Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Plasmid Construction
2.3. The Recombinant Protein Purification
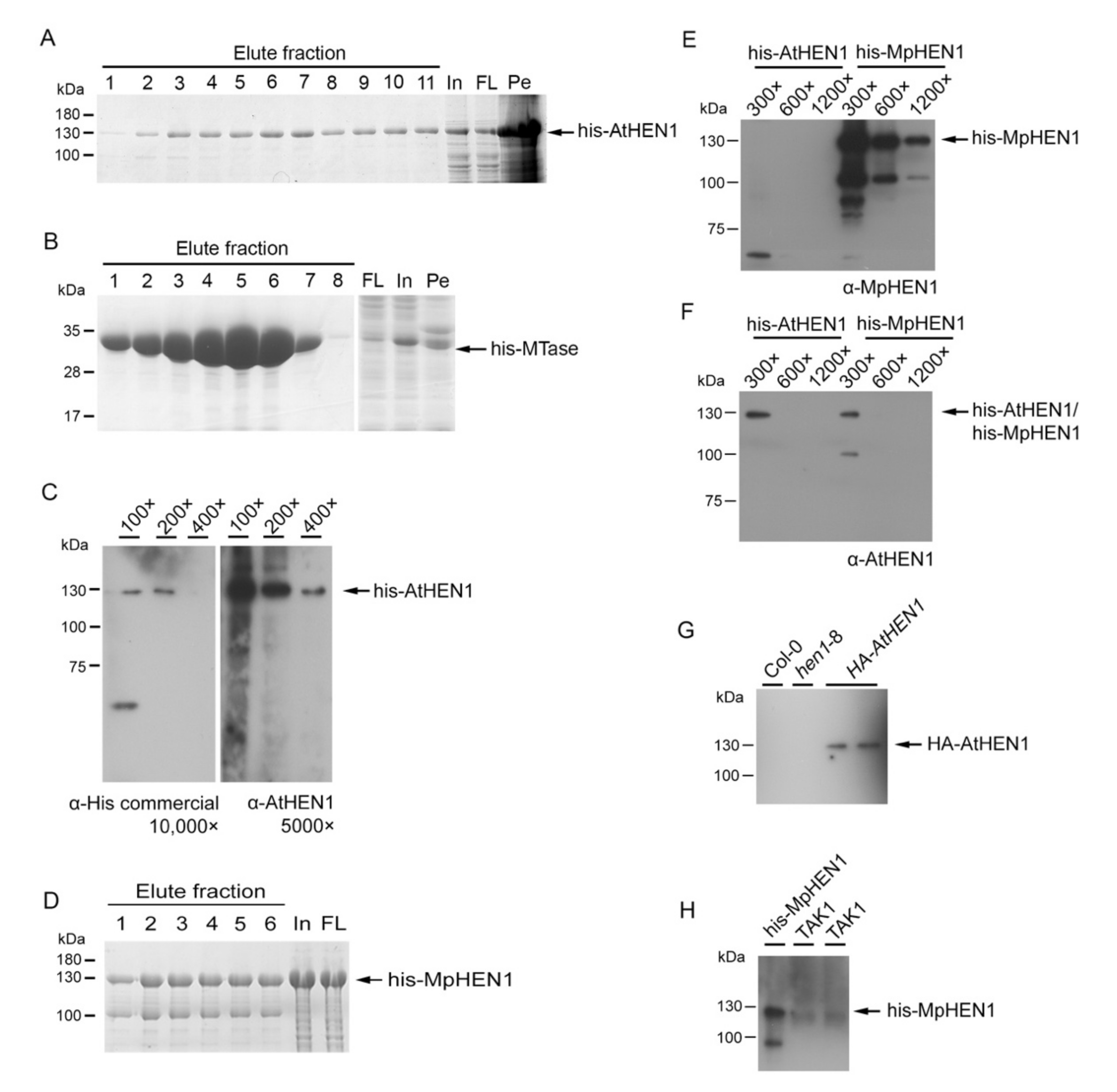
2.4. The α-AtHEN1 and α-MpHEN1 IgG Production
2.5. Western Blotting
2.6. In Vitro and In Vivo HEN1 Methylation Assay
2.7. β-Elimination
2.8. In Vitro Pull-Down and In Vivo IP
2.9. EMSA
2.10. MpHEN1 Structure Modeling
2.11. Sequence Identity, Similarity, and Phylogenetic Analysis
3. Results
3.1. HEN1 Antibody Production and Sensitivity Evaluation
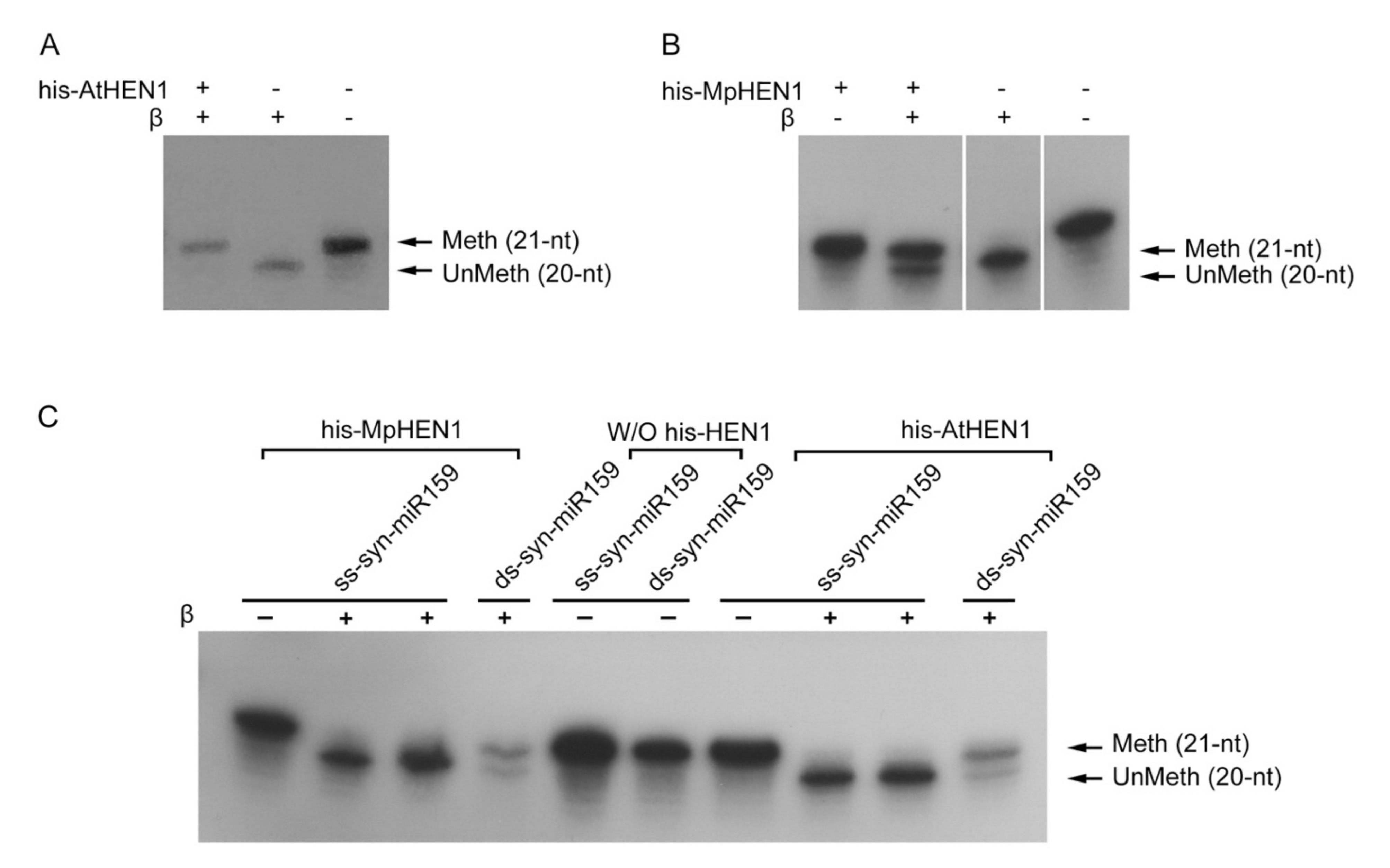
3.2. The Recombinant HEN1 Methylation Ability and Substrate Specificity
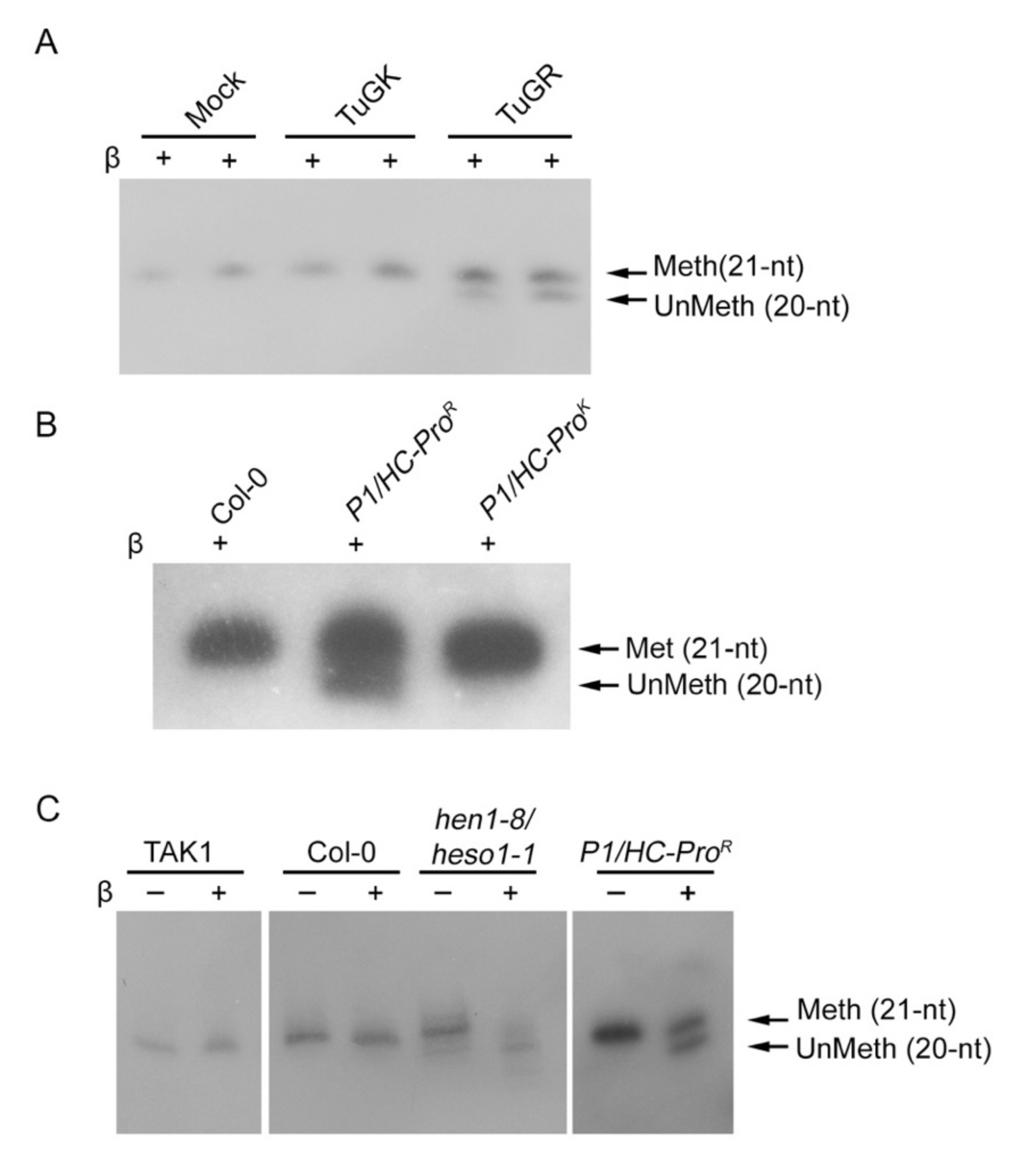
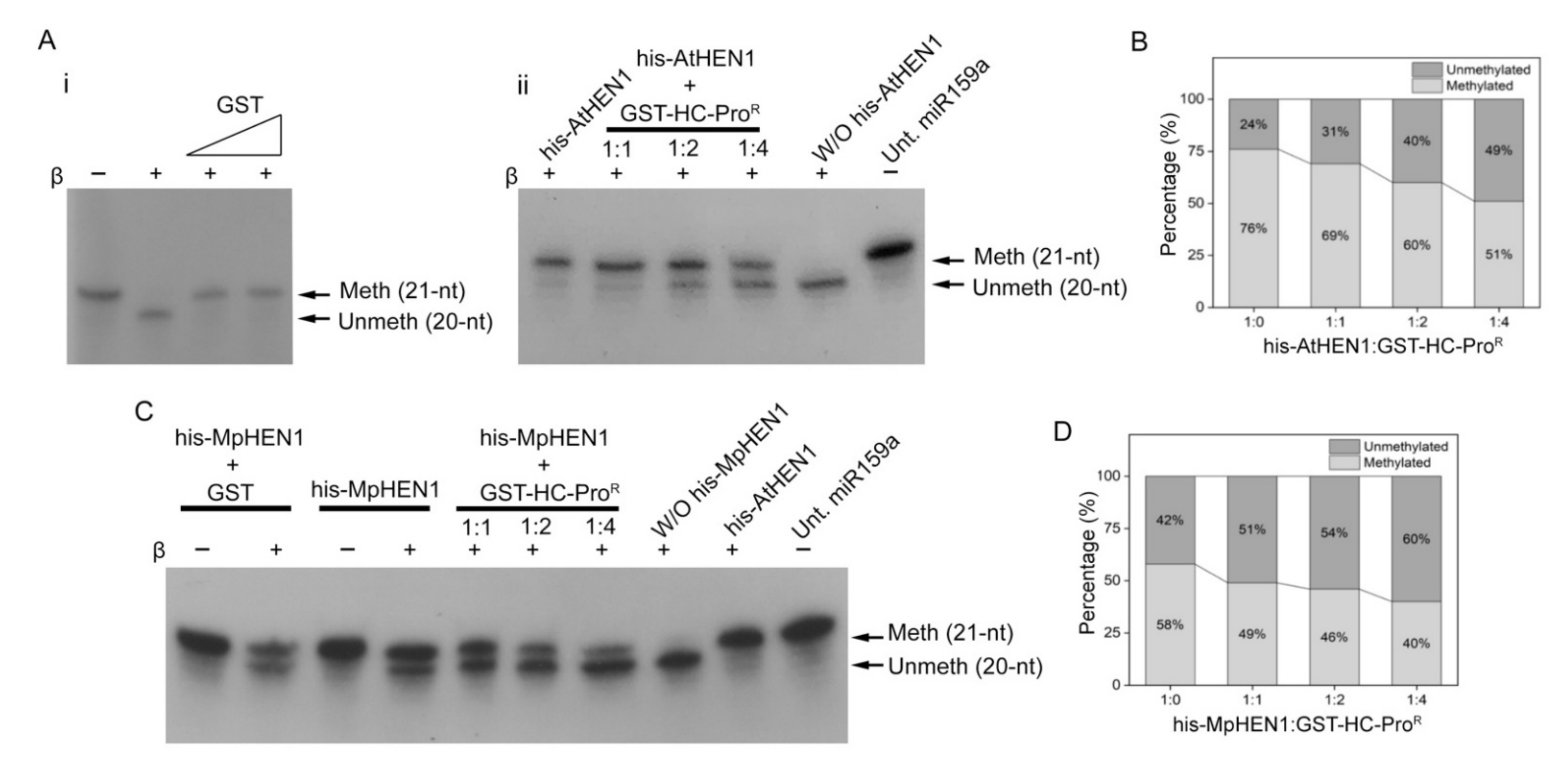
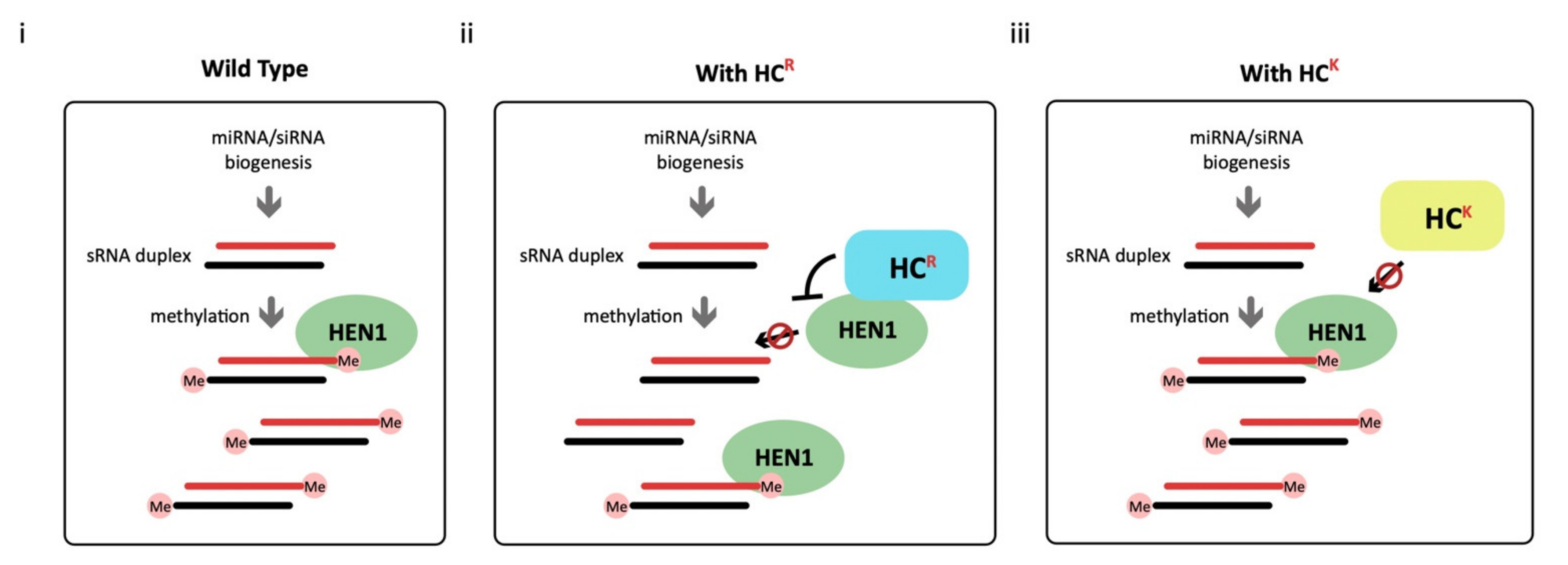
3.3. HC-Pro Suppressor of TuMV Inhibits HEN1 Activity
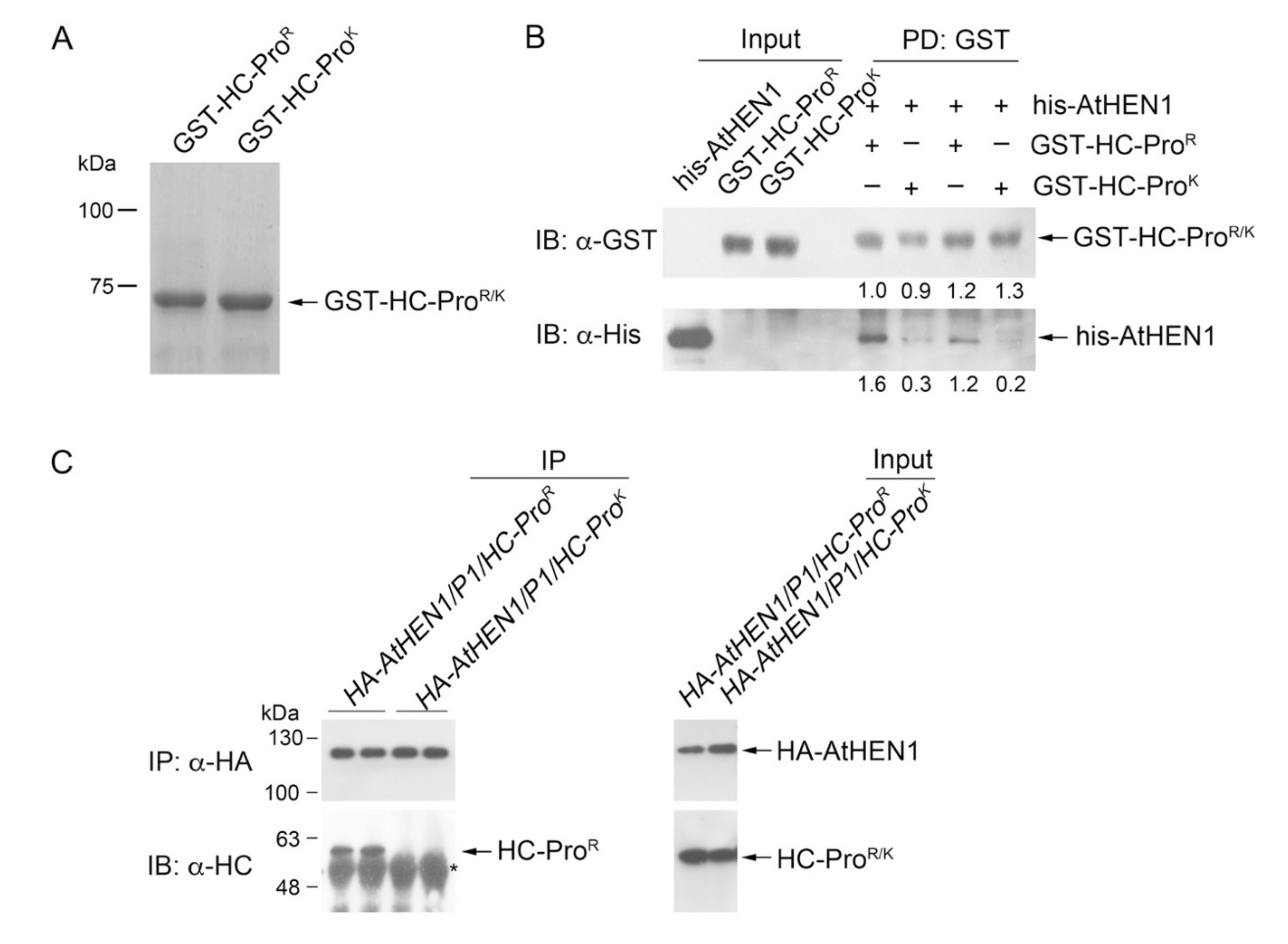
3.4. HC-ProR Binds HEN1 to Impair miRNA Methylation
3.5. HC-ProR Inhibits HEN1 Methyltransferase Activity In Vitro
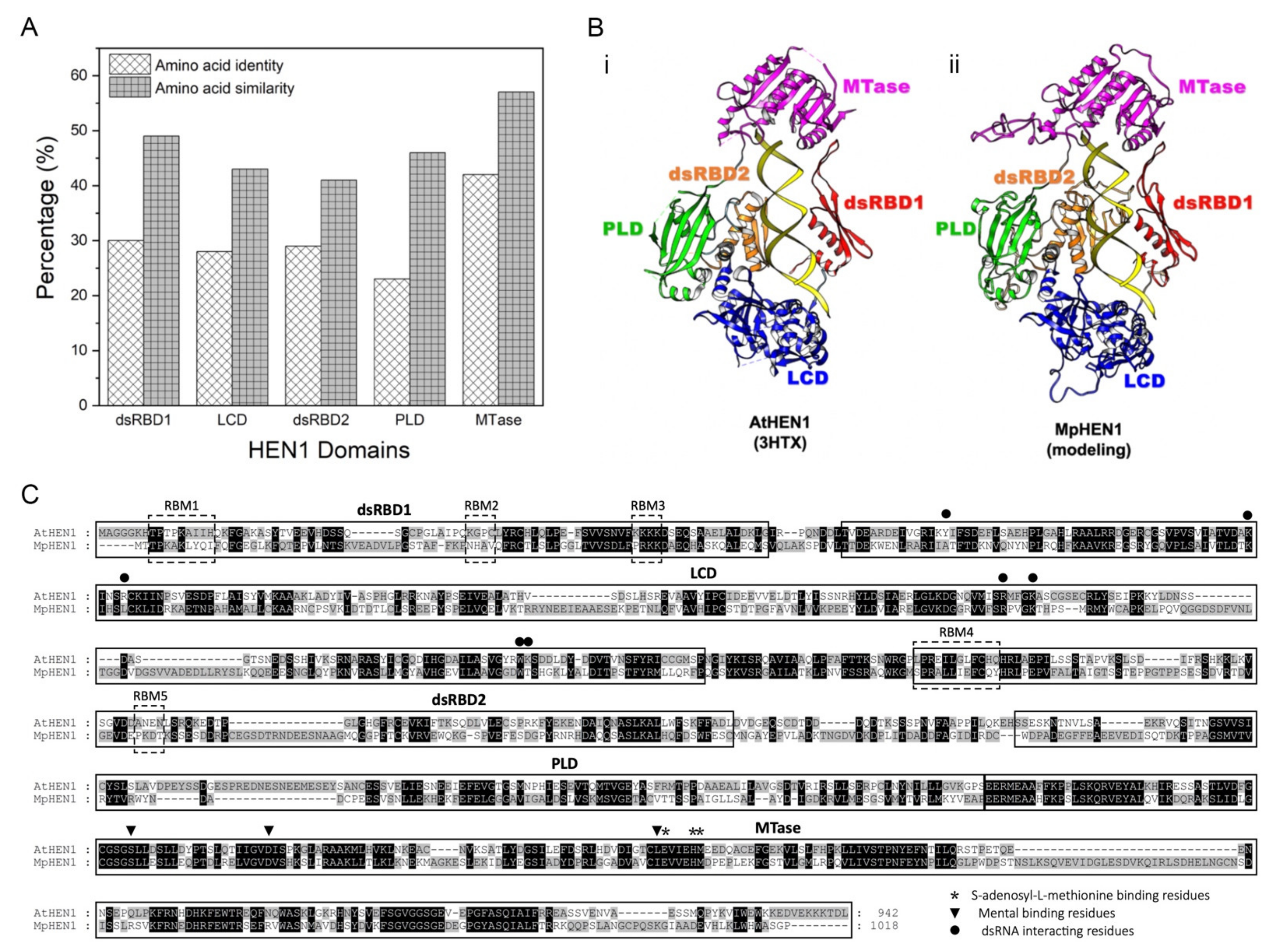
3.6. Functional Domain Comparison between AtHEN1 and MpHEN1
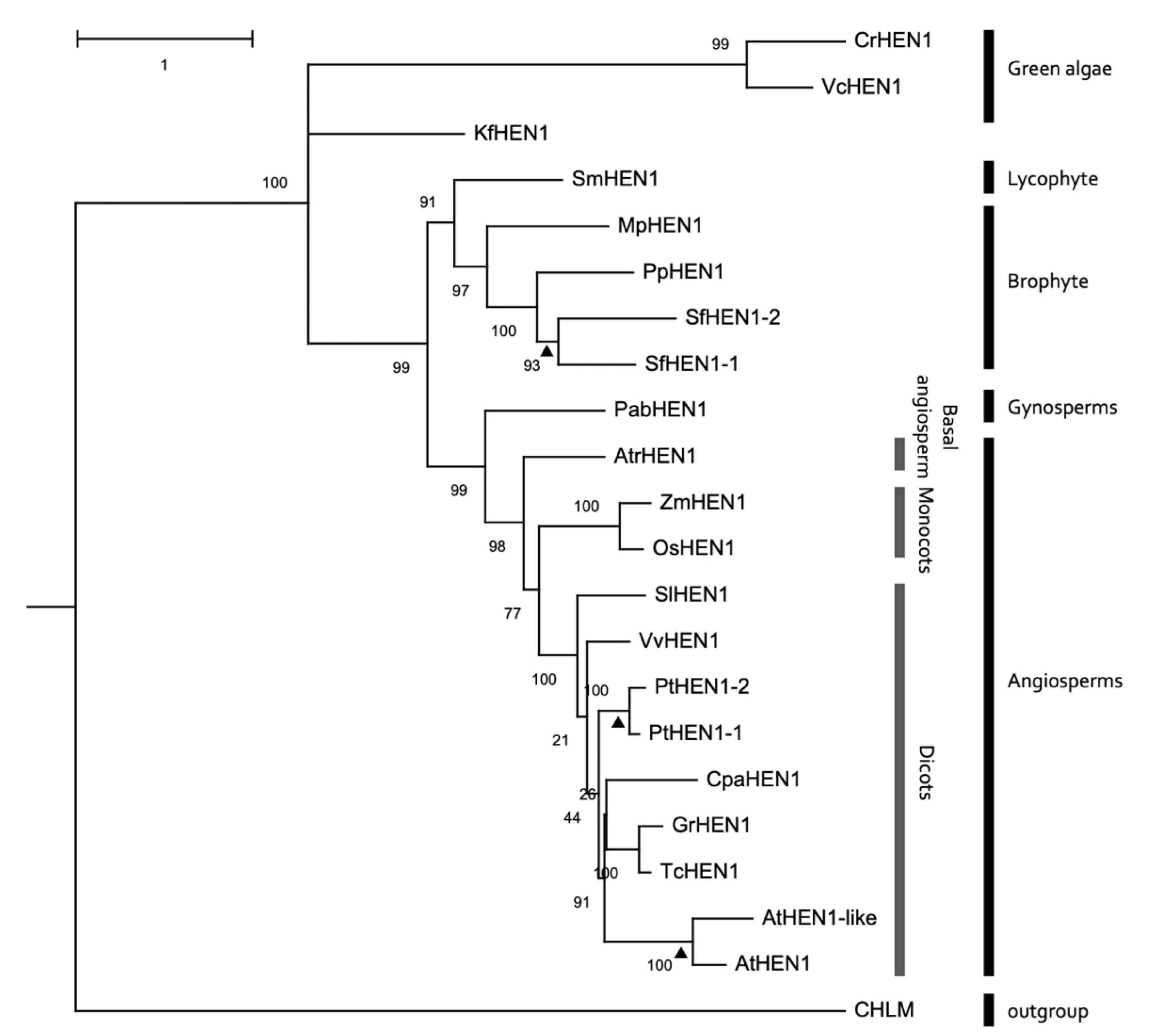
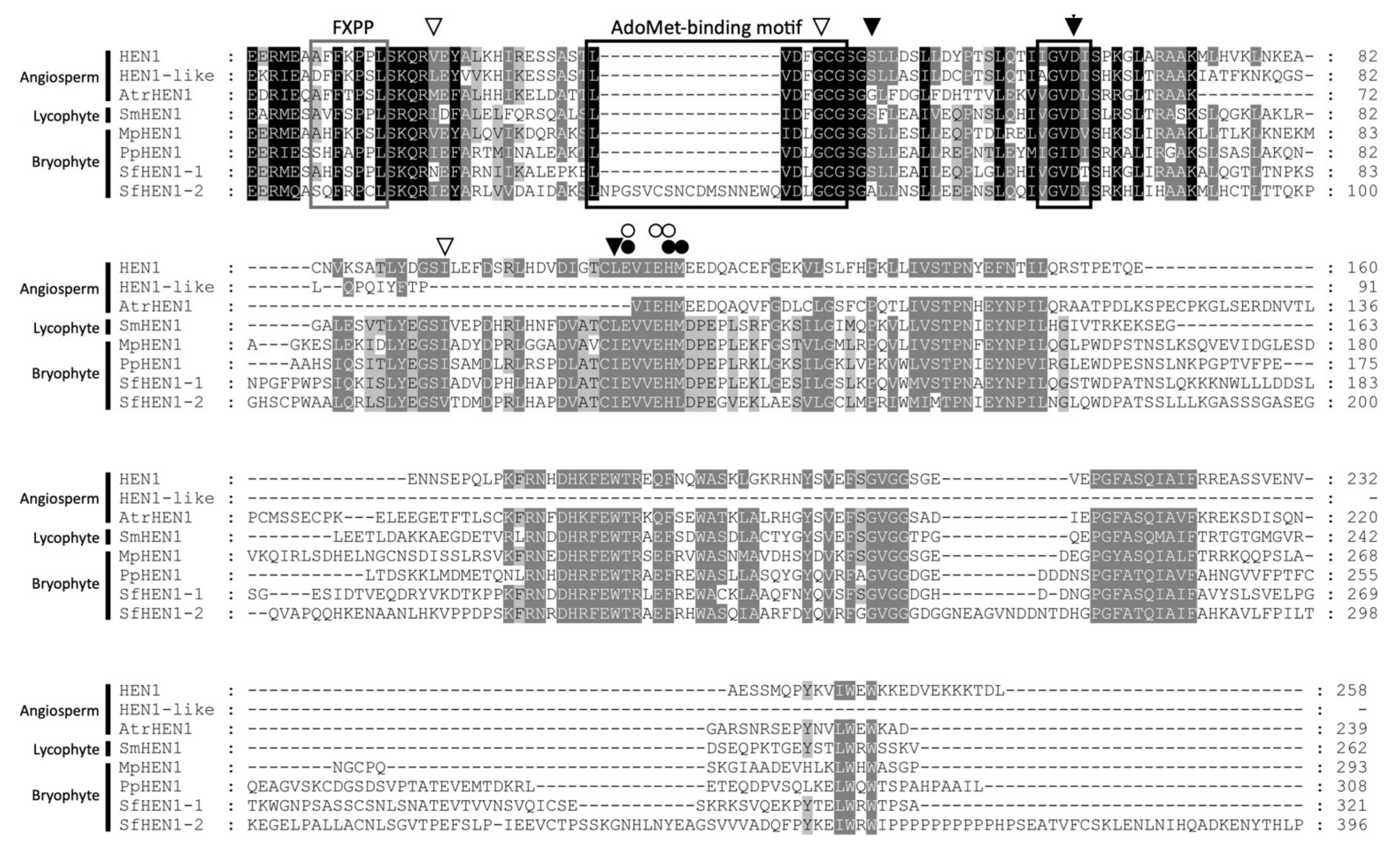
3.7. Conservation of HEN1 Orthologs in Plant Species
4. Discussion
4.1. The Serology of HEN1
4.2. HC-Pro Has a Common HEN1 Inhibition Ability in Bryophyte and Angiosperm
4.3. HEN1 Biology in Planta
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, Homeostasis, and Degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef] [Green Version]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell. Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Liu, M.F. Mechanisms of microRNA-mediated gene regulation. Sci. China C Life Sci. 2009, 52, 1111–1116. [Google Scholar] [CrossRef]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Ebright, Y.W.; Yu, B.; Chen, X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2’ OH of the 3’ terminal nucleotide. Nucleic Acids Res. 2006, 34, 667–675. [Google Scholar] [CrossRef]
- Yu, B.; Bi, L.; Zhai, J.; Agarwal, M.; Li, S.; Wu, Q.; Ding, S.W.; Meyers, B.C.; Vaucheret, H.; Chen, X. siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 2010, 38, 5844–5850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, J.; Zhao, Y.; Simon, S.A.; Huang, S.; Petsch, K.; Arikit, S.; Pillay, M.; Ji, L.; Xie, M.; Cao, X.; et al. Plant microRNAs display differential 3’ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 2013, 25, 2417–2428. [Google Scholar] [CrossRef]
- Tu, B.; Liu, L.; Xu, C.; Zhai, J.; Li, S.; Lopez, M.A.; Zhao, Y.; Yu, Y.; Ramachandran, V.; Ren, G.; et al. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 2015, 11, e1005119. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Dou, Y.; Chen, L.; Wang, J.; Jiang, N.; Guo, C.; Yao, Q.; Wang, C.; Liu, L.; et al. Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3’ to 5’ exoribonuclease Atrimmer 2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E6659–E6667. [Google Scholar] [CrossRef] [Green Version]
- Boutet, S.; Vazquez, F.; Liu, J.; Béclin, C.; Fagard, M.; Gratias, A.; Morel, J.B.; Crété, P.; Chen, X.; Vaucheret, H. Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003, 13, 843–848. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Ji, L.; Huang, Q.; Vassylyev, D.G.; Chen, X.; Ma, J.B. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 2009, 461, 823–827. [Google Scholar] [CrossRef]
- Baranauskė, S.; Mickutė, M.; Plotnikova, A.; Finke, A.; Venclovas, Č.; Klimašauskas, S.; Vilkaitis, G. Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. 2015, 43, 2802–2812. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Siomi, H.; Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2’-O-methylation of Piwi- interacting RNAs at their 3’ ends. Genes Dev. 2007, 21, 1603–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirino, Y.; Mourelatos, Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 2007, 13, 1397–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horwich, M.D.; Li, C.; Matranga, C.; Vagin, V.; Farley, G.; Wang, P.; Zamore, P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007, 17, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.; Shuman, S. Bacterial Hen1 is a 3’ terminal RNA ribose 2’-O-methyltransferase component of a bacterial RNA repair cassette. RNA 2010, 16, 316–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 2017, 171, 287–304.e15. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, A.; Terai, M.; Ishizaki, K.; Suetsugu, N.; Tsuboi, H.; Nishihama, R.; Yamato, K.T.; Wada, M.; Kohchi, T. Phototropin encoded by a single-copy gene mediates chloroplast photorelocation movements in the liverwort Marchantia polymorpha. Plant Physiol. 2014, 166, 411–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, M.; Kuniyoshi, T.; Yamamoto, H.; Sugimoto, K.; Ishizaki, K.; Kohchi, T.; Nishimura, Y.; Shikanai, T. Composition and physiological function of the chloroplast NADH dehydrogenase-like complex in Marchantia polymorpha. Plant J. 2012, 72, 683–693. [Google Scholar] [CrossRef]
- You, C.; Cui, J.; Wang, H.; Qi, X.; Kuo, L.Y.; Ma, H.; Gao, L.; Mo, B.; Chen, X. Conservation and divergence of small RNA pathways and microRNAs in land plants. Genome Biol. 2017, 18, 158. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Lu, C.W.; Shen, B.N.; Lee, G.Z.; Bowman, J.L.; Arteaga-Vazquez, M.A.; Liu, L.Y.; Hong, S.F.; Lo, C.F.; Su, G.M.; et al. Identification of miRNAs and Their Targets in the Liverwort Marchantia polymorpha by Integrating RNA-Seq and Degradome Analyses. Plant Cell Physiol. 2016, 57, 339–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.S.; Bowman, J.L. Micro RNAs in Marchantia polymorpha. New Phytol. 2018, 220, 409–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, Y.J.; Lin, P.C.; Yeh, S.D.; Hong, S.F.; Chua, N.H.; Liu, L.Y.; Lin, C.P.; Huang, Y.H.; Wu, H.W.; Chen, C.C.; et al. Genetic analyses of the FRNK motif function of Turnip mosaic virus uncover multiple and potentially interactive pathways of cross-protection. Mol. Plant-Microbe Interact. 2014, 27, 944–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.W.; Lin, S.S.; Chen, K.C.; Yeh, S.D.; Chua, N.H. Discriminating mutations of HC-Pro of zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol. Plant-Microbe Interact. 2010, 23, 17–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valli, A.A.; Gallo, A.; Rodamilans, B.; López-Moya, J.J.; García, J.A. The HCPro from the Potyviridae family: An enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 2018, 19, 744–763. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, A. RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PLoS Pathog. 2018, 14, e1007228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De, S.; Pollari, M.; Varjosalo, M.; Mäkinen, K. Association of host protein VARICOSE with HCPro within a multiprotein complex is crucial for RNA silencing suppression, translation, encapsidation and systemic spread of potato virus A infection. PLoS Pathog. 2020, 16, e1008956. [Google Scholar] [CrossRef]
- Lin, Y.Y.; Fang, M.M.; Lin, P.C.; Chiu, M.T.; Liu, L.Y.; Lin, C.P.; Lin, S.S. Improving initial infectivity of the Turnip mosaic virus (TuMV) infectious clone by an mini binary vector via agro-infiltration. Bot. Stud. 2013, 54, 22. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Chapman, E.J.; Yang, Z.; Carrington, J.C.; Chen, X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 2006, 580, 3117–3120. [Google Scholar] [CrossRef] [Green Version]
- Martínez, F.; Daròs, J.A. Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J. Virol. 2014, 88, 10725–10737. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, L.; Thilmony, R.; You, F.M.; Gu, Y.Q.; Coleman-Derr, D. PIECE 2.0: An update for the plant gene structure comparison and evolution database. Nucleic Acids Res. 2017, 45, 1015–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Liang, S.; Zhang, Z.; Liu, H.; Wang, S.; Pan, K.; Xu, J.; Ren, X.; Pei, S.; Yang, G. Genome assembly of Nannochloropsis oceanica provides evidence of host nucleus overthrow by the symbiont nucleus during speciation. Commun. Biol. 2019, 2, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Huang, R.H. Unique 2’-O-methylation by Hen1 in eukaryotic RNA interference and bacterial RNA repair. Biochemistry 2012, 51, 4087–4095. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, V.; Chen, X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 2008, 321, 1490–1492. [Google Scholar] [CrossRef] [Green Version]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell. 2003, 4, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Curr. Biol. 2005, 15, 1501–1507. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.; Zhang, F.; Shang, R.; Wang, X.; Chen, J.; Chou, J.J.; Ma, J.; Wu, L.; Huang, Y. Identification of substrates of the small RNA methyltransferase Hen1 in mouse spermatogonial stem cells and analysis of its methyl-transfer domain. J. Biol. Chem. 2018, 293, 9981–9994. [Google Scholar] [CrossRef] [Green Version]
- Schmid, M.W.; Giraldo-Fonseca, A.; Rövekamp, M.; Smetanin, D.; Bowman, J.L.; Grossniklaus, U. Extensive epigenetic reprogramming during the life cycle of Marchantia polymorpha. Genome Biol. 2018, 19, 9. [Google Scholar] [CrossRef]
- Hu, S.F.; Wei, W.L.; Hong, S.F.; Fang, R.Y.; Wu, H.Y.; Lin, P.C.; Sanobar, N.; Wang, H.P.; Sulistio, M.; Wu, C.T.; et al. Investigation of the effects of P1 on HC-pro-mediated gene silencing suppression through genetics and omics approaches. Bot. Stud. 2020, 61, 22. [Google Scholar] [CrossRef] [PubMed]
- Jamous, R.M.; Boonrod, K.; Fuellgrabe, M.W.; Ali-Shtayeh, M.S.; Krczal, G.; Wassenegger, M. The helper component-proteinase of the Zucchini yellow mosaic virus inhibits the Hua Enhancer 1 methyltransferase activity in vitro. J. Gen. Virol. 2011, 92 Pt 9, 2222–2226. [Google Scholar] [CrossRef]
- Leibman, D.; Wolf, D.; Saharan, V.; Zelcer, A.; Arazi, T.; Yoel, S.; Gaba, V.; Gal-On, A. A high level of transgenic viral small RNA is associated with broad potyvirus resistance in cucurbits. Mol. Plant-Microbe Interact. 2011, 24, 1220–1238. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Carbonell, A.; Hoyer, J.S.; Fahlgren, N.; Gilbert, K.B.; Takeda, A.; Giampetruzzi, A.; Garcia Ruiz, M.T.; McGinn, M.G.; Lowery, N.; et al. Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Pathog. 2015, 11, e1004755. [Google Scholar] [CrossRef] [PubMed]
- Anandalakshmi, R.; Pruss, G.J.; Ge, X.; Marathe, R.; Mallory, A.C.; Smith, T.H.; Vance, V.B. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 1998, 95, 13079–13084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajamäki, M.L.; Kelloniemi, J.; Alminaite, A.; Kekarainen, T.; Rabenstein, F.; Valkonen, J.P. A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology 2005, 342, 88–101. [Google Scholar] [CrossRef] [Green Version]
- Valli, A.; Martín-Hernández, A.M.; López-Moya, J.J.; García, J.A. RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 2006, 80, 10055–10063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasin, F.; Shan, H.; García, B.; Müller, M.; San León, D.; Ludman, M.; Fresno, D.H.; Fátyol, K.; Munné-Bosch, S.; Rodrigo, G.; et al. Abscisic Acid Connects Phytohormone Signaling with RNA Metabolic Pathways and Promotes an Antiviral Response that Is Evaded by a Self-Controlled RNA Virus. Plant Commun. 2020, 1, 100099. [Google Scholar] [CrossRef] [PubMed]
- Tena Fernández, F.; González, I.; Doblas, P.; Rodríguez, C.; Sahana, N.; Kaur, H.; Tenllado, F.; Praveen, S.; Canto, T. The influence of cis-acting P1 protein and translational elements on the expression of Potato virus Y helper-component proteinase (HCPro) in heterologous systems and its suppression of silencing activity. Mol. Plant Pathol. 2013, 14, 530–541. [Google Scholar] [CrossRef]
- Akbergenov, R.; Si-Ammour, A.; Blevins, T.; Amin, I.; Kutter, C.; Vanderschuren, H.; Zhang, P.; Gruissem, W.; Meins, F., Jr.; Hohn, T.; et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006, 34, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Billi, A.C.; Alessi, A.F.; Khivansara, V.; Han, T.; Freeberg, M.; Mitani, S.; Kim, J.K. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 2012, 8, e1002617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilkaitis, G.; Plotnikova, A.; Klimasauskas, S. Kinetic and functional analysis of the small RNA methyltransferase HEN1: The catalytic domain is essential for preferential modification of duplex RNA. RNA 2010, 16, 1935–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, W.; Li, J.; Song, R.; Messing, J.; Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002, 12, 1484–1495. [Google Scholar] [CrossRef] [Green Version]

| Similarity | AtHEN1 | AtHEN1-like | AtrHEN1 | MpHEN1 | PpHEN1 | SfHEN1-1 | SfHEN1-2 | SmHEN1 | |
|---|---|---|---|---|---|---|---|---|---|
| Identity | |||||||||
| AtHEN1 | - | 51.98 | 65.09 | 61.88 | 56.43 | 54.70 | 37.37 | 69.80 | |
| AtHEN1-like | 50.49 | - | 49.00 | 42.32 | 35.14 | 31.43 | 14.10 | 47.52 | |
| AtrHEN1 | 59.90 | 46.78 | - | 57.92 | 47.52 | 50.00 | 28.71 | 60.39 | |
| MpHEN1 | 54.95 | 39.35 | 52.22 | - | 59.40 | 63.11 | 42.32 | 63.86 | |
| PpHEN1 | 50.00 | 33.16 | 42.57 | 52.97 | - | 64.35 | 46.78 | 63.61 | |
| SfHEN1-1 | 48.51 | 30.44 | 44.80 | 57.17 | 58.91 | - | 47.27 | 58.66 | |
| SfHEN1-2 | 29.95 | 11.63 | 23.26 | 35.64 | 37.37 | 39.85 | - | 38.61 | |
| SmHEN1 | 62.62 | 44.80 | 54.20 | 57.17 | 56.43 | 52.47 | 30.69 | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanobar, N.; Lin, P.-C.; Pan, Z.-J.; Fang, R.-Y.; Tjita, V.; Chen, F.-F.; Wang, H.-C.; Tsai, H.-L.; Wu, S.-H.; Shen, T.-L.; et al. Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte. Viruses 2021, 13, 1837. https://doi.org/10.3390/v13091837
Sanobar N, Lin P-C, Pan Z-J, Fang R-Y, Tjita V, Chen F-F, Wang H-C, Tsai H-L, Wu S-H, Shen T-L, et al. Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte. Viruses. 2021; 13(9):1837. https://doi.org/10.3390/v13091837
Chicago/Turabian StyleSanobar, Neda, Pin-Chun Lin, Zhao-Jun Pan, Ru-Ying Fang, Veny Tjita, Fang-Fang Chen, Hao-Ching Wang, Huang-Lung Tsai, Shu-Hsing Wu, Tang-Long Shen, and et al. 2021. "Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte" Viruses 13, no. 9: 1837. https://doi.org/10.3390/v13091837
APA StyleSanobar, N., Lin, P.-C., Pan, Z.-J., Fang, R.-Y., Tjita, V., Chen, F.-F., Wang, H.-C., Tsai, H.-L., Wu, S.-H., Shen, T.-L., Chen, Y.-H., & Lin, S.-S. (2021). Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte. Viruses, 13(9), 1837. https://doi.org/10.3390/v13091837









