Estimating the Effectiveness of Shielding during Pregnancy against SARS-CoV-2 in New York City during the First Year of the COVID-19 Pandemic
Abstract
1. Introduction
2. Materials and Methods
2.1. Pregnant Patients Data
2.2. General Population Data
2.3. Exposure Inference in Pregnant Patients
2.4. Exposure Inference in General Population
3. Results
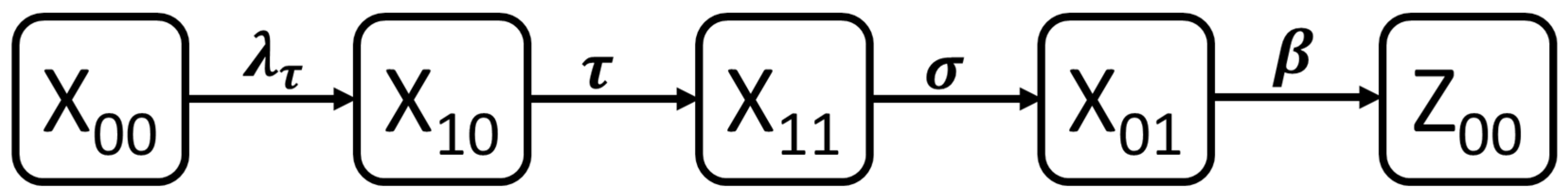
3.1. Dynamic Model of SARS-CoV-2 Infection
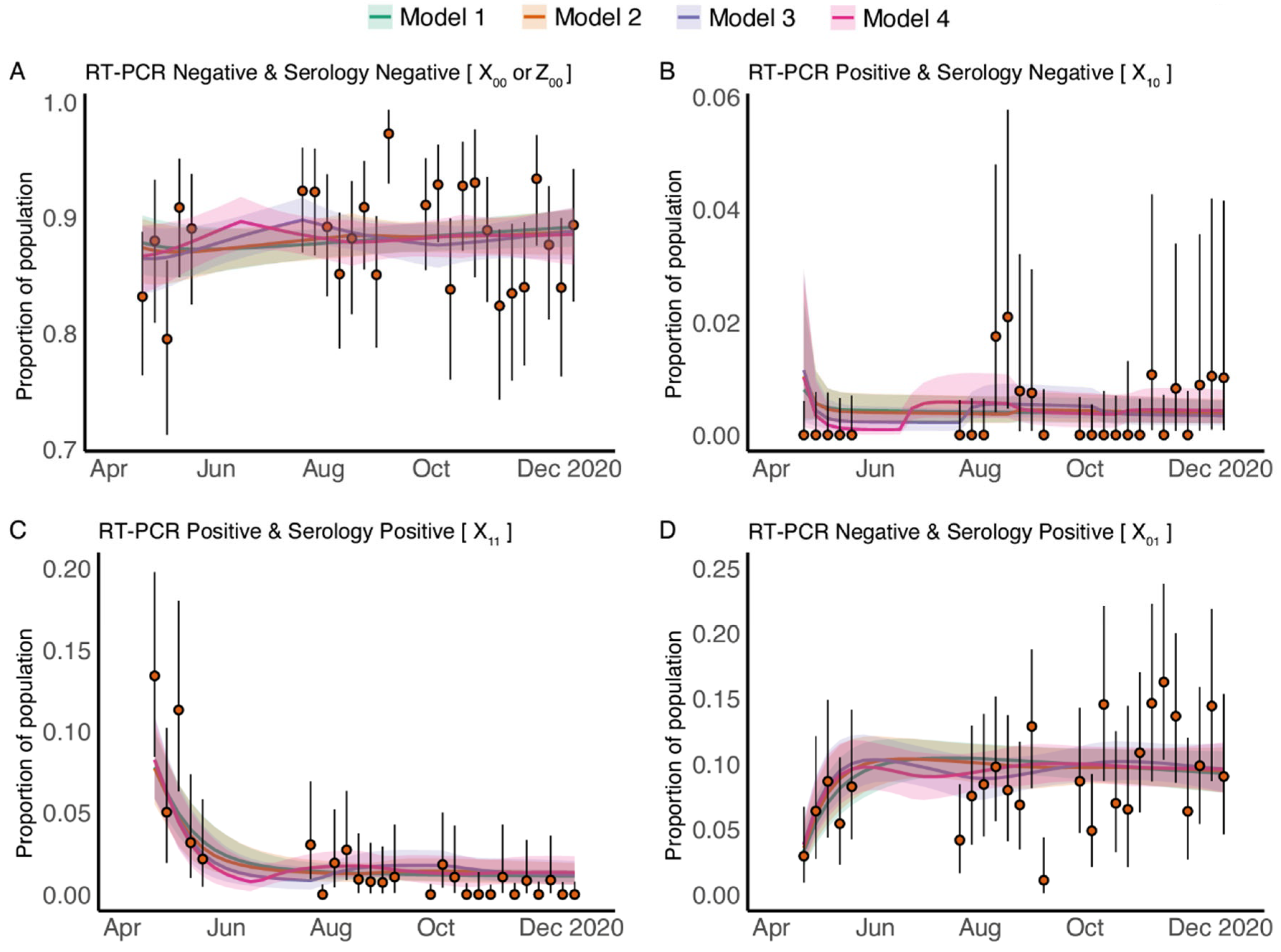
3.2. Longitudinal Cross-Sectional RT-PCR and Serology Results
3.3. Fitting Data from Pregnant Patients to the Dynamic Model
3.4. Transmission Parameters of COVID-19 in Pregnant Patients Are Estimated to Be Consistent with Those Estimated for General Population
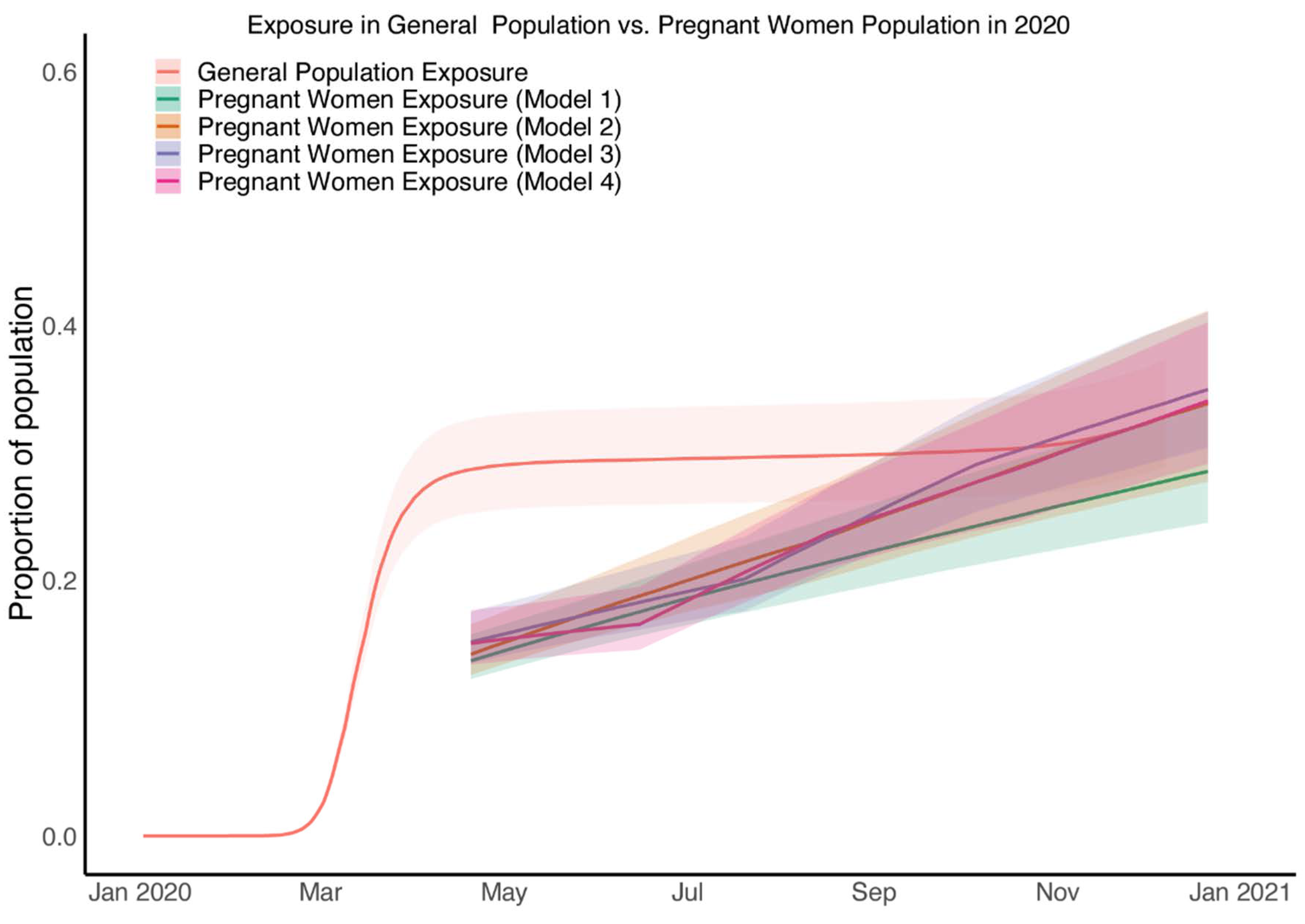
3.5. Estimated SARS-CoV-2 Exposure in Pregnant Patients Is Higher than Seropositivity Rates Would Suggest
3.6. SARS-CoV-2 Exposure in Pregnant Patients at the Time of Birth Rose from Half That of the General Population to Equal That of the General Population by Late 2020
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prabhu, M.; Cagino, K.; Matthews, K.C.; Friedlander, R.L.; Glynn, S.M.; Kubiak, J.M.; Yang, Y.J.; Zhao, Z.; Baergen, R.N.; DiPace, J.I.; et al. Pregnancy and postpartum outcomes in a universally tested population for SARS-CoV-2 in New York City: A prospective cohort study. BJOG Int. J. Obstet. Gynaecol. 2020, 127, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, J.M.; Murphy, E.A.; Yee, J.; Cagino, K.A.; Friedlander, R.L.; Glynn, S.M.; Matthews, K.C.; Jurkiewicz, M.; Sukhu, A.C.; Zhao, Z.; et al. Severe acute respiratory syndrome coronavirus 2 serology levels in pregnant women and their neonates. Am. J. Obstet. Gynecol. 2021, 225, 73.e1–73.e7. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.E.; Park, S.W.; Yang, W.; Vecchi, G.A.; Metcalf, C.J.E.; Grenfell, B.T. The impact of COVID-19 nonpharmaceutical interventions on the future dynamics of endemic infections. Proc. Natl. Acad. Sci. USA 2020, 117, 30547–30553. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Shaikh, M.; Hauck, K.; Miraldo, M. Impacts of introducing and lifting nonpharmaceutical interventions on COVID-19 daily growth rate and compliance in the United States. Proc. Natl. Acad. Sci. USA 2021, 118, e2021359118. [Google Scholar] [CrossRef] [PubMed]
- Brauner, J.M.; Mindermann, S.; Sharma, M.; Johnston, D.; Salvatier, J.; Gavenčiak, T.; Stephenson, A.B.; Leech, G.; Altman, G.; Mikulik, V.; et al. Inferring the effectiveness of government interventions against COVID-19. Science 2021, 371, eabd9338. [Google Scholar] [CrossRef] [PubMed]
- Flaxman, S.; Mishra, S.; Gandy, A.; Unwin, H.J.T.; Mellan, T.A.; Coupland, H.; Whittaker, C.; Zhu, H.; Berah, T.; Eaton, J.W.; et al. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature 2020, 584, 257–261. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. COVID-19 in Pregnant and Recently Pregnant People Guideline; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/pregnant-people.html#:~:text=of%20Severe%20Illness-,If%20you%20are%20pregnant%20or%20were%20recently%20pregnant%2C%20you%20are,one%20that%20causes%20COVID%2D19 (accessed on 20 April 2021).
- NHS. Pregnancy and coronavirus (COVID-19). Available online: https://www.nhs.uk/conditions/coronavirus-covid-19/people-at-higher-risk/pregnancy-and-coronavirus/ (accessed on 27 September 2022).
- Ortiz, E.I.; Castañeda, E.H.; De La Torre, A. Coronavirus (COVID 19) Infection in Pregnancy. Colomb. Med. 2020, 51, e4271. [Google Scholar] [CrossRef]
- Qiao, J. What are the risks of COVID-19 infection in pregnant women? Lancet 2020, 395, 760–762. [Google Scholar] [CrossRef]
- Chen, S.; Flegg, J.A.; White, L.J.; Aguas, R. Levels of SARS-CoV-2 population exposure are considerably higher than suggested by seroprevalence surveys. PLoS Comput. Biol. 2021, 17, e1009436. [Google Scholar] [CrossRef]
- Shioda, K.; Lau, M.S.; Kraay, A.N.; Nelson, K.N.; Siegler, A.J.; Sullivan, P.S.; Collins, M.H.; Weitz, J.S.; Lopman, B.A. Estimating the cumulative incidence of SARS-CoV-2 infection and the infection fatality ratio in light of waning antibodies. Epidemiology 2021, 32, 518. [Google Scholar] [CrossRef]
- Vousden, N.; Ramakrishnan, R.; Bunch, K.; Quigley, M.; Kurinczuk, J.; Knight, M. Severity of maternal infection and perinatal outcomes during periods in which Wildtype, Alpha and Delta SARS-CoV-2 variants were dominant: Data from the UK Obstetric Surveillance System national cohort. BMJ Med. 2022, 1, e000053. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. COVID Data Tracker; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022. Available online: https://covid.cdc.gov/covid-data-tracker (accessed on 20 April 2021).
- Havers, F.P.; Reed, C.; Lim, T.; Montgomery, J.M.; Klena, J.D.; Hall, A.J.; Fry, A.M.; Cannon, D.L.; Chiang, C.-F.; Gibbons, A.; et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23-May 12, 2020. JAMA Intern. Med. 2020, 180, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Stan Development Team, R. R Stan: The R Interface to Stan. 2018. Available online: http://mc-stan.org/ (accessed on 1 January 2020).
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Mallett, S.; Allen, A.J.; Graziadio, S.; Taylor, S.A.; Sakai, N.S.; Green, K.; Suklan, J.; Hyde, C.; Shinkins, B.; Zhelev, Z.; et al. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med. 2020, 18, 346. [Google Scholar] [CrossRef]
- Li, K.; Huang, B.; Wu, M.; Zhong, A.; Li, L.; Cai, Y.; Wang, Z.; Wu, L.; Zhu, M.; Li, J.; et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat. Commun. 2020, 11, 6044. [Google Scholar] [CrossRef]
- Wei, J.; Matthews, P.C.; Stoesser, N.; Maddox, T.; Lorenzi, L.; Studley, R.; Bell, J.I.; Newton, J.N.; Farrar, J.; Diamond, I.; et al. Anti-spike antibody response to natural SARS-CoV-2 infection in the general population. Nat. Commun. 2021, 12, 6250. [Google Scholar] [CrossRef] [PubMed]
- Sethuraman, N.; Jeremiah, S.S.; Ryo, A. Interpreting diagnostic tests for SARS-CoV-2. JAMA 2020, 323, 2249–2251. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Cruz, C.S.D.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef]
- Xiang, F.; Wang, X.; He, X.; Peng, Z.; Yang, B.; Zhang, J.; Zhou, Q.; Ye, H.; Ma, Y.; Li, H.; et al. Antibody Detection and Dynamic Characteristics in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 1930–1934. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.Y.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Seow, J.; Graham, C.; Merrick, B.; Acors, S.; Pickering, S.; Steel, K.J.; Hemmings, O.; O’Byrne, A.; Kouphou, N.; Pedro Galao, R.; et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020, 5, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Ibarrondo, F.J.; Fulcher, J.A.; Goodman-Meza, D.; Elliott, J.; Hofmann, C.; Hausner, M.A.; Ferbas, K.G.; Tobin, N.H.; Aldrovandi, G.M.; Yang, O.O. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020, 383, 1085–1087. [Google Scholar] [CrossRef]
- Long, Q.-X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Pu Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Jering, K.S.; Claggett, B.L.; Cunningham, J.W.; Rosenthal, N.; Vardeny, O.; Greene, M.F.; Solomon, S.D. Clinical Characteristics and Outcomes of Hospitalized Women Giving Birth With and Without COVID-19. JAMA Intern. Med. 2021, 181, 714–717. [Google Scholar] [CrossRef]
- Mullins, E.; Hudak, M.L.; Banerjee, J.; Getzlaff, T.; Townson, J.; Barnette, K.; Playle, R.; Perry, A.; Bourne, T.; Lees, C.C. Pregnancy and neonatal outcomes of COVID-19: Coreporting of common outcomes from PAN-COVID and AAP-SONPM registries. Ultrasound Obstet. Gynecol. 2021, 57, 573–581. [Google Scholar] [CrossRef]
- Smithgall, M.C.; Murphy, E.A.; Schatz-Siemers, N.; Matrai, C.; Tu, J.; Baergen, R.N.; Yang, Y.J. Placental pathology in women vaccinated and unvaccinated against SARS-CoV-2. Am. J. Obstet. Gynecol. 2022. [Google Scholar] [CrossRef]
- Yang, Y.J.; Murphy, E.A.; Singh, S.; Sukhu, A.C.; Wolfe, I.; Adurty, S.; Eng, D.; Yee, J.; Mohammed, I.; Zhao, Z.; et al. Association of Gestational Age at Coronavirus Disease 2019 (COVID-19) Vaccination, History of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection, and a Vaccine Booster Dose With Maternal and Umbilical Cord Antibody Levels at Delivery. Obstet. Gynecol. 2021, 139, 373–380. [Google Scholar] [CrossRef]
- Prabhu, M.; Murphy, E.A.; Sukhu, A.C.; Yee, J.; Singh, S.; Eng, D.; Zhao, Z.; Riley, L.E.; Yang, Y.J. Antibody Response to Coronavirus Disease 2019 (COVID-19) Messenger RNA Vaccination in Pregnant Women and Transplacental Passage Into Cord Blood. Obstet. Gynecol. 2021, 138, 278–280. [Google Scholar] [CrossRef]
- Shimabukuro, T.T.; Kim, S.Y.; Myers, T.R.; Moro, P.L.; Oduyebo, T.; Panagiotakopoulos, L.; Marquez, P.L.; Olson, C.K.; Liu, R.; Chang, K.T.; et al. Preliminary findings of mRNA Covid-19 vaccine safety in pregnant persons. N. Engl. J. Med. 2021, 384, 2273–2282. [Google Scholar] [CrossRef] [PubMed]

| Total | Women Giving Birth Prior to August 2020 | Women Giving Birth from August 2020 Onwards | Test | p-Value | |
|---|---|---|---|---|---|
| n = 2682 | n = 1781 | n = 901 | |||
| Ethnicity | Chi Square: 5.82 | 0.324 | |||
| Not Hispanic or Latino or Spanish Origin | 1769 (66%) | 1173 (65.9%) | 596 (66.1%) | ||
| Hispanic or Latino or Spanish Origin | 219 (8.2%) | 142 (8%) | 77 (8.5%) | ||
| African American | 1 (0%) | 1 (0.1%) | 0 (0%) | ||
| Multi-racial | 1 (0%) | 0 (0%) | 1 (0.1%) | ||
| Declined | 600 (22.4%) | 396 (22.2%) | 204 (22.6%) | ||
| Unknown | 92 (3.4%) | 69 (3.9%) | 23 (2.6%) | ||
| Race | Chi Square: 11.49 | 0.244 | |||
| White | 1346 (50.2%) | 876 (49.2%) | 470 (52.2%) | ||
| Asian | 336 (12.5%) | 224 (12.6%) | 112 (12.4%) | ||
| Black or African American | 169 (6.3%) | 118 (6.6%) | 51 (5.7%) | ||
| American Indian or Alaska Nation | 6 (0.2%) | 2 (0.1%) | 4 (0.4%) | ||
| Nat. Hawaiian/Oth. Pacific Island | 3 (0.1%) | 2 (0.1%) | 1 (0.1%) | ||
| Ashkenazi Jewish | 2 (0.1%) | 1 (0.1%) | 1 (0.1%) | ||
| Multiple races reported | 15 (0.6%) | 7 (0.4%) | 8 (0.9%) | ||
| Other combinations not described | 258 (9.6%) | 170 (9.5%) | 88 (9.8%) | ||
| Declined | 464 (17.3%) | 319 (17.9%) | 145 (16.1%) | ||
| Unknown | 83 (3.1%) | 62 (3.5%) | 21 (2.3%) | ||
| Mom Age (SD) years | t-test: −0.47 | 0.636 | |||
| 34.4 (5.0) | 34.4 (5.0) | 34.5 (5.0) | |||
| Gestational Age at delivery (SD) weeks | t-test: 1.53 | 0.126 | |||
| 38.8 (2.1) | 38.8 (2.0) | 38.7 (2.4) |
| Variables | Definition |
|---|---|
| proportion of naïve population who are both RT-PCR and serology negative and never exposed | |
| proportion of early phase infected population who are RT-PCR positive but serology negative | |
| proportion of middle-phase infected population who are both RT-PCR and serology positive | |
| proportion of late-phase infected population who are RT-PCR negative but serology positive | |
| proportion of past infected population who are both RT-PCR and serology negative but previously exposed |
| Parameter (Unit) | Definition | Model | Median | 5% | 95% | |
|---|---|---|---|---|---|---|
| (days) | average time lag between virus detectability and antibody detectability | 1 | 7 | 4 | 18 | |
| 2 | 5 | 3 | 16 | |||
| 3 | 5 | 3 | 10 | |||
| 4 | 6 | 4 | 13 | |||
| (days) | average time lag between antibody detectability and virus undetectability | 1 | 22 | 14 | 37 | |
| 2 | 18 | 11 | 32 | |||
| 3 | 17 | 11 | 27 | |||
| 4 | 18 | 12 | 28 | |||
| (days) | average time lag between seroconversion and seroreversion among pregnant patients | 1 | 152 | 84 | 336 | |
| 2 | 118 | 64 | 270 | |||
| 3 | 110 | 65 | 208 | |||
| 4 | 117 | 66 | 240 | |||
| (−) | proportion of patients who were giving birth and not exposed by 20 April 2020 | 1 | 0.87 | 0.79 | 0.90 | |
| 2 | 0.86 | 0.76 | 0.90 | |||
| 3 | 0.86 | 0.74 | 0.89 | |||
| 4 | 0.85 | 0.74 | 0.89 | |||
| (−) | force of infection | 1 | 0.0052 | 0.0022 | 0.010 | |
| (−) | 2 | 0.0063 | 0.0028 | 0.013 | ||
| (−) | 0.0079 | 0.0025 | 0.0182 | |||
| (−) | 3 | 0.0041 | 0.0052 | 0.019 | ||
| (−) | 0.011 | 0.0052 | 0.019 | |||
| (−) | 0.0077 | 0.0030 | 0.019 | |||
| (−) | 4 | 0.00013 | 0.000088 | 0.00072 | ||
| (−) | 0.0095 | 0.0051 | 0.016 | |||
| (−) | 0.0070 | 0.0013 | 0.0178 | |||
| (−) | 0.0083 | 0.0033 | 0.019 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Murphy, E.A.; Pendergrass, A.G.; Sukhu, A.C.; Eng, D.; Jurkiewicz, M.; Mohammed, I.; Rand, S.; White, L.J.; Hupert, N.; et al. Estimating the Effectiveness of Shielding during Pregnancy against SARS-CoV-2 in New York City during the First Year of the COVID-19 Pandemic. Viruses 2022, 14, 2408. https://doi.org/10.3390/v14112408
Chen S, Murphy EA, Pendergrass AG, Sukhu AC, Eng D, Jurkiewicz M, Mohammed I, Rand S, White LJ, Hupert N, et al. Estimating the Effectiveness of Shielding during Pregnancy against SARS-CoV-2 in New York City during the First Year of the COVID-19 Pandemic. Viruses. 2022; 14(11):2408. https://doi.org/10.3390/v14112408
Chicago/Turabian StyleChen, Siyu, Elisabeth A. Murphy, Angeline G. Pendergrass, Ashley C. Sukhu, Dorothy Eng, Magdalena Jurkiewicz, Iman Mohammed, Sophie Rand, Lisa J. White, Nathaniel Hupert, and et al. 2022. "Estimating the Effectiveness of Shielding during Pregnancy against SARS-CoV-2 in New York City during the First Year of the COVID-19 Pandemic" Viruses 14, no. 11: 2408. https://doi.org/10.3390/v14112408
APA StyleChen, S., Murphy, E. A., Pendergrass, A. G., Sukhu, A. C., Eng, D., Jurkiewicz, M., Mohammed, I., Rand, S., White, L. J., Hupert, N., & Yang, Y. J. (2022). Estimating the Effectiveness of Shielding during Pregnancy against SARS-CoV-2 in New York City during the First Year of the COVID-19 Pandemic. Viruses, 14(11), 2408. https://doi.org/10.3390/v14112408





