West Nile Virus Lineage 1 in Italy: Newly Introduced or a Re-Occurrence of a Previously Circulating Strain?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bird Conditions and Laboratory Analyses
2.2. Virus Strain and Laboratory Tests (IZSAM)
2.3. Phylogenetic Analyses
3. Results
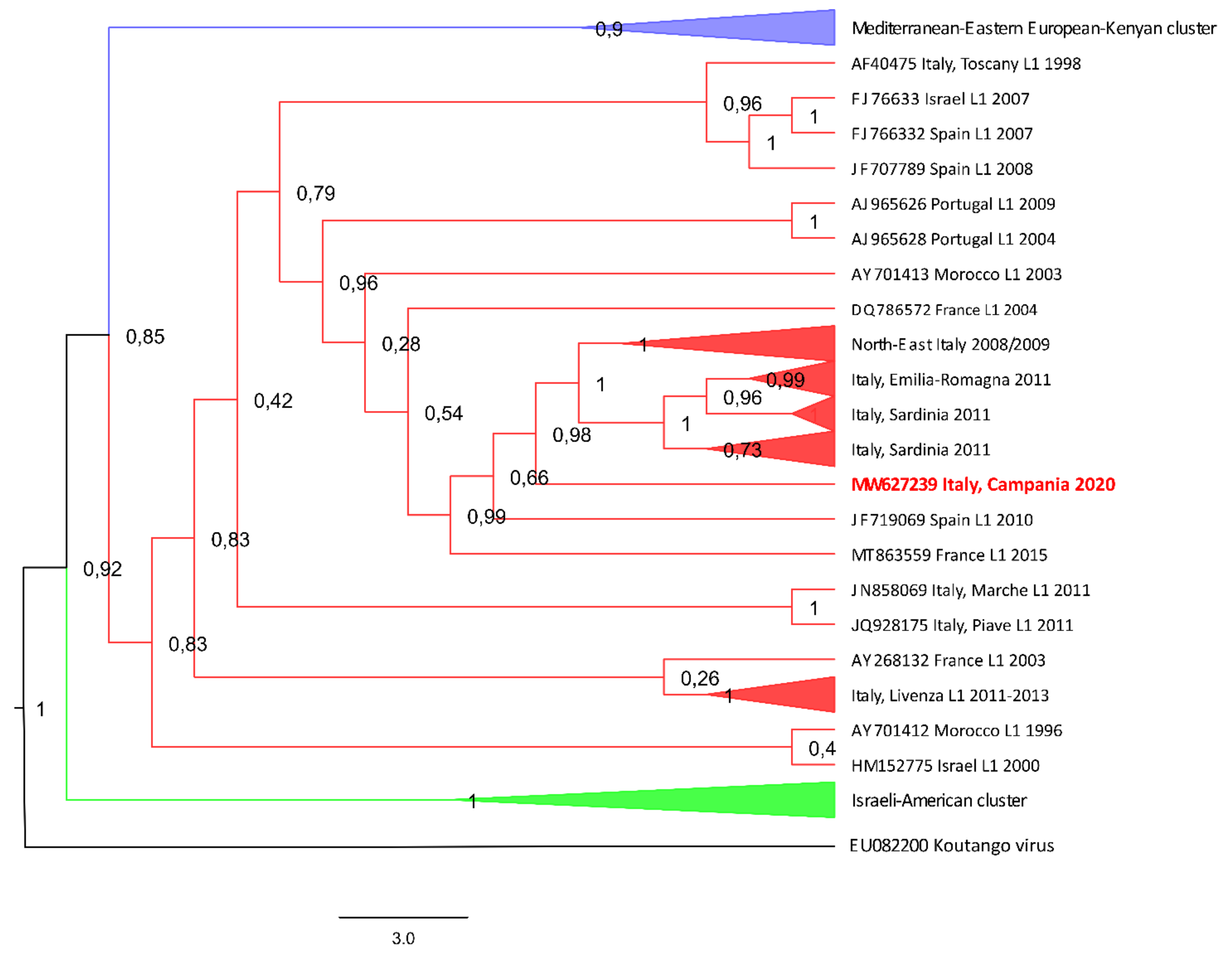
3.1. Strain Characterization and Phylogenetic Analysis
3.1.1. WNV-L1 Detection
3.1.2. Phylogenetic Tree Inferred with Maximum-Likelihood Analysis
3.1.3. Full-Length Polyprotein Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Llopis, I.V.; Rossi, L.; Di Gennaro, A.; Mosca, A.; Teodori, L.; Tomassone, L.; Grego, E.; Monaco, F.; Lorusso, A.; Savini, G. Further Circulation of West Nile and Usutu Viruses in Wild Birds in Italy. Infect. Genet. Evol. 2015, 32, 292–297. [Google Scholar] [CrossRef]
- Marini, G.; Rosà, R.; Pugliese, A.; Rizzoli, A.; Rizzo, C.; Russo, F.; Montarsi, F.; Capelli, G. West Nile Virus Transmission and Human Infection Risk in Veneto (Italy): A Modelling Analysis. Sci. Rep. 2018, 8, 14005. [Google Scholar] [CrossRef]
- Venter, M.; Swanepoel, R. West Nile Virus Lineage 2 as a Cause of Zoonotic Neurological Disease in Humans and Horses in Southern Africa. Vector-Borne Zoonotic Dis. 2010, 10, 659–664. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Ramírez, E.; Llorente, F.; Del Amo, J.; Fall, G.; Lubisi, A.; Lecollinet, S.; Vázquez, A.; Jiménez-Clavero, M.Á. Pathogenicity Evaluation of Twelve West Nile Virus Strains Belonging to Four Lineages from Five Continents in a Mouse Model: Discrimination between Three Pathogenicity Categories. J. Gen. Virol. 2017, 98, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.; Jimenez-Clavero, M.A.; Leblond, A.; Durand, B.; Nowotny, N.; Leparc-Goffart, I.; Zientara, S.; Jourdain, E.; Lecollinet, S. Flaviviruses in Europe: Complex Circulation Patterns and Their Consequences for the Diagnosis and Control of West Nile Disease. Int. J. Environ. Res. Public Health 2013, 10, 6049–6083. [Google Scholar] [CrossRef] [Green Version]
- Epidemiological History and Phylogeography of West Nile Virus Lineage 2—Abstract—Europe PMC. Available online: https://europepmc.org/article/med/23542457 (accessed on 22 June 2020).
- Fall, G.; Di Paola, N.; Faye, M.; Dia, M.; Freire, C.C.d.M.; Loucoubar, C.; Zanotto, P.M.d.A.; Faye, O.; Sall, A.A. Biological and Phylogenetic Characteristics of West African Lineages of West Nile Virus. PLoS Negl. Trop. Dis. 2017, 11, e0006078. [Google Scholar] [CrossRef]
- Beck, C.; Leparc Goffart, I.; Franke, F.; Gonzalez, G.; Dumarest, M.; Lowenski, S.; Blanchard, Y.; Lucas, P.; de Lamballerie, X.; Grard, G.; et al. Contrasted Epidemiological Patterns of West Nile Virus Lineages 1 and 2 Infections in France from 2015 to 2019. Pathogens 2020, 9, 908. [Google Scholar] [CrossRef] [PubMed]
- Filipe, A.R.; Pinto, M.R. Survey for Antibodies to Arboviruses in Serum of Animals from Southern Portugal. Am. J. Trop. Med. Hyg. 1969, 18, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Joubert, L.; Oudar, J.; Hannoun, C.; Beytout, D.; Corniou, B.; Guillon, J.C.; Panthier, R. Epidemiology of the West Nile virus: Study of a focus in Camargue. IV. Meningo-encephalomyelitis of the horse. Ann. Inst. Pasteur 1970, 118, 239–247. [Google Scholar]
- Malkinson, M.; Banet, C.; Weisman, Y. Intercontinental Spread of West Nile Virus by Wild Birds—Recent Epidemiological Findings in Israeli Livestock and Birds. In Proceedings of the 2nd International Conference on Emerging Zoonoses, Strasbourg, France, 5–9 November 1998; Volume 2. [Google Scholar]
- Cantile, C.; Di Guardo, G.; Eleni, C.; Arispici, M. Clinical and Neuropathological Features of West Nile Virus Equine Encephalomyelitis in Italy. Equine Vet. J. 2000, 32, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Autorino, G.L.; Battisti, A.; Deubel, V.; Ferrari, G.; Forletta, R.; Giovannini, A.; Lelli, R.; Murri, S.; Scicluna, M.T. West Nile Virus Epidemic in Horses, Tuscany Region, Italy. Emerg. Infect. Dis. 2002, 8, 1372–1378. [Google Scholar] [CrossRef] [PubMed]
- Murgue, B.; Murri, S.; Triki, H.; Deubel, V.; Zeller, H.G. West Nile in the Mediterranean Basin: 1950–2000. Ann. N. Y. Acad. Sci. 2001, 951, 117–126. [Google Scholar] [CrossRef]
- Steinman, A.; Banet, C.; Sutton, G.A.; Yadin, H.; Hadar, S.; Brill, A. Clinical Signs of West Nile Virus Encephalomyelitis in Horses during the Outbreak in Israel in 2000. Vet. Rec. 2002, 151, 47–49. [Google Scholar] [CrossRef]
- Schuffenecker, I.; Peyrefitte, C.N.; el Harrak, M.; Murri, S.; Leblond, A.; Zeller, H.G. West Nile Virus in Morocco, 2003. Emerg. Infect. Dis. 2005, 11, 306–309. [Google Scholar] [CrossRef]
- Cernescu, C.; Nedelcu, N.-I.; Tardei, G.; Ruta, S.; Tsai, T.F. Continued Transmission of West Nile Virus to Humans in Southeastern Romania, 1997–1998. J. Infect. Dis. 2000, 181, 710–712. [Google Scholar] [CrossRef]
- Platonov, A.E.; Shipulin, G.A.; Shipulina, O.Y.; Tyutyunnik, E.N.; Frolochkina, T.I.; Lanciotti, R.S.; Yazyshina, S.; Platonova, O.V.; Obukhov, I.L.; Zhukov, A.N.; et al. Outbreak of West Nile Virus Infection, Volgograd Region, Russia, 1999. Emerg. Infect. Dis. 2001, 7, 128–132. [Google Scholar] [CrossRef]
- Ceianu, C.S.; Ungureanu, A.; Nicolescu, G.; Cernescu, C.; Nitescu, L.; Tardei, G.; Petrescu, A.; Pitigoi, D.; Martin, D.; Ciulacu-Purcarea, V. West Nile Virus Surveillance in Romania: 1997–2000. Viral Immunol. 2001, 14, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N.; Brault, A.C.; Gallian, P.; Lemasson, J.-J.; Murgue, B.; Murri, S.; Pastorino, B.; Zeller, H.; de Chesse, R.; de Micco, P.; et al. Evolutionary Relationship between Old World West Nile Virus Strains. Evidence for Viral Gene Flow between Africa, the Middle East, and Europe. Virology 2003, 315, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Campbell, G.L.; Ceianu, C.S.; Savage, H.M. Epidemic West Nile Encephalitis in Romania: Waiting for History to Repeat Itself. Ann. N. Y. Acad. Sci. 2001, 951, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Del Giudice, P.; Schuffenecker, I.; Vandenbos, F.; Counillon, E.; Zellet, H. Human West Nile Virus, France. Emerg. Infect. Dis. 2004, 10, 1885–1886. [Google Scholar] [CrossRef]
- Johnson, N.; Fernández de Marco, M.; Giovannini, A.; Ippoliti, C.; Danzetta, M.L.; Svartz, G.; Erster, O.; Groschup, M.H.; Ziegler, U.; Mirazimi, A.; et al. Emerging Mosquito-Borne Threats and the Response from European and Eastern Mediterranean Countries. Int. J. Environ. Res. Public Health 2018, 15, 2775. [Google Scholar] [CrossRef] [Green Version]
- Bakonyi, T.; Ivanics, É.; Erdélyi, K.; Ursu, K.; Ferenczi, E.; Weissenböck, H.; Nowotny, N. Lineage 1 and 2 Strains of Encephalitic West Nile Virus, Central Europe. Emerg. Infect. Dis. 2006, 12, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Bakonyi, T.; Ferenczi, E.; Erdélyi, K.; Kutasi, O.; Csörgő, T.; Seidel, B.; Weissenböck, H.; Brugger, K.; Bán, E.; Nowotny, N. Explosive Spread of a Neuroinvasive Lineage 2 West Nile Virus in Central Europe, 2008/2009. Vet. Microbiol. 2013, 165, 61–70. [Google Scholar] [CrossRef]
- Papa, A.; Danis, K.; Baka, A.; Bakas, A.; Dougas, G.; Lytras, T.; Theocharopoulos, G.; Chrysagis, D.; Vassiliadou, E.; Kamaria, F.; et al. Ongoing Outbreak of West Nile Virus Infections in Humans in Greece, July–August 2010. Eurosurveillance 2010, 15, 19644. [Google Scholar] [CrossRef] [PubMed]
- Napp, S.; Petrić, D.; Busquets, N. West Nile Virus and Other Mosquito-Borne Viruses Present in Eastern Europe. Pathog. Glob. Health 2018, 112, 233–248. [Google Scholar] [CrossRef]
- Merdić, E.; Perić, L.; Pandak, N.; Kurolt, I.C.; Turić, N.; Vignjević, G.; Stolfa, I.; Milas, J.; Bogojević, M.S.; Markotić, A. West Nile Virus Outbreak in Humans in Croatia, 2012. Coll. Antropol. 2013, 37, 943–947. [Google Scholar]
- Savini, G.; Capelli, G.; Monaco, F.; Polci, A.; Russo, F.; Di Gennaro, A.; Marini, V.; Teodori, L.; Montarsi, F.; Pinoni, C.; et al. Evidence of West Nile Virus Lineage 2 Circulation in Northern Italy. Vet. Microbiol. 2012, 158, 267–273. [Google Scholar] [CrossRef]
- De Heus, P.; Kolodziejek, J.; Camp, J.V.; Dimmel, K.; Bagó, Z.; Hubálek, Z.; van den Hoven, R.; Cavalleri, J.-M.V.; Nowotny, N. Emergence of West Nile Virus Lineage 2 in Europe: Characteristics of the First Seven Cases of West Nile Neuroinvasive Disease in Horses in Austria. Transbound. Emerg. Dis. 2020, 67, 1189–1197. [Google Scholar] [CrossRef]
- Magurano, F.; Remoli, M.E.; Baggieri, M.; Fortuna, C.; Marchi, A.; Fiorentini, C.; Bucci, P.; Benedetti, E.; Ciufolini, M.G.; Rizzo, C.; et al. Circulation of West Nile Virus Lineage 1 and 2 during an Outbreak in Italy. Clin. Microbiol. Infect. 2012, 18, E545–E547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzo, C.; Napoli, C.; Venturi, G.; Pupella, S.; Lombardini, L.; Calistri, P.; Monaco, F.; Cagarelli, R.; Angelini, P.; Bellini, R.; et al. West Nile Virus Transmission: Results from the Integrated Surveillance System in Italy, 2008 to 2015. Eurosurveillance 2016, 21, 30340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zehender, G.; Veo, C.; Ebranati, E.; Carta, V.; Rovida, F.; Percivalle, E.; Moreno, A.; Lelli, D.; Calzolari, M.; Lavazza, A.; et al. Reconstructing the Recent West Nile Virus Lineage 2 Epidemic in Europe and Italy Using Discrete and Continuous Phylogeography. PLoS ONE 2017, 12, e0179679. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, A.; Sánchez-Seco, M.P.; Ruiz, S.; Molero, F.; Hernández, L.; Moreno, J.; Magallanes, A.; Tejedor, C.G.; Tenorio, A. Putative New Lineage of West Nile Virus, Spain. Emerg. Infect. Dis. 2010, 16, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Martín-Acebes, M.A.; Saiz, J.-C. West Nile Virus: A Re-Emerging Pathogen Revisited. World J. Virol. 2012, 1, 51–70. [Google Scholar] [CrossRef] [PubMed]
- Benjelloun, A.; El Harrak, M.; Calistri, P.; Loutfi, C.; Kabbaj, H.; Conte, A.; Ippoliti, C.; Danzetta, M.L.; Belkadi, B. Seroprevalence of West Nile Virus in Horses in Different Moroccan Regions. Vet. Med. Sci. 2017, 3, 198–207. [Google Scholar] [CrossRef] [Green Version]
- Amdouni, J.; Monaco, F.; Portanti, O.; Sghaier, S.; Conte, A.; Hassine, T.B.; Polci, A.; Valleriani, F.; Gennaro, A.D.; Zoueri, M.; et al. Detection of Enzootic Circulation of a New Strain of West Nile Virus Lineage 1 in Sentinel Chickens in the North of Tunisia. Acta Trop. 2020, 202, 105223. [Google Scholar] [CrossRef]
- Assaid, N.; Mousson, L.; Moutailler, S.; Arich, S.; Akarid, K.; Monier, M.; Beck, C.; Lecollinet, S.; Failloux, A.-B.; Sarih, M. Evidence of Circulation of West Nile Virus in Culex Pipiens Mosquitoes and Horses in Morocco. Acta Trop. 2020, 205, 105414. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Barriga, D.; Aguilera-Sepúlveda, P.; Guerrero-Carvajal, F.; Llorente, F.; Reina, D.; Pérez-Martín, J.E.; Jiménez-Clavero, M.Á.; Frontera, E. West Nile and Usutu Virus Infections in Wild Birds Admitted to Rehabilitation Centres in Extremadura, Western Spain, 2017–2019. Vet. Microbiol. 2021, 255, 109020. [Google Scholar] [CrossRef]
- Conte, A.; Candeloro, L.; Ippoliti, C.; Monaco, F.; Massis, F.D.; Bruno, R.; Sabatino, D.D.; Danzetta, M.L.; Benjelloun, A.; Belkadi, B.; et al. Spatio-Temporal Identification of Areas Suitable for West Nile Disease in the Mediterranean Basin and Central Europe. PLoS ONE 2015, 10, e0146024. [Google Scholar] [CrossRef]
- Cotar, A.I.; Fălcuță, E.; Dinu, S.; Necula, A.; Bîrluțiu, V.; Ceianu, C.S.; Prioteasa, F.L. West Nile Virus Lineage 2 in Romania, 2015–2016: Co-Circulation and Strain Replacement. Parasit. Vectors 2018, 11, 562. [Google Scholar] [CrossRef] [Green Version]
- Ozkul, A.; Ergunay, K.; Koysuren, A.; Alkan, F.; Arsava, E.M.; Tezcan, S.; Emekdas, G.; Hacioglu, S.; Turan, M.; Us, D. Concurrent Occurrence of Human and Equine West Nile Virus Infections in Central Anatolia, Turkey: The First Evidence for Circulation of Lineage 1 Viruses. Int. J. Infect. Dis. 2013, 17, e546–e551. [Google Scholar] [CrossRef] [Green Version]
- Ergünay, K.; Litzba, N.; Brinkmann, A.; Günay, F.; Sarıkaya, Y.; Kar, S.; Örsten, S.; Öter, K.; Domingo, C.; Erisoz Kasap, Ö.; et al. Co-Circulation of West Nile Virus and Distinct Insect-Specific Flaviviruses in Turkey. Parasit. Vectors 2017, 10, 149. [Google Scholar] [CrossRef] [Green Version]
- Richter, J.; Tryfonos, C.; Tourvas, A.; Floridou, D.; Paphitou, N.I.; Christodoulou, C. Complete Genome Sequence of West Nile Virus (WNV) from the First Human Case of Neuroinvasive WNV Infection in Cyprus. Genome Announc. 2017, 5, e01110-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Ruiz, N.; del Carmen Montaño-Remacha, M.; Durán-Pla, E.; Pérez-Ruiz, M.; Navarro-Marí, J.M.; Salamanca-Rivera, C.; Miranda, B.; Oyonarte-Gómez, S.; Ruiz-Fernández, J. West Nile Virus Outbreak in Humans and Epidemiological Surveillance, West Andalusia, Spain, 2016. Eurosurveillance 2018, 23, 17-00261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yıldırım, Y.; Yılmaz, V.; Yazıcı, K.; Öziç, C.; Ozkul, A.; Çağırgan, A.A. Phylogenetic Analysis of West Nile Virus: First Report of Lineage 1 in Donkey in Turkey. Trop. Anim. Health Prod. 2021, 53, 453. [Google Scholar] [CrossRef] [PubMed]
- Akıner, M.M.; Öztürk, M.; Başer, A.B.; Günay, F.; Hacıoğlu, S.; Brinkmann, A.; Emanet, N.; Alten, B.; Özkul, A.; Nitsche, A.; et al. Arboviral Screening of Invasive Aedes Species in Northeastern Turkey: West Nile Virus Circulation and Detection of Insect-Only Viruses. PLoS Negl. Trop. Dis. 2019, 13, e0007334. [Google Scholar] [CrossRef]
- Lustig, Y.; Gosinov, R.; Zuckerman, N.; Glazer, Y.; Orshan, L.; Sofer, D.; Schwartz, E.; Schvartz, G.; Farnoushi, Y.; Lublin, A.; et al. Epidemiologic and Phylogenetic Analysis of the 2018 West Nile Virus (WNV) Outbreak in Israel Demonstrates Human Infection of WNV Lineage I. Eurosurveillance 2019, 24, 1800662. [Google Scholar] [CrossRef]
- Schvartz, G.; Farnoushi, Y.; Berkowitz, A.; Edery, N.; Hahn, S.; Steinman, A.; Lublin, A.; Erster, O. Molecular Characterization of the Re-Emerging West Nile Virus in Avian Species and Equids in Israel, 2018, and Pathological Description of the Disease. Parasit. Vectors 2020, 13, 528. [Google Scholar] [CrossRef]
- Petrović, T.; Šekler, M.; Petrić, D.; Vidanović, D.; Debeljak, Z.; Lazić, G.; Lupulović, D.; Kavran, M.; Samojlović, M.; Ignjatović Ćupina, A.; et al. Intensive West Nile Virus Circulation in Serbia in 2018—Results of Integrated Surveillance Program. Pathogens 2021, 10, 1294. [Google Scholar] [CrossRef] [PubMed]
- Savini, G.; Monaco, F.; Calistri, P.; Lelli, R. Phylogenetic Analysis of West Nile Virus Isolated in Italy in 2008. Eurosurveillance 2008, 13, 19048. [Google Scholar] [CrossRef] [PubMed]
- Monaco, F.; Goffredo, M.; Briguglio, P.; Pinoni, C.; Polci, A.; Iannetti, S.; Marruchella, G.; Di Francesco, G.; Di Gennaro, A.P.; Pais, M.; et al. The 2011 West Nile Disease Outbreak in Sardinia Region, Italy. Vet. Ital. 2015, 51, 5–16. [Google Scholar] [CrossRef]
- Monaco, F.; Savini, G.; Calistri, P.; Polci, A.; Pinoni, C.; Bruno, R.; Lelli, R. 2009 West Nile Disease Epidemic in Italy: First Evidence of Overwintering in Western Europe? Res. Vet. Sci. 2011, 91, 321–326. [Google Scholar] [CrossRef]
- Paternoster, G.; Tomassone, L.; Tamba, M.; Chiari, M.; Lavazza, A.; Piazzi, M.; Favretto, A.R.; Balduzzi, G.; Pautasso, A.; Vogler, B.R. The Degree of One Health Implementation in the West Nile Virus Integrated Surveillance in Northern Italy, 2016. Front. Public Health 2017, 5, 236. [Google Scholar] [CrossRef] [Green Version]
- Calistri, P.; Giovannini, A.; Savini, G.; Monaco, F.; Bonfanti, L.; Ceolin, C.; Terregino, C.; Tamba, M.; Cordioli, P.; Lelli, R. West Nile Virus Transmission in 2008 in North-Eastern Italy. Zoonoses Public Health 2010, 57, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Barzon, L.; Pacenti, M.; Franchin, E.; Squarzon, L.; Lavezzo, E.; Toppo, S.; Martello, T.; Cattai, M.; Cusinato, R.; Palù, G. Novel West Nile Virus Lineage 1a Full Genome Sequences from Human Cases of Infection in North-Eastern Italy, 2011. Clin. Microbiol. Infect. 2012, 18, E541–E544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barzon, L.; Papa, A.; Lavezzo, E.; Franchin, E.; Pacenti, M.; Sinigaglia, A.; Masi, G.; Trevisan, M.; Squarzon, L.; Toppo, S.; et al. Phylogenetic Characterization of Central/Southern European Lineage 2 West Nile Virus: Analysis of Human Outbreaks in Italy and Greece, 2013–2014. Clin. Microbiol. Infect. 2015, 21, 1122.e1–1122.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagnarelli, P.; Marinelli, K.; Trotta, D.; Monachetti, A.; Tavio, M.; Del Gobbo, R.; Capobianchi, M.; Menzo, S.; Nicoletti, L.; Magurano, F.; et al. Human Case of Autochthonous West Nile Virus Lineage 2 Infection in Italy, September 2011. Eurosurveillance 2011, 16, 20002. [Google Scholar] [CrossRef]
- Barzon, L.; Pacenti, M.; Franchin, E.; Squarzon, L.; Lavezzo, E.; Cattai, M.; Cusinato, R.; Palù, G. The Complex Epidemiological Scenario of West Nile Virus in Italy. Int. J. Environ. Res. Public Health 2013, 10, 4669–4689. [Google Scholar] [CrossRef] [Green Version]
- Del Amo, J.; Sotelo, E.; Fernández-Pinero, J.; Gallardo, C.; Llorente, F.; Agüero, M.; Jiménez-Clavero, M.A. A Novel Quantitative Multiplex Real-Time RT-PCR for the Simultaneous Detection and Differentiation of West Nile Virus Lineages 1 and 2, and of Usutu Virus. J. Virol. Methods 2013, 189, 321–327. [Google Scholar] [CrossRef]
- Vázquez, A.; Herrero, L.; Negredo, A.; Hernández, L.; Sánchez-Seco, M.P.; Tenorio, A. Real Time PCR Assay for Detection of All Known Lineages of West Nile Virus. J. Virol. Methods 2016, 236, 266–270. [Google Scholar] [CrossRef]
- Marcacci, M.; De Luca, E.; Zaccaria, G.; Di Tommaso, M.; Mangone, I.; Aste, G.; Savini, G.; Boari, A.; Lorusso, A. Genome Characterization of Feline Morbillivirus from Italy. J. Virol. Methods 2016, 234, 160–163. [Google Scholar] [CrossRef]
- Aprea, G.; Scattolini, S.; D’Angelantonio, D.; Chiaverini, A.; Di Lollo, V.; Olivieri, S.; Marcacci, M.; Mangone, I.; Salucci, S.; Antoci, S.; et al. Whole Genome Sequencing Characterization of HEV3-e and HEV3-f Subtypes among the Wild Boar Population in the Abruzzo Region, Italy: First Report. Microorganisms 2020, 8, 1393. [Google Scholar] [CrossRef] [PubMed]
- Djikeng, A.; Halpin, R.; Kuzmickas, R.; Depasse, J.; Feldblyum, J.; Sengamalay, N.; Afonso, C.; Zhang, X.; Anderson, N.G.; Ghedin, E.; et al. Viral Genome Sequencing by Random Priming Methods. BMC Genom. 2008, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Cito, F.; Pasquale, A.D.; Cammà, C.; Cito, P. The Italian Information System for the Collection and Analysis of Complete Genome Sequence of Pathogens Isolated from Animal, Food and Environment. Int. J. Infect. Dis. 2018, 73, 296–297. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Sotelo, E.; Fernández-Pinero, J.; Llorente, F.; Vázquez, A.; Moreno, A.; Agüero, M.; Cordioli, P.; Tenorio, A.; Jiménez-Clavero, M.Á. Phylogenetic Relationships of Western Mediterranean West Nile Virus Strains (1996–2010) Using Full-Length Genome Sequences: Single or Multiple Introductions? J. Gen. Virol. 2011, 92, 2512–2522. [Google Scholar] [CrossRef]
- Monaco, F.; Lelli, R.; Teodori, L.; Pinoni, C.; Di Gennaro, A.; Polci, A.; Calistri, P.; Savini, G. Re-Emergence of West Nile Virus in Italy. Zoonoses Public Health 2010, 57, 476–486. [Google Scholar] [CrossRef]
- Sotelo, E.; Fernandez-Pinero, J.; Llorente, F.; Agüero, M.; Hoefle, U.; Blanco, J.M.; Jiménez-Clavero, M.A. Characterization of West Nile Virus Isolates from Spain: New Insights into the Distinct West Nile Virus Eco-Epidemiology in the Western Mediterranean. Virology 2009, 395, 289–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesko, K.N.; Ebel, G.D. West Nile Virus Population Genetics and Evolution. Infect. Genet. Evol. 2012, 12, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Kaneko, C.; Kawakami, R.; Hasebe, R.; Sawa, H.; Yoshii, K.; Kariwa, H. Amino Acid 159 of the Envelope Protein Affects Viral Replication and T-Cell Infiltration by West Nile Virus in Intracranial Infection. Sci. Rep. 2020, 10, 7168. [Google Scholar] [CrossRef] [PubMed]
- Marfin, A.A.; Petersen, L.R.; Eidson, M.; Miller, J.; Hadler, J.; Farello, C.; Werner, B.; Campbell, G.L.; Layton, M.; Smith, P.; et al. Widespread West Nile Virus Activity, Eastern United States, 2000. Emerg. Infect. Dis. 2001, 7, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Banet-Noach, C.; Simanov, L.; Malkinson, M. Direct (Non-Vector) Transmission of West Nile Virus in Geese. Avian Pathol. J. WVPA 2003, 32, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Gamino, V.; Höfle, U. Pathology and Tissue Tropism of Natural West Nile Virus Infection in Birds: A Review. Vet. Res. 2013, 44, 39. [Google Scholar] [CrossRef] [Green Version]
- Komar, N.; Langevin, S.; Hinten, S.; Nemeth, N.; Edwards, E.; Hettler, D.; Davis, B.; Bowen, R.; Bunning, M. Experimental Infection of North American Birds with the New York 1999 Strain of West Nile Virus. Emerg. Infect. Dis. 2003, 9, 311–322. [Google Scholar] [CrossRef]
- Pérez-Ramírez, E.; Llorente, F.; Jiménez-Clavero, M.Á. Experimental Infections of Wild Birds with West Nile Virus. Viruses 2014, 6, 752–781. [Google Scholar] [CrossRef] [Green Version]
- Nemeth, N.M.; Kratz, G.E.; Bates, R.; Scherpelz, J.A.; Bowen, R.A.; Komar, N. Clinical Evaluation and Outcomes of Naturally Acquired West Nile Virus Infection in Raptors. J. Zoo Wildl. Med. 2009, 40, 51–63. [Google Scholar] [CrossRef]
- Ziegler, U.; Angenvoort, J.; Fischer, D.; Fast, C.; Eiden, M.; Rodriguez, A.V.; Revilla-Fernández, S.; Nowotny, N.; de la Fuente, J.G.; Lierz, M.; et al. Pathogenesis of West Nile Virus Lineage 1 and 2 in Experimentally Infected Large Falcons. Vet. Microbiol. 2013, 161, 263–273. [Google Scholar] [CrossRef]
- Vidaña, B.; Busquets, N.; Napp, S.; Pérez-Ramírez, E.; Jiménez-Clavero, M.Á.; Johnson, N. The Role of Birds of Prey in West Nile Virus Epidemiology. Vaccines 2020, 8, 550. [Google Scholar] [CrossRef]
- Höfle, U.; Blanco, J.M.; Crespo, E.; Naranjo, V.; Jiménez-Clavero, M.A.; Sanchez, A.; de la Fuente, J.; Gortazar, C. West Nile Virus in the Endangered Spanish Imperial Eagle. Vet. Microbiol. 2008, 129, 171–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Clavero, M.A.; Sotelo, E.; Fernandez-Pinero, J.; Llorente, F.; Blanco, J.M.; Rodriguez-Ramos, J.; Perez-Ramirez, E.; Höfle, U. West Nile Virus in Golden Eagles, Spain, 2007. Emerg. Infect. Dis. 2008, 14, 1489–1491. [Google Scholar] [CrossRef] [Green Version]
- Garmendia, A.E.; Van Kruiningen, H.J.; French, R.A.; Anderson, J.F.; Andreadis, T.G.; Kumar, A.; West, A.B. Recovery and Identification of West Nile Virus from a Hawk in Winter. J. Clin. Microbiol. 2000, 38, 3110–3111. [Google Scholar] [CrossRef] [Green Version]
- Wünschmann, A.; Shivers, J.; Bender, J.; Carroll, L.; Fuller, S.; Saggese, M.; van Wettere, A.; Redig, P. Pathologic Findings in Red-Tailed Hawks (Buteo jamaicensis) and Cooper’s Hawks (Accipiter cooper) Naturally Infected with West Nile Virus. Avian Dis. 2004, 48, 570–580. [Google Scholar] [CrossRef]
- Wünschmann, A.; Shivers, J.; Bender, J.; Carroll, L.; Fuller, S.; Saggese, M.; van Wettere, A.; Redig, P. Pathologic and Immunohistochemical Findings in Goshawks (Accipiter gentilis) and Great Horned Owls (Bubo virginianus) Naturally Infected with West Nile Virus. Avian Dis. 2005, 49, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Donadieu, E.; Bahuon, C.; Lowenski, S.; Zientara, S.; Coulpier, M.; Lecollinet, S. Differential Virulence and Pathogenesis of West Nile Viruses. Viruses 2013, 5, 2856–2880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiz, J.-C.; Martín-Acebes, M.A.; Blázquez, A.B.; Escribano-Romero, E.; Poderoso, T.; Jiménez de Oya, N. Pathogenicity and Virulence of West Nile Virus Revisited Eight Decades after Its First Isolation. Virulence 2021, 12, 1145–1173. [Google Scholar] [CrossRef]
- Gray, R.R.; Veras, N.M.C.; Santos, L.A.; Salemi, M. Evolutionary Characterization of the West Nile Virus Complete Genome. Mol. Phylogenet. Evol. 2010, 56, 195–200. [Google Scholar] [CrossRef]
- Fredericksen, B.L.; Smith, M.; Katze, M.G.; Shi, P.-Y.; Gale, M. The Host Response to West Nile Virus Infection Limits Viral Spread through the Activation of the Interferon Regulatory Factor 3 Pathway. J. Virol. 2004, 78, 7737–7747. [Google Scholar] [CrossRef] [Green Version]
- Kinney, R.M.; Huang, C.Y.-H.; Whiteman, M.C.; Bowen, R.A.; Langevin, S.A.; Miller, B.R.; Brault, A.C. Avian Virulence and Thermostable Replication of the North American Strain of West Nile Virus. J. Gen. Virol. 2006, 87, 3611–3622. [Google Scholar] [CrossRef] [PubMed]
- Brault, A.C.; Huang, C.Y.-H.; Langevin, S.A.; Kinney, R.M.; Bowen, R.A.; Ramey, W.N.; Panella, N.A.; Holmes, E.C.; Powers, A.M.; Miller, B.R. A Single Positively Selected West Nile Viral Mutation Confers Increased Virogenesis in American Crows. Nat. Genet. 2007, 39, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Montagnaro, S.; Piantedosi, D.; Ciarcia, R.; Loponte, R.; Veneziano, V.; Fusco, G.; Amoroso, M.G.; Ferrara, G.; Damiano, S.; Iovane, G.; et al. Serological Evidence of Mosquito-Borne Flaviviruses Circulation in Hunting Dogs in Campania Region, Italy. Vector-Borne Zoonotic Dis. 2019, 19, 142–147. [Google Scholar] [CrossRef] [PubMed]
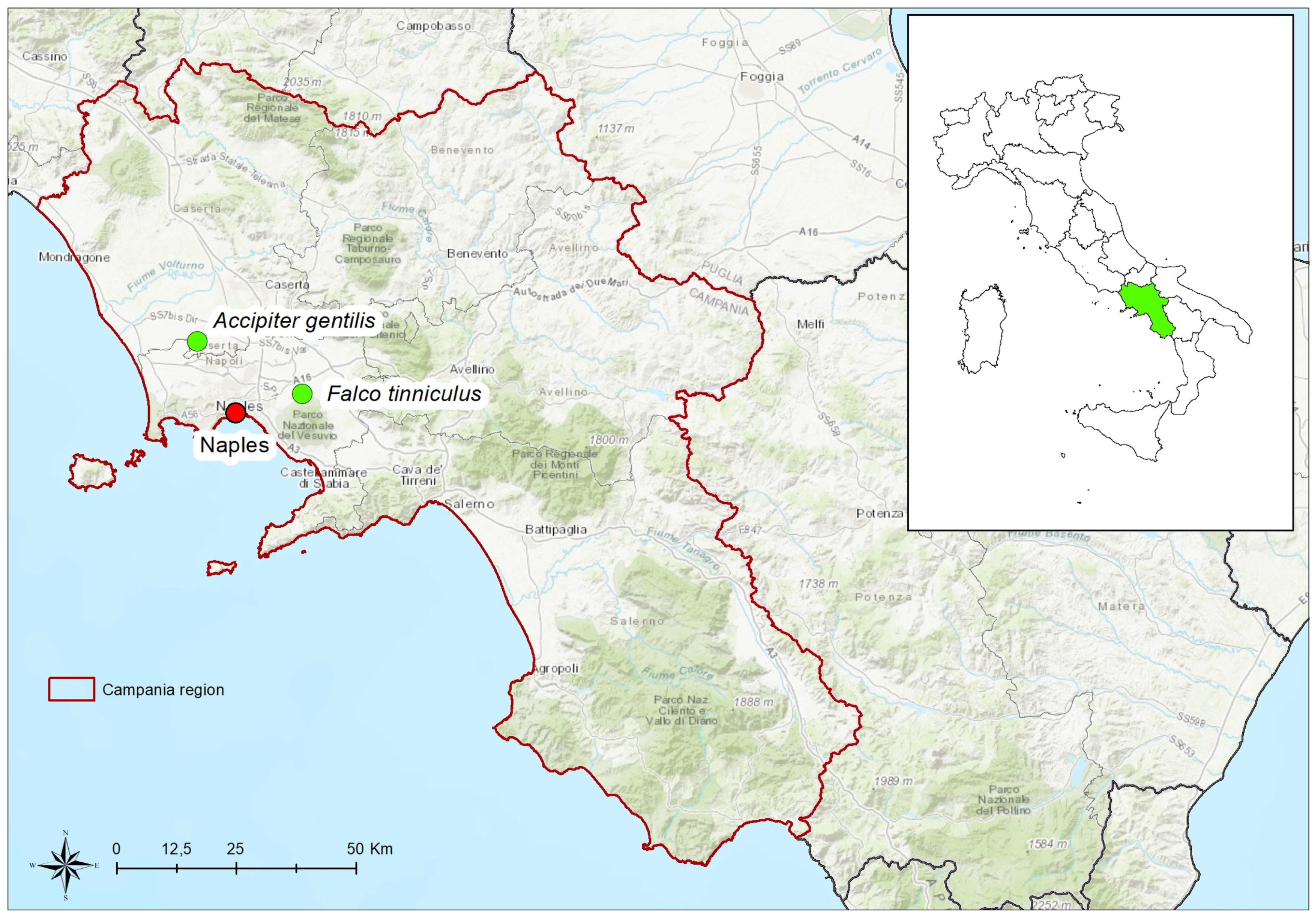

| Strain Number | Viral Species | Isolation Material | Host | Country | Year of Isolation | Accession Number |
|---|---|---|---|---|---|---|
| TE.362447.2020 | WNV L1 | Homogenate | Northern goshawk | Italy | 2020 | MW627239 |
| TE.15803.2008 | WNV L1 | - | Magpie | Italy | 2008 | FJ483548 |
| TE.15217.2008 | WNV L1 | - | Magpie | Italy | 2008 | FJ483549 |
| TE.229892.2008 | WNV L1 | - | Magpie | Italy | 2008 | KU573077 |
| Ita09 | WNV L1 | Blood | Human | Italy | 2009 | GU011992 |
| Italy/2009/J-225677 | WNV L1 | C636 cells P1, Vero cells P3 | Eurasian jay | Italy | 2009 | JF719068 |
| Italy/2009/FIN | WNV L1 | - | Human | Italy | 2009 | KF234080 |
| Italy/2008/J-242853 | WNV L1 | C636 cells P1, Vero cells P3 | Eurasian jay | Italy | 2008 | JF719065 |
| Italy/2009/G-223184 | WNV L1 | C636 cells P1, Vero cells P3 | Gull | Italy | 2009 | JF719067 |
| Italy/2008/M-203204 | WNV L1 | C636 cells P1, Vero cells P3 | Magpie | Italy | 2008 | JF719066 |
| 204913/2009 | WNV L1 | - | Culex pipiens mosquito | Italy | 2009 | KU573078 |
| TE.14444.2011 | WNV L1 | Organ pool | Magpie | Italy | 2011 | MW835356 |
| TE.17196.2011 | WNV L1 | Organ pool | Owl | Italy | 2011 | MW835357 |
| TE.17208.2011 | WNV L1 | Organ pool | Crow | Italy | 2011 | MW835358 |
| TE.20224/1.2011 | WNV L1 | Plasma | Chicken | Italy | 2011 | MW835359 |
| TE.21370.2011 | WNV L1 | Plasma | Horse | Italy | 2011 | MW835361 |
| TE.20875.2011 | WNV L1 | Organ pool | Eurasian jay | Italy | 2011 | MW835360 |
| TE.23237.2011 | WNV L1 | Plasma | Chicken | Italy | 2011 | MW835363 |
| TE.20224/8.2011 | WNV L1 | Plasma | Chicken | Italy | 2011 | Under publication |
| TE.21412.2011 | WNV L1 | Brain | Owl | Italy | 2011 | MW835362 |
| 04.05 | WNV L1 | Brain | Horse | Morocco | 2003 | AY701413 |
| PT5.2 | WNV L1 | - | - | Portugal | 2004 | AJ965628 |
| PT6.16 | WNV L1 | - | - | Portugal | 2009 | AJ965626 |
| Spain/2010/H-1b | WNV L1 | Brain | Horse | Spain | 2010 | JF719069 |
| WN Italy 1998-equine | WNV L1 | - | Equine | Italy | 1998 | AF404757 |
| 96-111 | WNV L1 | Brain | Equine | Morocco | 1996 | AY701412 |
| PaAn001 | WNV L1 | - | - | France | 2003 | AY268132 |
| France 405/04 | WNV L1 | Brain | House sparrow | France | 2004 | DQ786572 |
| WNV_0304h_ISR00 | WNV L1 | - | Human | Israel | 2000 | HM152775 |
| GE-2o/V | WNV L1 | Vero cells | Golden eagle | Spain | 2007 | FJ766332 |
| GE-1b/B | WNV L1 | BSR cells | Golden eagle | Israel | 2007 | FJ766331 |
| HU6365/08 | WNV L1 | - | Culex perexiguus mosquito | Spain | 2008 | JF707789 |
| RO97-50 | WNV L1 | - | Culex pipiens mosquito | Romania | 1996 | AF260969 |
| KN3829 | WNV L1 | - | Culex univittatus mosquito | USA | 2003 | AY262283 |
| VLG-4 | WNV L1 | Brain | Human | Russia | 1999 | AF317203 |
| Tomsk/bird/2006/A4 | WNV L1 | - | Blyth’s reed warbler | Russia | 2006 | MN149538 |
| LEIV-Vlg99-27889 | WNV L1 | Brain | Human | Russia | 1999 | AY277252 |
| LEIV-Vlg00-27924 | WNV L1 | Blood | Human | Russia | 2000 | AY278442 |
| Italy/2012/Livenza/37.1 | WNV L1 | Urine | Human | Italy | 2012 | KC954092 |
| Italy/2012/Livenza/31.1 | WNV L1 | Culture viral isolate from blood | Human | Italy | 2012 | JX556213 |
| Italy/2013/Livenza/35.1 | WNV L1 | Plasma | Human | Italy | 2013 | KF647253 |
| Italy/2011/Livenza | WNV L1 | Plasma | Human | Italy | 2011 | JQ928174 |
| Italy/2013/Livenza/37.1 | WNV L1 | Urine | Human | Italy | 2013 | KF823807 |
| Akela/France/2015 | WNV L1 | - | Human | France | 2015 | MT863559 |
| Italy/2011/AN-1 | WNV L1 | Urine | Human | Italy | 2011 | JN858069 |
| Italy/2011/Piave | WNV L1 | Urine | Human | Italy | 2011 | JQ928175 |
| WNV_Cy2016 | WNV L1 | Urine | Human | Cyprus | 2016 | MF797870 |
| T2 | WNV L1 | Blood | Equine | Turkey | 2011 | KJ958922 |
| ArB310/67 | WNV L1 | - | - | Central African Republic | 1967 | GQ851608 |
| IBAN7019 | WNV L1 | - | - | Nigeria | 1965 | GQ851607 |
| ArD27875 | WNV L1 | - | Mosquito | Senegal | 1979 | GQ851606 |
| PaH001 | WNV L1 | - | - | France | 2003 | AY268133 |
| NY99-flamingo382-99 | WNV L1 | Chicken embryo | Flamingo | USA | 1999 | AF196835 |
| ABB-B13 | WNV L1 | - | Mouse | Spain | 2007 | KC407667 |
| NY99 | WNV L1 | Vero cell P2 | - | USA | 1999 | NC009942 |
| NY99iso-1 | WNV L1 | Vero cell E6 | - | Japan | 2007 | FJ411043 |
| NY99-crow-V76/1 | WNV L1 | - | American crow | USA | 1999 | FJ151394 |
| 3356K VP2 | WNV L1 | Kidney | American crow | USA | 2000 | EF657887 |
| WNV-1/US/BID-V6527/2001 | WNV L1 | Kidney and Spleen | American crow | USA | 2001 | KJ501343 |
| WNV-1/US/BID-V6506/2002 | WNV L1 | Kidney and Spleen | American crow | USA | 2002 | KJ501489 |
| Ast99-901 | WNV L1 | Blood | Human | Russia | 1999 | AY278441 |
| KN3829 | WNV L1 | - | Culex univittatus mosquito | Kenya | 1998 | AF146082 |
| RO97-50 | WNV L1 | - | - | Romania | 1997 | AF130362 |
| SEN-ArD93548 | WNV L1 | - | Mosquito | Senegal | 1993 | AF001570 |
| Dak Ar D 5443 | KOUTANGO VIRUS | - | - | Senegal | 2013 | EU082200 |
| Viral Protein | Amino Acid Position | MW627239 ITA 2020 | AF404757 ITA 1998 | JF719066 ITA 2008 | FJ766332 SPA 2007 | JF719065 ITA 2008 | FJ483548 ITA 2008 | MW835362 ITA 2011 | KF647253 ITA 2013 | NC_009942 USA 1999 | JF719069 SPA 2010 | MT863559 FRA 2015 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | 34 | M | M | M | V | M | M | M | M | M | M | M |
| 100 | S | S | S | S | S | S | S | S | S | S | S | |
| prM | 72 | S | S | S | S | S | S | S | S | S | S | S |
| M | 36 | I | I | I | I | I | I | I | I | I | I | I |
| E | 35 | S | S | S | S | S | S | S | S | S | S | S |
| 51 | T | A | A | T | A | A | A | A | A | A | A | |
| 76 | T | T | T | T | T | T | T | T | T | T | T | |
| 88 | P | P | P | S | P | P | P | P | P | P | P | |
| 126 | T | T | T | T | T | T | T | T | I | T | T | |
| 153 | G | G | G | G | G | G | G | G | G | G | G | |
| 159 | I | I | I | I | I | I | I | I | V | I | I | |
| 278 | T | T | T | T | T | T | T | T | T | T | T | |
| 312 | L | L | L | L | L | L | L | L | L | L | L | |
| 442 | V | V | V | V | V | V | V | V | V | V | V | |
| NS1 | 17 | S | S | S | S | S | S | S | S | S | S | S |
| 35 | Y | Y | Y | H | Y | Y | Y | Y | Y | Y | Y | |
| 45 | I | I | I | I | I | I | I | I | I | I | I | |
| 70 | S | S | S | S | S | S | S | S | A | S | S | |
| 94 | E | E | E | E | E | E | E | E | E | E | E | |
| 138 | P | P | P | P | P | P | P | P | P | P | P | |
| 141 | K | K | K | K | K | K | K | K | K | K | K | |
| 188 | V | V | V | V | V | V | V | V | V | V | V | |
| 208 | D | D | D | H | D | D | D | D | D | D | D | |
| 288 | S | S | S | S | S | S | S | S | S | S | S | |
| 289 | E | E | E | G | E | E | E | E | E | E | E | |
| NS2A | 85 | I | I | V | I | V | V | I | I | I | I | I |
| 104 | N | N | N | N | N | N | N | N | N | N | N | |
| 119 | H | H | H | H | H | H | H | H | H | H | H | |
| 128 | E | E | E | E | E | E | E | E | E | E | E | |
| 138 | V | V | V | V | V | V | V | V | V | V | V | |
| 165 | G | G | G | G | G | G | G | G | G | G | G | |
| NS2B | 82 | D | D | D | D | D | D | D | D | D | D | D |
| 83 | G | G | G | G | G | G | G | G | G | G | G | |
| 103 | A | A | A | A | A | A | A | V | V | A | A | |
| 120 | V | I | V | V | V | I | V | V | V | V | V | |
| NS3 | 46 | F | F | F | F | F | F | F | F | F | F | F |
| 244 | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | H | |
| 249 | T | T | T | P | T | P | T | P | P | T | T | |
| 356 | I | I | I | I | I | I | I | I | T | I | I | |
| 496 | L | L | L | L | L | L | L | L | L | L | L | |
| 503 | N | N | N | N | N | N | N | N | N | N | N | |
| 521 | D | D | D | D | D | D | D | D | D | D | D | |
| NS4A | 85 | V | V | I | V | I | I | V | V | A | V | V |
| 100 | P | P | S | P | S | S | P | P | P | P | P | |
| 122 | P | P | P | P | P | P | P | P | P | P | P | |
| NS5 | 53 | H | H | H | H | H | H | H | H | H | H | H |
| 54 | P | P | P | P | P | P | P | P | P | P | P | |
| 257 | D | D | D | D | D | D | D | D | D | D | D | |
| 258 | V | V | A | V | A | A | A | V | V | V | V | |
| 280 | K | K | K | K | K | K | K | K | K | K | K | |
| 372 | V | V | V | V | V | V | V | V | V | V | V | |
| 374 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | |
| 422 | R | R | K | R | K | K | K | R | R | R | R | |
| 426 | E | E | E | A | E | E | E | E | E | E | E | |
| 436 | M | M | M | I | M | M | M | M | M | M | M | |
| 526 | T | T | T | T | T | T | T | T | T | T | T | |
| 653 | F | F | F | F | F | F | F | F | F | F | F | |
| 681 | T | T | T | T | T | T | T | T | T | T | T |
| MW627239 ITA 2020 | AF404757 ITA 1998 | JF719066 ITA 2008 | JF719065 ITA 2008 | FJ483548 ITA 2008 | MW835362 ITA 2011 | KF647253 ITA 2013 | NC_009942 USA 1999 | FJ766332 SPA 2007 | JF719069 SPA 2010 | |
|---|---|---|---|---|---|---|---|---|---|---|
| MW627239 ITA 2020 | - | 98.5% | 98.31% | 98.33% | 98.33% | 98.02% | 97.67% | 95.80% | 97.96% | 98.27% |
| AF404757 ITA 1998 | 99.85% | - | 98.50% | 98.53% | 98.55% | 98.18% | 98.54% | 96.44% | 98.93% | 98.51% |
| JF719066 ITA 2008 | 99.68% | 99.77% | - | 99.95% | 99.41% | 98.93% | 98.13% | 96.04% | 98.37% | 98.75% |
| JF719065 ITA 2008 | 99.71% | 99.80% | 99.91% | - | 99.94% | 98.95% | 98.15% | 96.07% | 98.41% | 98.78% |
| FJ483548 ITA 2008 | 99.68% | 99.83% | 99.88% | 99.91% | - | 98.93% | 98.15% | 96.10% | 98.39% | 98.76% |
| MW835362 ITA 2011 | 99.68% | 99.77% | 99.71% | 99.74% | 99.71% | - | 97.85% | 95.76% | 98.00% | 98.43% |
| KF647253 ITA 2013 | 99.74% | 99.83% | 99.65% | 99.68% | 99.71% | 99.65% | - | 96.03% | 98.34% | 98.11% |
| NC_009942 USA 1999 | 99.62% | 99.71% | 99.56% | 99.59% | 99.62% | 99.53% | 99.71% | - | 96.29% | 96.16% |
| FJ766332 SPA 2007 | 99.65% | 99.68% | 99.50% | 99.53% | 99.56% | 98.75% | 99.62% | 99.50% | - | 98.35% |
| JF719069 SPA 2010 | 99.65% | 99.74% | 99.56% | 99.59% | 99.56% | 99.39% | 99.62% | 99.50% | 99.48% | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mencattelli, G.; Iapaolo, F.; Monaco, F.; Fusco, G.; de Martinis, C.; Portanti, O.; Di Gennaro, A.; Curini, V.; Polci, A.; Berjaoui, S.; et al. West Nile Virus Lineage 1 in Italy: Newly Introduced or a Re-Occurrence of a Previously Circulating Strain? Viruses 2022, 14, 64. https://doi.org/10.3390/v14010064
Mencattelli G, Iapaolo F, Monaco F, Fusco G, de Martinis C, Portanti O, Di Gennaro A, Curini V, Polci A, Berjaoui S, et al. West Nile Virus Lineage 1 in Italy: Newly Introduced or a Re-Occurrence of a Previously Circulating Strain? Viruses. 2022; 14(1):64. https://doi.org/10.3390/v14010064
Chicago/Turabian StyleMencattelli, Giulia, Federica Iapaolo, Federica Monaco, Giovanna Fusco, Claudio de Martinis, Ottavio Portanti, Annapia Di Gennaro, Valentina Curini, Andrea Polci, Shadia Berjaoui, and et al. 2022. "West Nile Virus Lineage 1 in Italy: Newly Introduced or a Re-Occurrence of a Previously Circulating Strain?" Viruses 14, no. 1: 64. https://doi.org/10.3390/v14010064
APA StyleMencattelli, G., Iapaolo, F., Monaco, F., Fusco, G., de Martinis, C., Portanti, O., Di Gennaro, A., Curini, V., Polci, A., Berjaoui, S., Di Felice, E., Rosà, R., Rizzoli, A., & Savini, G. (2022). West Nile Virus Lineage 1 in Italy: Newly Introduced or a Re-Occurrence of a Previously Circulating Strain? Viruses, 14(1), 64. https://doi.org/10.3390/v14010064










