 2022, 35, 496–499. [Google Scholar] [CrossRef]
- Van Truong Thi, B.; Pham Khanh, N.H.; Namikawa, R.; Miki, K.; Kondo, A.; Dang Thi, P.T.; Kamei, K. Genomic characterization of Ralstonia solanacearum phage varphiRS138 of the family Siphoviridae. Arch. Virol. 2016, 161, 483–486. [Google Scholar] [CrossRef]
- Liao, M. Genomic characterization of the novel Ralstonia phage RPSC1. Arch. Virol. 2018, 163, 1969–1971. [Google Scholar] [CrossRef]
- Effantin, G.; Fujiwara, A.; Kawasaki, T.; Yamada, T.; Schoehn, G. High Resolution Structure of the Mature Capsid of Ralstonia solanacearum Bacteriophage ϕRSA1 by Cryo-Electron Microscopy. Int. J. Mol. Sci. 2021, 22, 11053. [Google Scholar] [CrossRef]
- Neumann, E.; Kawasaki, T.; Effantin, G.; Estrozi, L.F.; Chatchawankanphanich, O.; Yamada, T.; Schoehn, G. 3D structure of three jumbo phage heads. J. Gen. Virol. 2020, 101, 1219–1226. [Google Scholar] [CrossRef]
- Bhunchoth, A.; Blanc-Mathieu, R.; Mihara, T.; Nishimura, Y.; Askora, A.; Phironrit, N.; Leksomboon, C.; Chatchawankanphanich, O.; Kawasaki, T.; Nakano, M.; et al. Two asian jumbo phages, varphiRSL2 and varphiRSF1, infect Ralstonia solanacearum and show common features of varphiKZ-related phages. Virology 2016, 494, 56–66. [Google Scholar] [CrossRef]
- Effantin, G.; Hamasaki, R.; Kawasaki, T.; Bacia, M.; Moriscot, C.; Weissenhorn, W.; Yamada, T.; Schoehn, G. Cryo-electron microscopy three-dimensional structure of the jumbo phage PhiRSL1 infecting the phytopathogen Ralstonia solanacearum. Structure 2013, 21, 298–305. [Google Scholar] [CrossRef]
- Lee, S.Y.; Thapa Magar, R.; Kim, H.J.; Choi, K.; Lee, S.W. Complete Genome Sequence of a Novel Bacteriophage RpY1 Infecting Ralstonia solanacearum Strains. Curr. Microbiol. 2021, 78, 2044–2050. [Google Scholar] [CrossRef]
- Ahmad, A.A.; Elhalag, K.M.; Addy, H.S.; Nasr-Eldin, M.A.; Hussien, A.S.; Huang, Q. Sequencing, genome analysis and host range of a novel Ralstonia phage, RsoP1EGY, isolated in Egypt. Arch. Virol. 2018, 163, 2271–2274. [Google Scholar] [CrossRef]
- Li, X.; Mooney, P.; Zheng, S.; Booth, C.R.; Braunfeld, M.B.; Gubbens, S.; Agard, D.A.; Cheng, Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 2013, 10, 584–590. [Google Scholar] [CrossRef]
- Kimanius, D.; Forsberg, B.O.; Scheres, S.H.; Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 2016, 5, e18722. [Google Scholar] [CrossRef]
- Kivioja, T.; Ravantti, J.; Verkhovsky, A.; Ukkonen, E.; Bamford, D. Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 2000, 131, 126–134. [Google Scholar] [CrossRef]
- Li, X.; Zhou, N.; Chen, W.; Zhu, B.; Wang, X.; Xu, B.; Wang, J.; Liu, H.; Cheng, L. Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV. J. Mol. Biol. 2017, 429, 79–87. [Google Scholar] [CrossRef]
- Li, X.; Liu, H.; Cheng, L. Symmetry-mismatch reconstruction of genomes and associated proteins within icosahedral viruses using cryo-EM. Biophys. Rep. 2016, 2, 25–32. [Google Scholar] [CrossRef]
- Fuller, S.D.; Butcher, S.J.; Cheng, R.H.; Baker, T.S. Three-dimensional reconstruction of icosahedral particles--the uncommon line. J. Struct. Biol. 1996, 116, 48–55. [Google Scholar] [CrossRef]
- Thuman-Commike, P.A.; Chiu, W. Improved common line-based icosahedral particle image orientation estimation algorithms. Ultramicroscopy 1997, 68, 231–255. [Google Scholar] [CrossRef]
- Chen, S.; McMullan, G.; Faruqi, A.R.; Murshudov, G.N.; Short, J.M.; Scheres, S.H.; Henderson, R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 2013, 135, 24–35. [Google Scholar] [CrossRef]
- Buchan, D.W.; Minneci, F.; Nugent, T.C.; Bryson, K.; Jones, D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, W349–W357. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Rosenthal, P.B.; Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003, 333, 721–745. [Google Scholar] [CrossRef]
- Wang, C.; Zeng, J.; Wang, J. Structural basis of bacteriophage lambda capsid maturation. Structure 2022, 30, 637–645.e633. [Google Scholar] [CrossRef]
- Hardy, J.M.; Dunstan, R.A.; Grinter, R.; Belousoff, M.J.; Wang, J.; Pickard, D.; Venugopal, H.; Dougan, G.; Lithgow, T.; Coulibaly, F. The architecture and stabilisation of flagellotropic tailed bacteriophages. Nat. Commun. 2020, 11, 3748. [Google Scholar] [CrossRef]
- Kamiya, R.; Uchiyama, J.; Matsuzaki, S.; Murata, K.; Iwasaki, K.; Miyazaki, N. Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy. Structure 2022, 30, 300–312.e303. [Google Scholar] [CrossRef]
- Newcomer, R.L.; Schrad, J.R.; Gilcrease, E.B.; Casjens, S.R.; Feig, M.; Teschke, C.M.; Alexandrescu, A.T.; Parent, K.N. The phage L capsid decoration protein has a novel OB-fold and an unusual capsid binding strategy. Elife 2019, 8, e45345. [Google Scholar] [CrossRef]
- Fang, Q.; Tang, W.C.; Fokine, A.; Mahalingam, M.; Shao, Q.; Rossmann, M.G.; Rao, V.B. Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly. Proc. Natl. Acad. Sci. USA 2022, 119, e2203272119. [Google Scholar] [CrossRef]
- Wilson, D.P. Protruding Features of Viral Capsids Are Clustered on Icosahedral Great Circles. PLoS ONE 2016, 11, e0152319. [Google Scholar] [CrossRef][Green Version]
- Helgstrand, C.; Wikoff, W.R.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Liljas, L. The refined structure of a protein catenane: The HK97 bacteriophage capsid at 3.44 A resolution. J. Mol. Biol. 2003, 334, 885–899. [Google Scholar] [CrossRef]
- Bayfield, O.W.; Klimuk, E.; Winkler, D.C.; Hesketh, E.L.; Chechik, M.; Cheng, N.; Dykeman, E.C.; Minakhin, L.; Ranson, N.A.; Severinov, K.; et al. Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc. Natl. Acad. Sci. USA 2019, 116, 3556–3561. [Google Scholar] [CrossRef]
- Chaban, Y.; Lurz, R.; Brasiles, S.; Cornilleau, C.; Karreman, M.; Zinn-Justin, S.; Tavares, P.; Orlova, E.V. Structural rearrangements in the phage head-to-tail interface during assembly and infection. Proc. Natl. Acad. Sci. USA 2015, 112, 7009–7014. [Google Scholar] [CrossRef]
- Arisaka, F. Assembly and infection process of bacteriophage T4. Chaos 2005, 15, 047502. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Chen, W.; Xiao, H.; Yang, F.; Li, X.; Song, J.; Cheng, L.; Liu, H. A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9. Viruses 2022, 14, 2431. https://doi.org/10.3390/v14112431
Zheng J, Chen W, Xiao H, Yang F, Li X, Song J, Cheng L, Liu H. A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9. Viruses. 2022; 14(11):2431. https://doi.org/10.3390/v14112431
Chicago/Turabian StyleZheng, Jing, Wenyuan Chen, Hao Xiao, Fan Yang, Xiaowu Li, Jingdong Song, Lingpeng Cheng, and Hongrong Liu. 2022. "A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9" Viruses 14, no. 11: 2431. https://doi.org/10.3390/v14112431
APA StyleZheng, J., Chen, W., Xiao, H., Yang, F., Li, X., Song, J., Cheng, L., & Liu, H. (2022). A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9. Viruses, 14(11), 2431. https://doi.org/10.3390/v14112431




