Designing Lentiviral Vectors for Gene Therapy of Genetic Diseases
Abstract
:1. Introduction
2. The Retroviral Genome
3. Retroviral Integration: A Key Aspect of Viral Vector Design
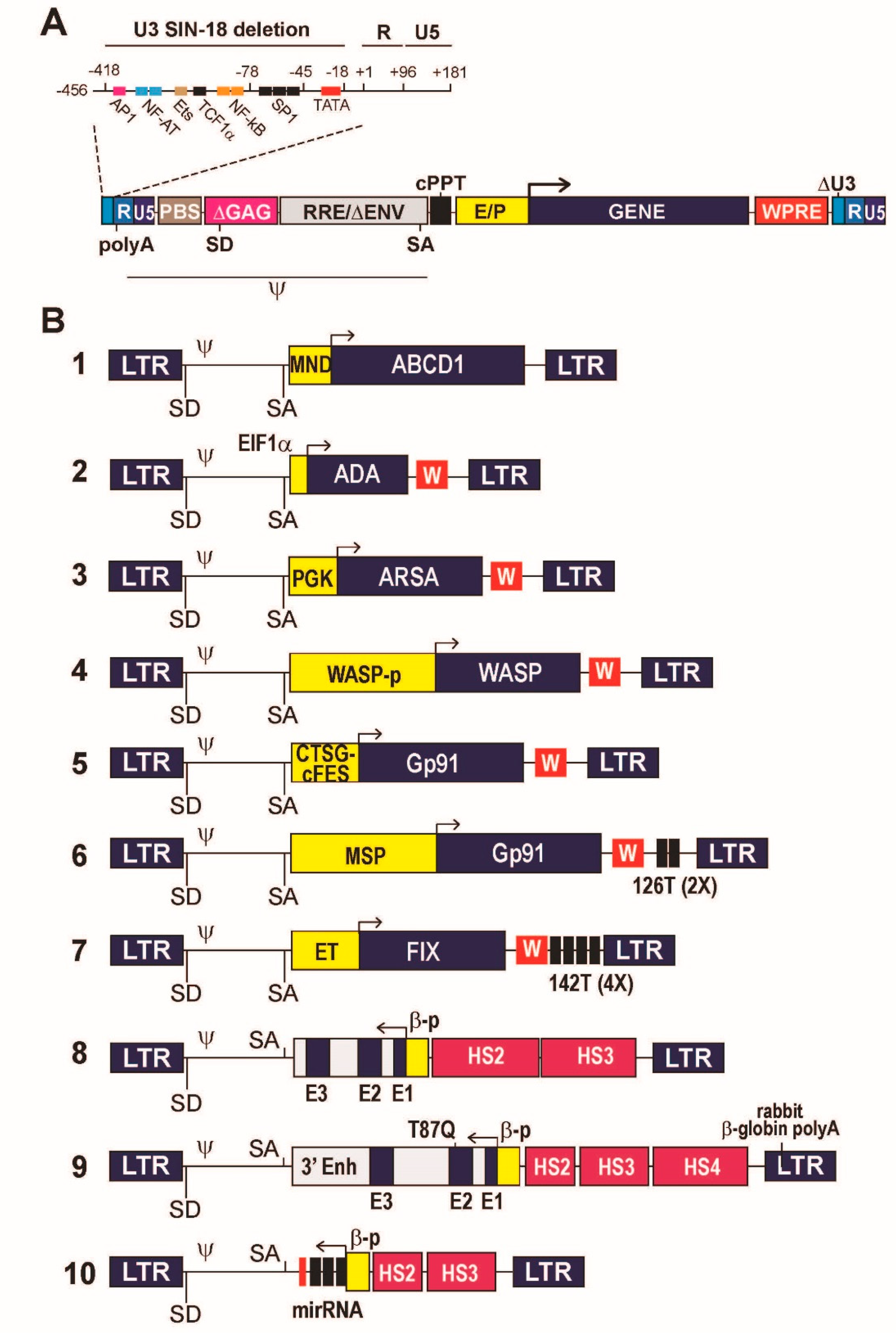
4. Designing a Lentiviral Vector
5. Designing a Transgene Expression Cassette
5.1. Constitutive Gene Expression
5.2. Cell Type-Specific Gene Expression
5.3. Regulation of Gene Expression In Vivo
5.4. Expression of Intron-Containing, Complex Gene Expression Cassettes
5.5. Impact of Expression Cassette Design on Vector Titer, Performance and Biosafety
5.6. Expression of Regulatory RNAs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tucci, F.; Scaramuzza, S.; Aiuti, A.; Mortellaro, A. Update on Clinical Ex Vivo Hematopoietic Stem Cell Gene Therapy for Inherited Monogenic Diseases. Mol. Ther. 2021, 29, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Aiuti, A.; Roncarolo, M.G.; Naldini, L. Gene therapy for ADA-SCID, the first marketing approval of an ex vivo gene therapy in Europe: Paving the road for the next generation of advanced therapy medicinal products. EMBO Mol. Med. 2017, 9, 737–740. [Google Scholar] [CrossRef]
- Williams, D.A.; Thrasher, A.J. Concise review: Lessons learned from clinical trials of gene therapy in monogenic immunodeficiency diseases. Stem Cells Transl. Med. 2014, 3, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.N. HIV Capsid and Integration Targeting. Viruses 2021, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Poletti, V.; Mavilio, F. Interactions between Retroviruses and the Host Cell Genome. Mol. Ther. Methods Clin. Dev. 2018, 8, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef]
- Aiuti, A.; Biasco, L.; Scaramuzza, S.; Ferrua, F.; Cicalese, M.P.; Baricordi, C.; Dionisio, F.; Calabria, A.; Giannelli, S.; Castiello, M.C.; et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 2013, 341, 1233151. [Google Scholar] [CrossRef] [Green Version]
- Hacein-Bey Abina, S.; Gaspar, H.B.; Blondeau, J.; Caccavelli, L.; Charrier, S.; Buckland, K.; Picard, C.; Six, E.; Himoudi, N.; Gilmour, K.; et al. Outcomes following gene therapy in patients with severe Wiskott-Aldrich syndrome. JAMA 2015, 313, 1550–1563. [Google Scholar] [CrossRef]
- Kohn, D.B.; Booth, C.; Kang, E.M.; Pai, S.Y.; Shaw, K.L.; Santilli, G.; Armant, M.; Buckland, K.F.; Choi, U.; De Ravin, S.S.; et al. Lentiviral gene therapy for X-linked chronic granulomatous disease. Nat. Med. 2020, 26, 200–206. [Google Scholar] [CrossRef]
- Kohn, D.B.; Booth, C.; Shaw, K.L.; Xu-Bayford, J.; Garabedian, E.; Trevisan, V.; Carbonaro-Sarracino, D.A.; Soni, K.; Terrazas, D.; Snell, K.; et al. Autologous Ex Vivo Lentiviral Gene Therapy for Adenosine Deaminase Deficiency. N. Engl. J. Med. 2021, 384, 2002–2013. [Google Scholar] [CrossRef]
- Ribeil, J.A.; Hacein-Bey-Abina, S.; Payen, E.; Magnani, A.; Semeraro, M.; Magrin, E.; Caccavelli, L.; Neven, B.; Bourget, P.; El Nemer, W.; et al. Gene Therapy in a Patient with Sickle Cell Disease. N. Engl. J. Med. 2017, 376, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Walters, M.C.; Kwiatkowski, J.; Rasko, J.E.J.; Ribeil, J.A.; Hongeng, S.; Magrin, E.; Schiller, G.J.; Payen, E.; Semeraro, M.; et al. Gene Therapy in Patients with Transfusion-Dependent beta-Thalassemia. N. Engl. J. Med. 2018, 378, 1479–1493. [Google Scholar] [CrossRef]
- Rio, P.; Navarro, S.; Wang, W.; Sanchez-Dominguez, R.; Pujol, R.M.; Segovia, J.C.; Bogliolo, M.; Merino, E.; Wu, N.; Salgado, R.; et al. Successful engraftment of gene-corrected hematopoietic stem cells in non-conditioned patients with Fanconi anemia. Nat. Med. 2019, 25, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Cartier, N.; Hacein-Bey-Abina, S.; Bartholomae, C.C.; Veres, G.; Schmidt, M.; Kutschera, I.; Vidaud, M.; Abel, U.; Dal-Cortivo, L.; Caccavelli, L.; et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009, 326, 818–823. [Google Scholar] [CrossRef] [Green Version]
- Biffi, A.; Montini, E.; Lorioli, L.; Cesani, M.; Fumagalli, F.; Plati, T.; Baldoli, C.; Martino, S.; Calabria, A.; Canale, S.; et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013, 341, 1233158. [Google Scholar] [CrossRef] [Green Version]
- Sessa, M.; Lorioli, L.; Fumagalli, F.; Acquati, S.; Redaelli, D.; Baldoli, C.; Canale, S.; Lopez, I.D.; Morena, F.; Calabria, A.; et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: An ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet 2016, 388, 476–487. [Google Scholar] [CrossRef]
- Eichler, F.; Duncan, C.; Musolino, P.L.; Orchard, P.J.; De Oliveira, S.; Thrasher, A.J.; Armant, M.; Dansereau, C.; Lund, T.C.; Miller, W.P.; et al. Hematopoietic Stem-Cell Gene Therapy for Cerebral Adrenoleukodystrophy. N. Engl. J. Med. 2017, 377, 1630–1638. [Google Scholar] [CrossRef] [Green Version]
- Coffin, J.M.; Huges, S.H.; Varmus, H.E. Retroviruses; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Vigna, E.; Naldini, L. Lentiviral vectors: Excellent tools for experimental gene transfer and promising candidates for gene therapy. J. Gene Med. 2000, 2, 308–316. [Google Scholar] [CrossRef]
- Maetzig, T.; Galla, M.; Baum, C.; Schambach, A. Gammaretroviral vectors: Biology, technology and application. Viruses 2011, 3, 677–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schott, J.W.; Morgan, M.; Galla, M.; Schambach, A. Viral and Synthetic RNA Vector Technologies and Applications. Mol. Ther. 2016, 24, 1513–1527. [Google Scholar] [CrossRef] [Green Version]
- Scoca, V.; Di Nunzio, F. The HIV-1 Capsid: From Structural Component to Key Factor for Host Nuclear Invasion. Viruses 2021, 13, 273. [Google Scholar] [CrossRef] [PubMed]
- Ocwieja, K.E.; Brady, T.L.; Ronen, K.; Huegel, A.; Roth, S.L.; Schaller, T.; James, L.C.; Towers, G.J.; Young, J.A.; Chanda, S.K.; et al. HIV integration targeting: A pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 2011, 7, e1001313. [Google Scholar] [CrossRef] [Green Version]
- Di Nunzio, F.; Fricke, T.; Miccio, A.; Valle-Casuso, J.C.; Perez, P.; Souque, P.; Rizzi, E.; Severgnini, M.; Mavilio, F.; Charneau, P.; et al. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 2013, 440, 8–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buffone, C.; Martinez-Lopez, A.; Fricke, T.; Opp, S.; Severgnini, M.; Cifola, I.; Petiti, L.; Frabetti, S.; Skorupka, K.; Zadrozny, K.K.; et al. Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells. J. Virol. 2018, 92, e00648-18. [Google Scholar] [CrossRef] [Green Version]
- Marini, B.; Kertesz-Farkas, A.; Ali, H.; Lucic, B.; Lisek, K.; Manganaro, L.; Pongor, S.; Luzzati, R.; Recchia, A.; Mavilio, F.; et al. Nuclear architecture dictates HIV-1 integration site selection. Nature 2015, 521, 227–231. [Google Scholar] [CrossRef]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef]
- Wang, G.P.; Ciuffi, A.; Leipzig, J.; Berry, C.C.; Bushman, F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007, 17, 1186–1194. [Google Scholar] [CrossRef] [Green Version]
- Cattoglio, C.; Pellin, D.; Rizzi, E.; Maruggi, G.; Corti, G.; Miselli, F.; Sartori, D.; Guffanti, A.; Di Serio, C.; Ambrosi, A.; et al. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 2010, 116, 5507–5517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Larue, R.C.; Plumb, M.R.; Malani, N.; Male, F.; Slaughter, A.; Kessl, J.J.; Shkriabai, N.; Coward, E.; Aiyer, S.S.; et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. USA 2013, 110, 12036–12041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rijck, J.; de Kogel, C.; Demeulemeester, J.; Vets, S.; El Ashkar, S.; Malani, N.; Bushman, F.D.; Landuyt, B.; Husson, S.J.; Busschots, K.; et al. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013, 5, 886–894. [Google Scholar] [CrossRef] [Green Version]
- Zufferey, R.; Dull, T.; Mandel, R.J.; Bukovsky, A.; Quiroz, D.; Naldini, L.; Trono, D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998, 72, 9873–9880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zufferey, R.; Nagy, D.; Mandel, R.J.; Naldini, L.; Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997, 15, 871–875. [Google Scholar] [CrossRef]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998, 72, 8463–8471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Harrison, G.P.; Miele, G.; Hunter, E.; Lever, A.M. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 1998, 72, 5886–5896. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Iwakuma, T.; Chang, L.J. Contributions of viral splice sites and cis-regulatory elements to lentivirus vector function. J. Virol. 1999, 73, 6171–6176. [Google Scholar] [CrossRef] [Green Version]
- Naldini, L.; Blomer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In Vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ory, D.S.; Neugeboren, B.A.; Mulligan, R.C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 1996, 93, 11400–11406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornetta, K.; Duffy, L.; Turtle, C.J.; Jensen, M.; Forman, S.; Binder-Scholl, G.; Fry, T.; Chew, A.; Maloney, D.G.; June, C.H. Absence of Replication-Competent Lentivirus in the Clinic: Analysis of Infused T Cell Products. Mol. Ther. 2018, 26, 280–288. [Google Scholar] [CrossRef] [Green Version]
- Trono, D. Lentiviral vectors: Turning a deadly foe into a therapeutic agent. Gene Ther. 2000, 7, 20–23. [Google Scholar] [CrossRef] [Green Version]
- Naldini, L. Ex Vivo gene transfer and correction for cell-based therapies. Nat. Rev. 2011, 12, 301–315. [Google Scholar] [CrossRef]
- Sandrin, V.; Boson, B.; Salmon, P.; Gay, W.; Negre, D.; Le Grand, R.; Trono, D.; Cosset, F.L. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 2002, 100, 823–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relander, T.; Johansson, M.; Olsson, K.; Ikeda, Y.; Takeuchi, Y.; Collins, M.; Richter, J. Gene transfer to repopulating human CD34+ cells using amphotropic-, GALV-, or RD114-pseudotyped HIV-1-based vectors from stable producer cells. Mol. Ther. 2005, 11, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Guerrero, A.; Cosset, F.L.; Verhoeyen, E. Lentiviral Vector Pseudotypes: Precious Tools to Improve Gene Modification of Hematopoietic Cells for Research and Gene Therapy. Viruses 2020, 12, 1016. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.; Ott, M.G.; Schultze-Strasser, S.; Jauch, A.; Burwinkel, B.; Kinner, A.; Schmidt, M.; Kramer, A.; Schwable, J.; Glimm, H.; et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010, 16, 198–204. [Google Scholar] [CrossRef] [Green Version]
- Maruggi, G.; Porcellini, S.; Facchini, G.; Perna, S.K.; Cattoglio, C.; Sartori, D.; Ambrosi, A.; Schambach, A.; Baum, C.; Bonini, C.; et al. Transcriptional Enhancers Induce Insertional Gene Deregulation Independently From the Vector Type and Design. Mol. Ther. 2009, 17, 851–856. [Google Scholar] [CrossRef] [Green Version]
- Cesana, D.; Ranzani, M.; Volpin, M.; Bartholomae, C.; Duros, C.; Artus, A.; Merella, S.; Benedicenti, F.; Sergi Sergi, L.; Sanvito, F.; et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther 2014, 22, 774–785. [Google Scholar] [CrossRef] [Green Version]
- Booth, C.; Gaspar, H.B.; Thrasher, A.J. Treating Immunodeficiency through HSC Gene Therapy. Trends Mol. Med. 2016, 22, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Poletti, V.; Biffi, A. Gene-Based Approaches to Inherited Neurometabolic Diseases. Hum. Gene Ther. 2019, 30, 1222–1235. [Google Scholar] [CrossRef]
- Poletti, V.; Charrier, S.; Corre, G.; Gjata, B.; Vignaud, A.; Zhang, F.; Rothe, M.; Schambach, A.; Gaspar, H.B.; Thrasher, A.J.; et al. Preclinical Development of a Lentiviral Vector for Gene Therapy of X-Linked Severe Combined Immunodeficiency. Mol. Ther. Methods Clin. Dev. 2018, 9, 257–269. [Google Scholar] [CrossRef] [Green Version]
- Zufferey, R.; Donello, J.E.; Trono, D.; Hope, T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999, 73, 2886–2892. [Google Scholar] [CrossRef] [Green Version]
- Higashimoto, T.; Urbinati, F.; Perumbeti, A.; Jiang, G.; Zarzuela, A.; Chang, L.J.; Kohn, D.B.; Malik, P. The woodchuck hepatitis virus post-transcriptional regulatory element reduces readthrough transcription from retroviral vectors. Gene Ther. 2007, 14, 1298–1304. [Google Scholar] [CrossRef] [Green Version]
- Charrier, S.; Lagresle-Peyrou, C.; Poletti, V.; Rothe, M.; Cedrone, G.; Gjata, B.; Mavilio, F.; Fischer, A.; Schambach, A.; de Villartay, J.P.; et al. Biosafety Studies of a Clinically Applicable Lentiviral Vector for the Gene Therapy of Artemis-SCID. Mol. Ther. Methods Clin. Dev. 2019, 15, 232–245. [Google Scholar] [CrossRef] [Green Version]
- Braun, C.J.; Boztug, K.; Paruzynski, A.; Witzel, M.; Schwarzer, A.; Rothe, M.; Modlich, U.; Beier, R.; Gohring, G.; Steinemann, D.; et al. Gene therapy for Wiskott-Aldrich syndrome--long-term efficacy and genotoxicity. Sci. Transl. Med. 2014, 6, 227ra33. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.C.; Fox, T.; Chakraverty, R.; Tendeiro, R.; Snell, K.; Rivat, C.; Grace, S.; Gilmour, K.; Workman, S.; Buckland, K.; et al. Gene therapy for Wiskott-Aldrich syndrome in a severely affected adult. Blood 2017, 130, 1327–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrua, F.; Cicalese, M.P.; Galimberti, S.; Giannelli, S.; Dionisio, F.; Barzaghi, F.; Migliavacca, M.; Bernardo, M.E.; Calbi, V.; Assanelli, A.A.; et al. Lentiviral haemopoietic stem/progenitor cell gene therapy for treatment of Wiskott-Aldrich syndrome: Interim results of a non-randomised, open-label, phase 1/2 clinical study. Lancet Haematol. 2019, 6, e239–e253. [Google Scholar] [CrossRef] [Green Version]
- Biasco, L.; Rothe, M.; Buning, H.; Schambach, A. Analyzing the Genotoxicity of Retroviral Vectors in Hematopoietic Cell Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 21–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scala, S.; Basso-Ricci, L.; Dionisio, F.; Pellin, D.; Giannelli, S.; Salerio, F.A.; Leonardelli, L.; Cicalese, M.P.; Ferrua, F.; Aiuti, A.; et al. Dynamics of genetically engineered hematopoietic stem and progenitor cells after autologous transplantation in humans. Nat. Med. 2018, 24, 1683–1690. [Google Scholar] [CrossRef]
- Santilli, G.; Almarza, E.; Brendel, C.; Choi, U.; Beilin, C.; Blundell, M.P.; Haria, S.; Parsley, K.L.; Kinnon, C.; Malech, H.L.; et al. Biochemical correction of X-CGD by a novel chimeric promoter regulating high levels of transgene expression in myeloid cells. Mol. Ther. 2011, 19, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Brendel, C.; Rothe, M.; Santilli, G.; Charrier, S.; Stein, S.; Kunkel, H.; Abriss, D.; Muller-Kuller, U.; Gaspar, B.; Modlich, U.; et al. Non-Clinical Efficacy and Safety Studies on G1XCGD, a Lentiviral Vector for Ex Vivo Gene Therapy of X-Linked Chronic Granulomatous Disease. Hum. Gene Ther. Clin. Dev. 2018, 29, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.G.; Schmidt, M.; Schwarzwaelder, K.; Stein, S.; Siler, U.; Koehl, U.; Glimm, H.; Kuhlcke, K.; Schilz, A.; Kunkel, H.; et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006, 12, 401–409. [Google Scholar] [CrossRef]
- Chiriaco, M.; Farinelli, G.; Capo, V.; Zonari, E.; Scaramuzza, S.; Di Matteo, G.; Sergi, L.S.; Migliavacca, M.; Hernandez, R.J.; Bombelli, F.; et al. Dual-regulated lentiviral vector for gene therapy of X-linked chronic granulomatosis. Mol. Ther. 2014, 22, 1472–1483. [Google Scholar] [CrossRef] [Green Version]
- Brown, B.D.; Gentner, B.; Cantore, A.; Colleoni, S.; Amendola, M.; Zingale, A.; Baccarini, A.; Lazzari, G.; Galli, C.; Naldini, L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007, 25, 1457–1467. [Google Scholar] [CrossRef]
- Goudy, K.S.; Annoni, A.; Naldini, L.; Roncarolo, M.G. Manipulating Immune Tolerance with Micro-RNA Regulated Gene Therapy. Front. Microbiol. 2011, 2, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, B.D.; Cantore, A.; Annoni, A.; Sergi, L.S.; Lombardo, A.; Della Valle, P.; D’Angelo, A.; Naldini, L. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood 2007, 110, 4144–4152. [Google Scholar] [CrossRef] [Green Version]
- Cantore, A.; Nair, N.; Della Valle, P.; Di Matteo, M.; Matrai, J.; Sanvito, F.; Brombin, C.; Di Serio, C.; D’Angelo, A.; Chuah, M.; et al. Hyperfunctional coagulation factor IX improves the efficacy of gene therapy in hemophilic mice. Blood 2012, 120, 4517–4520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantore, A.; Ranzani, M.; Bartholomae, C.C.; Volpin, M.; Valle, P.D.; Sanvito, F.; Sergi, L.S.; Gallina, P.; Benedicenti, F.; Bellinger, D.; et al. Liver-directed lentiviral gene therapy in a dog model of hemophilia B. Sci. Transl. Med. 2015, 7, 277ra28. [Google Scholar] [CrossRef] [Green Version]
- Matsui, H.; Hegadorn, C.; Ozelo, M.; Burnett, E.; Tuttle, A.; Labelle, A.; McCray, P.B., Jr.; Naldini, L.; Brown, B.; Hough, C.; et al. A microRNA-regulated and GP64-pseudotyped lentiviral vector mediates stable expression of FVIII in a murine model of Hemophilia A. Mol. Ther. 2011, 19, 723–730. [Google Scholar] [CrossRef]
- Schmitt, F.; Remy, S.; Dariel, A.; Flageul, M.; Pichard, V.; Boni, S.; Usal, C.; Myara, A.; Laplanche, S.; Anegon, I.; et al. Lentiviral vectors that express UGT1A1 in liver and contain miR-142 target sequences normalize hyperbilirubinemia in Gunn rats. Gastroenterology 2010, 139, 999–1007. [Google Scholar] [CrossRef] [Green Version]
- Papapetrou, E.P.; Kovalovsky, D.; Beloeil, L.; Sant’angelo, D.; Sadelain, M. Harnessing endogenous miR-181a to segregate transgenic antigen receptor expression in developing versus post-thymic T cells in murine hematopoietic chimeras. J. Clin. Investig. 2009, 119, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Gentner, B.; Schira, G.; Giustacchini, A.; Amendola, M.; Brown, B.D.; Ponzoni, M.; Naldini, L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods 2009, 6, 63–66. [Google Scholar] [CrossRef]
- Lechman, E.R.; Gentner, B.; van Galen, P.; Giustacchini, A.; Saini, M.; Boccalatte, F.E.; Hiramatsu, H.; Restuccia, U.; Bachi, A.; Voisin, V.; et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 2012, 11, 799–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, G.; Cavazzana, M.; Mavilio, F. Gene Therapy Approaches to Hemoglobinopathies. Hematol. Oncol. Clin. N. Am. 2017, 31, 835–852. [Google Scholar] [CrossRef] [PubMed]
- Weatherall, D.J. The Thalassemias. In The Molecular Basis of Blood Diseases, 2nd ed.; Stamatoyannopoulos, G., Nienhuis, A.W., Majerus, P.W., Varmus, H., Eds.; Saunders: Philadelphia, PA, USA, 1994; pp. 157–205. [Google Scholar]
- Stamatoyannopoulos, G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005, 33, 259–271. [Google Scholar] [CrossRef] [Green Version]
- Marktel, S.; Scaramuzza, S.; Cicalese, M.P.; Giglio, F.; Galimberti, S.; Lidonnici, M.R.; Calbi, V.; Assanelli, A.; Bernardo, M.E.; Rossi, C.; et al. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ss-thalassemia. Nat. Med. 2019, 25, 234–241. [Google Scholar] [CrossRef]
- Han, J.; Tam, K.; Ma, F.; Tam, C.; Aleshe, B.; Wang, X.; Quintos, J.P.; Morselli, M.; Pellegrini, M.; Hollis, R.P.; et al. Beta-Globin Lentiviral Vectors Have Reduced Titers due to Incomplete Vector RNA Genomes and Lowered Virion Production. Stem Cell Rep. 2021, 16, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Walters, M.C.; Mapara, M.Y.; Kwiatkowski, J.L.; Krishnamurti, L.; Aygun, B.; Kasow, K.A.; Rifkin-Zenenberg, S.; Schmidt, M.; DelCarpini, J. Resolution of Serious Vaso-Occlusive Pain Crises and Reduction in Patient-Reported Pain Intensity: Results from the Ongoing Phase 1/2 HGB-206 Group C Study of LentiGlobin for Sickle Cell Disease (bb1111) Gene Therapy. Blood 2020, 136 (Suppl. 1), 16–17. [Google Scholar]
- Weber, L.; Poletti, V.; Magrin, E.; Antoniani, C.; Martin, S.; Bayard, C.; Sadek, H.; Felix, T.; Meneghini, V.; Antoniou, M.N.; et al. An Optimized Lentiviral Vector Efficiently Corrects the Human Sickle Cell Disease Phenotype. Mol. Ther. Methods Clin. Dev. 2018, 10, 268–280. [Google Scholar] [CrossRef] [Green Version]
- Urbinati, F.; Campo Fernandez, B.; Masiuk, K.E.; Poletti, V.; Hollis, R.P.; Koziol, C.; Kaufman, M.L.; Brown, D.; Mavilio, F.; Kohn, D.B. Gene Therapy for Sickle Cell Disease: A Lentiviral Vector Comparison Study. Hum. Gene Ther. 2018, 29, 1153–1166. [Google Scholar] [CrossRef] [PubMed]
- Moiani, A.; Paleari, Y.; Sartori, D.; Mezzadra, R.; Miccio, A.; Cattoglio, C.; Cocchiarella, F.; Lidonnici, M.R.; Ferrari, G.; Mavilio, F. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J. Clin. Investig. 2012, 122, 1653–1666. [Google Scholar] [CrossRef] [Green Version]
- Cesana, D.; Sgualdino, J.; Rudilosso, L.; Merella, S.; Naldini, L.; Montini, E. Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J. Clin. Investig. 2012, 122, 1667–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heckl, D.; Schwarzer, A.; Haemmerle, R.; Steinemann, D.; Rudolph, C.; Skawran, B.; Knoess, S.; Krause, J.; Li, Z.; Schlegelberger, B.; et al. Lentiviral vector induced insertional haploinsufficiency of Ebf1 causes murine leukemia. Mol. Ther. 2012, 20, 1187–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavazzana-Calvo, M.; Payen, E.; Negre, O.; Wang, G.; Hehir, K.; Fusil, F.; Down, J.; Denaro, M.; Brady, T.; Westerman, K.; et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 2010, 467, 318–322. [Google Scholar] [CrossRef]
- Bauer, D.E.; Orkin, S.H. Hemoglobin switching’s surprise: The versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr. Opin. Genet. Dev. 2015, 33, 62–70. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Hargreaves, V.V.; Zhu, Q.; Kurland, J.V.; Hong, J.; Kim, W.; Sher, F.; Macias-Trevino, C.; Rogers, J.M.; Kurita, R.; et al. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell 2018, 173, 430–442.e17. [Google Scholar] [CrossRef] [Green Version]
- Hoban, M.D.; Orkin, S.H.; Bauer, D.E. Genetic treatment of a molecular disorder: Gene therapy approaches to sickle cell disease. Blood 2016, 127, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Brendel, C.; Guda, S.; Renella, R.; Bauer, D.E.; Canver, M.C.; Kim, Y.J.; Heeney, M.M.; Klatt, D.; Fogel, J.; Milsom, M.D.; et al. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J. Clin. Investig. 2016, 126, 3868–3878. [Google Scholar] [CrossRef] [Green Version]
- Esrick, E.B.; Lehmann, L.E.; Biffi, A.; Achebe, M.; Brendel, C.; Ciuculescu, M.F.; Daley, H.; MacKinnon, B.; Morris, E.; Federico, A.; et al. Post-Transcriptional Genetic Silencing of BCL11A to Treat Sickle Cell Disease. N. Engl. J. Med. 2021, 384, 205–215. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poletti, V.; Mavilio, F. Designing Lentiviral Vectors for Gene Therapy of Genetic Diseases. Viruses 2021, 13, 1526. https://doi.org/10.3390/v13081526
Poletti V, Mavilio F. Designing Lentiviral Vectors for Gene Therapy of Genetic Diseases. Viruses. 2021; 13(8):1526. https://doi.org/10.3390/v13081526
Chicago/Turabian StylePoletti, Valentina, and Fulvio Mavilio. 2021. "Designing Lentiviral Vectors for Gene Therapy of Genetic Diseases" Viruses 13, no. 8: 1526. https://doi.org/10.3390/v13081526
APA StylePoletti, V., & Mavilio, F. (2021). Designing Lentiviral Vectors for Gene Therapy of Genetic Diseases. Viruses, 13(8), 1526. https://doi.org/10.3390/v13081526





