Structural Analysis of the Novel Variants of SARS-CoV-2 and Forecasting in North America
Abstract
1. Introduction
2. Materials and Methods
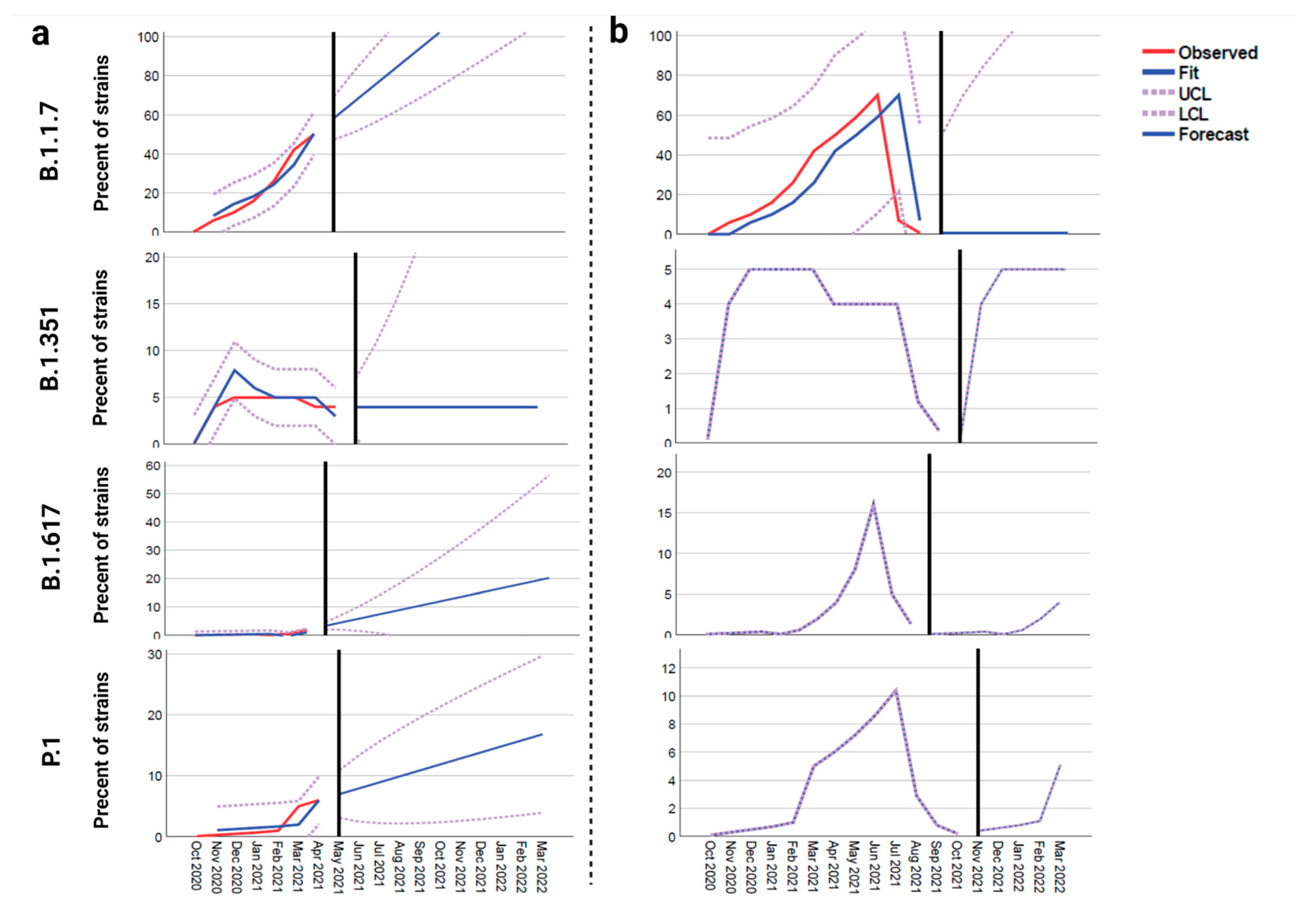
2.1. Time Series Forecasting
2.2. Construction of the Computational Model
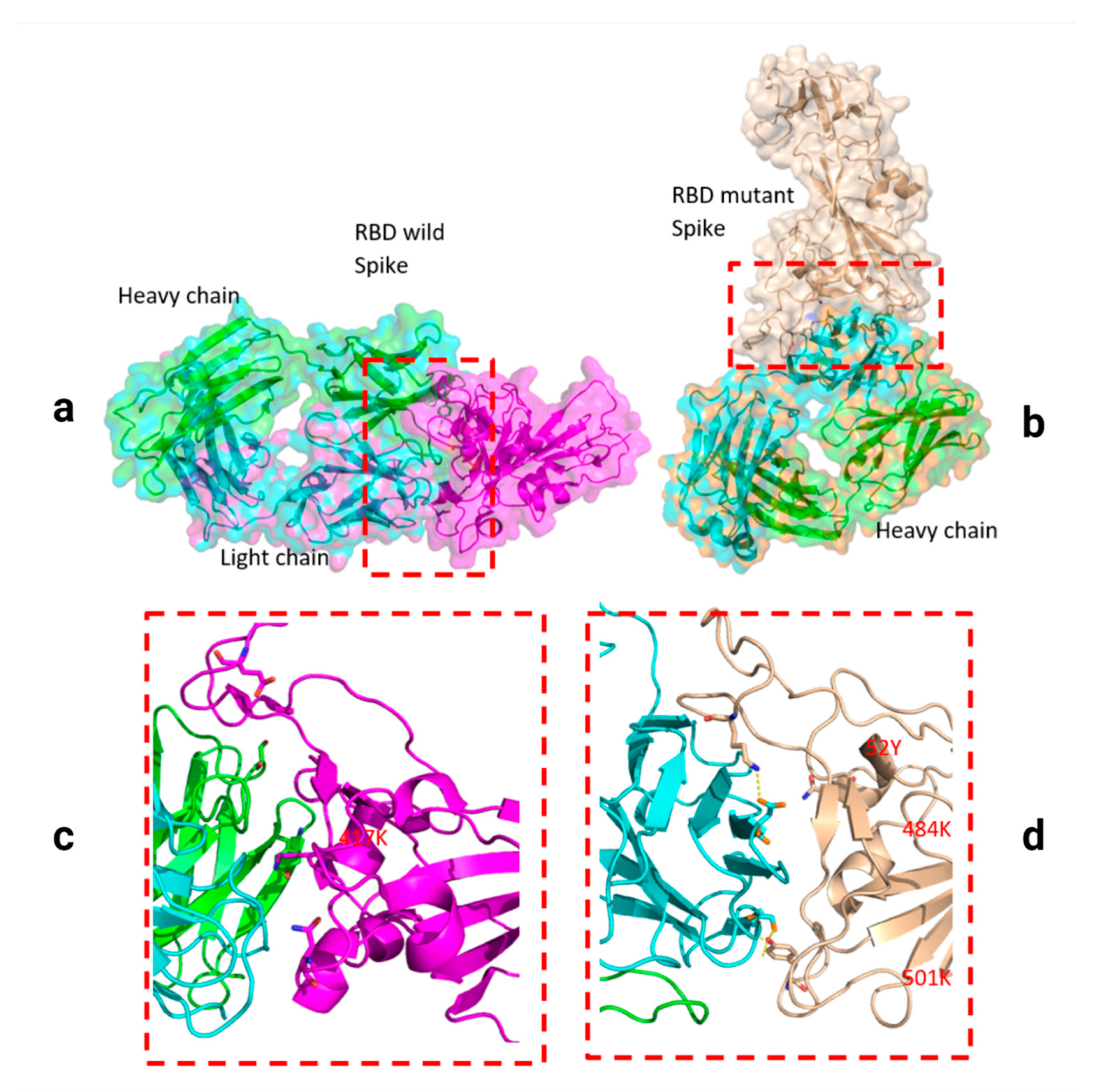
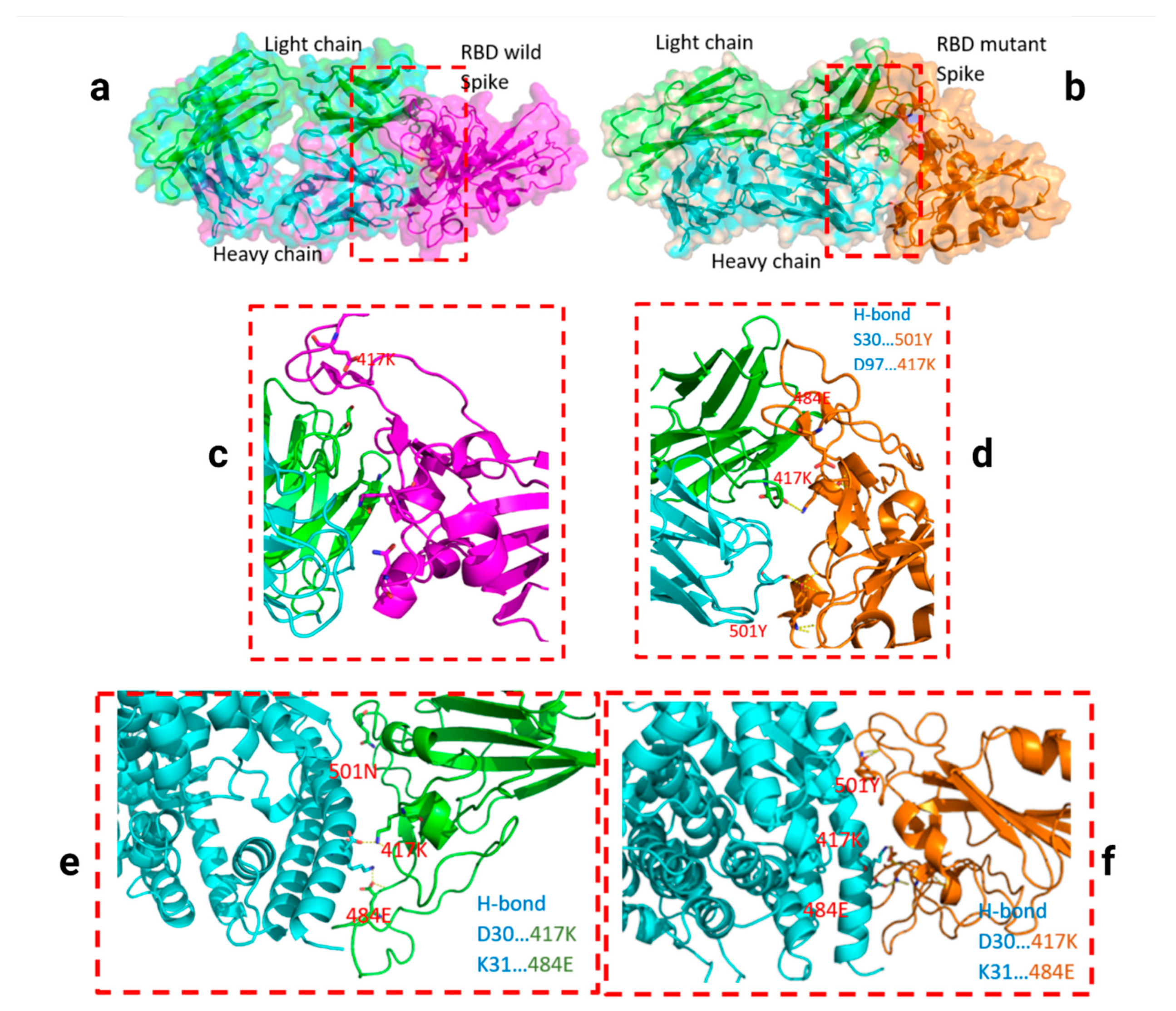
2.3. Analyzed Structures
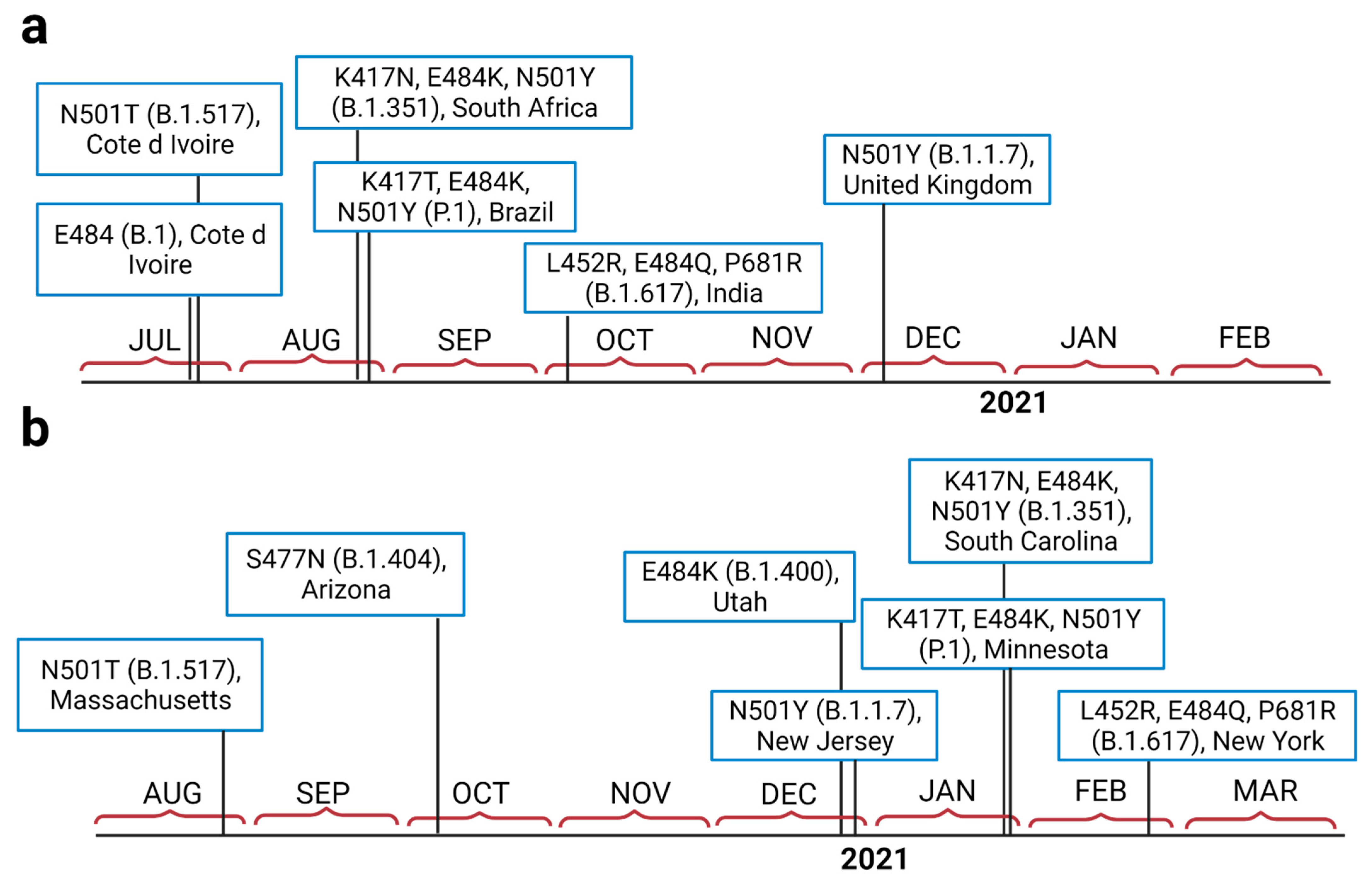
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020, 49, 141–154. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.M.; Wysocki, J.; Batlle, D. Interaction of SARS-CoV-2 and other Coronavirus with ACE (Angiotensin-Converting Enzyme)-2 as their main receptor: Therapeutic implications. Hypertension 2020, 76, 1339–1349. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2020, 19, 141–154. [Google Scholar]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv 2020. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. Mol. Immunol. 2020, 182, 812–827. [Google Scholar] [CrossRef]
- Mwenda, M.; Saasa, N.; Sinyange, N.; Busby, G.; Chipimo, P.J.; Hendry, J.; Kapona, O.; Yingst, S.; Hines, J.Z.; Minchella, P. Detection of B. 1.351 SARS-CoV-2 variant strain—Zambia, December 2020. MMWR 2021, 70, 280–282. [Google Scholar]
- Walensky, R.P.; Walke, H.T.; Fauci, A.S. SARS-CoV-2 variants of concern in the United States—Challenges and opportunities. JAMA 2021, 325, 1037–1038. [Google Scholar] [CrossRef]
- Available online: https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584 (accessed on 10 January 2021).
- Hirotsu, Y.; Omata, M. Discovery of SARS-CoV-2 strain of P. 1 lineage harboring K417T/E484K/N501Y by whole genome sequencing in the city, Japan. MedRxiv 2021. [Google Scholar] [CrossRef]
- Firestone, M.; Lorentz, A.; Meyer, S.; Wang, X.; Como-Sabetti, K.; Vetter, S.; Smith, K.; Holzbauer, S.; Beaudoin, A.; Garfin, J.; et al. First identified cases of SARS-CoV-2 variant P.1 in the United States—Minnesota, January 2021. MMWR 2021, 70, 278–279. [Google Scholar]
- Organization, W.H. SARS-CoV-2 Variant-United Kingdom of Great Britain and Northern Ireland. Disease Outbreak News, 2 May 2021. [Google Scholar]
- Ghaddar, M. Emergence and fast spread of B. 1.1. 7 lineage in Lebanon. MedRxiv 2021. [Google Scholar] [CrossRef]
- Washington, N.L.; Gangavarapu, K.; Zeller, M.; Bolze, A.; Cirulli, E.T.; Barrett, K.M.S.; Larsen, B.B.; Anderson, C.; White, S.; Cassens, T. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B. 1.1. 7 in the United States. MedRxiv 2021. [Google Scholar] [CrossRef]
- Umair, M.; Ikram, A.; Salman, M.; Alam, M.M.; Badar, N.; Rehman, Z.; Tamim, S.; Khurshid, A.; Ahad, A.; Ahmad, H. Importation of SARS-CoV-2 variant B. 1.1. 7 in Pakistan. J. Med. Virol. 2021, 93, 2623–2625. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, L.; Pullano, G.; Sabbatini, C.E.; Lévy-Bruhl, D.; Colizza, V. Impact of January 2021 social distancing measures on SARS-CoV-2 B. 1.1. 7 circulation in France. Euro. Surveill. 2021, 26, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Agwa, S.H.; Elghazaly, H.; El-Nakeep, S.; Moustafa, A.; El-Sayed, M.H.; Hafez, H.; Matboli, M.; Saad, M.; Elsayed, S.M.; Abd Elsamie, A.M. Laboratory based Retrospective Study to determine the start of SARS-CoV-2 in Patients with Severe Acute Respiratory Illness in Egypt at El-Demerdash tertiary hospitals. Res. Sq. 2020, 2, 1–17. [Google Scholar]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L. Emergence of SARS-CoV-2 B. 1.1. 7 lineage—United States, 29 December 2020–12 January 2021. MMWR 2021, 70, 95–99. [Google Scholar] [PubMed]
- Confirmed Cases of COVID-19 Variants Identified in UK. Available online: www.gov.uk (accessed on 28 April 2021).
- Expert Reaction to Cases of Variant B.1.617 (the Indian Variant) Being Investigated in the UK. Science Media Centre, 28 April 2021.
- Volz, E.; Mishra, S.; Chand, M.; Barrett, J.C.; Johnson, R.; Geidelberg, L.; Hinsley, W.R.; Laydon, D.J.; Dabrera, G.; O’Toole, Á. Transmission of SARS-CoV-2 Lineage, B. 1.1. 7 in England: Insights from linking epidemiological and genetic data. MedRxiv 2021. [Google Scholar] [CrossRef]
- Chand, M. Investigation of Novel SARS-COV-2 Variant: Variant of Concern 202012/01; Public Health England: London, UK, 2020.
- Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 9 December 2020).
- Yi, C.; Sun, X.; Ye, J.; Ding, L.; Liu, M.; Yang, Z.; Lu, X.; Zhang, Y.; Ma, L.; Gu, W. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020, 17, 621–630. [Google Scholar] [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef]
- Kadkhoda, K. Herd Immunity to COVID-19 Alluring and Elusive. Am. J. Clin. Pathol. 2021, 155, 471–472. [Google Scholar] [CrossRef]
- Hurlburt, N.K.; Seydoux, E.; Wan, Y.-H.; Edara, V.V.; Stuart, A.B.; Feng, J.; Suthar, M.S.; McGuire, A.T.; Stamatatos, L.; Pancera, M. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 2020, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Seydoux, E.; Homad, L.J.; MacCamy, A.J.; Parks, K.R.; Hurlburt, N.K.; Jennewein, M.F.; Akins, N.R.; Stuart, A.B.; Wan, Y.-H.; Feng, J. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 2020, 53, 98–105.e5. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Panjkovich, A.; Svergun, D.I. SASpy: A PyMOL plugin for manipulation and refinement of hybrid models against small angle X-ray scattering data. Bioinformatics 2016, 32, 2062–2064. [Google Scholar] [CrossRef] [PubMed]
- Vahed, M.; Calcagno, T.M.; Quinonez, E.; Mirsaeidi, M. Impacts of 203/204: RG> KR mutation in the N protein of SARS-CoV-2. BioRxiv 2021. [Google Scholar] [CrossRef]
- Vahed, M.; Sweeney, A.; Shirasawa, H.; Vahed, M. The initial stage of structural transformation of Aβ42 peptides from the human and mole rat in the presence of Fe2+ and Fe3+: Related to Alzheimer’s disease. Comput. Biol. Chem. 2019, 83, 107128. [Google Scholar] [CrossRef]
- Vahed, M.; Neya, S.; Matsuzaki, K.; Hoshino, T. Simulation study on complex conformations of Aβ42 peptides on a GM1 ganglioside-containing lipid membrane. Chem. Pharm. Bull. 2018, 66, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Vangone, A.; Spinelli, R.; Scarano, V.; Cavallo, L.; Oliva, R. COCOMAPS: A web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics 2011, 27, 2915–2916. [Google Scholar] [CrossRef]
- Vahed, M.; Ahmadian, G.; Ameri, N.; Vahed, M. G-rich VEGF aptamer as a potential inhibitor of chitin trafficking signal in emerging opportunistic yeast infection. Comput. Biol. Chem. 2019, 80, 168–176. [Google Scholar] [CrossRef]
- About Variants of the Virus That Causes COVID-19, February 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant.html (accessed on 2 May 2021).
- Available online: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-emerging-variants.html (accessed on 28 January 2021).
- Zhang, W.; Davis, B.D.; Chen, S.S.; Martinez, J.M.S.; Plummer, J.T.; Vail, E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA 2021, 325, 1324–1326. [Google Scholar] [CrossRef]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Subedi, S.; Olson, R.; Davis, J.J.; Saavedra, M.O.; Yerramilli, P.; Pruitt, L.; Reppond, K. Sequence analysis of 20,453 SARS-CoV-2 genomes from the Houston Metropolitan Area identifies the emergence and widespread distribution of multiple isolates of all major variants of concern. Am. J. Pathol 2021, accepted; in press. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.; Buzko, O.; Spilman, P.R.; Niazi, K.; Rabizadeh, S.; Soon-Shiong, P.R. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y. V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. BioRxiv 2021. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020, 9, e61312. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Lambson, B.E.; Vermeulen, M.; van den Berg, K.; Rossouw, T.; Boswell, M. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Xia, H.; Zhang, X.; Fontes-Garfias, C.R.; Swanson, K.A.; Cai, H.; Sarkar, R.; Chen, W.; Cutler, M. Neutralizing activity of BNT162b2-elicited serum—Preliminary report. N. Engl. J. Med. 2021, 384, 1466–1468. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.fda.gov/news-events/press-announcements/fda-issues-emergency-use-authorization-third-covid-19-vaccine (accessed on 27 February 2020).
- Koirala, A.; Joo, Y.J.; Khatami, A.; Chiu, C.; Britton, P.N. Vaccines for COVID-19: The current state of play. Paediatr. Respir. Rev. 2020, 35, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://investors.modernatx.com/news-releases/news-release-details/moderna-covid-19-vaccine-retains-neutralizing-activity-against (accessed on 25 January 2021).
- Available online: https://www.wits.ac.za/covid19/covid19-news/latest/oxford-covid-19-vaccine-trial-results.html (accessed on 7 February 2021).
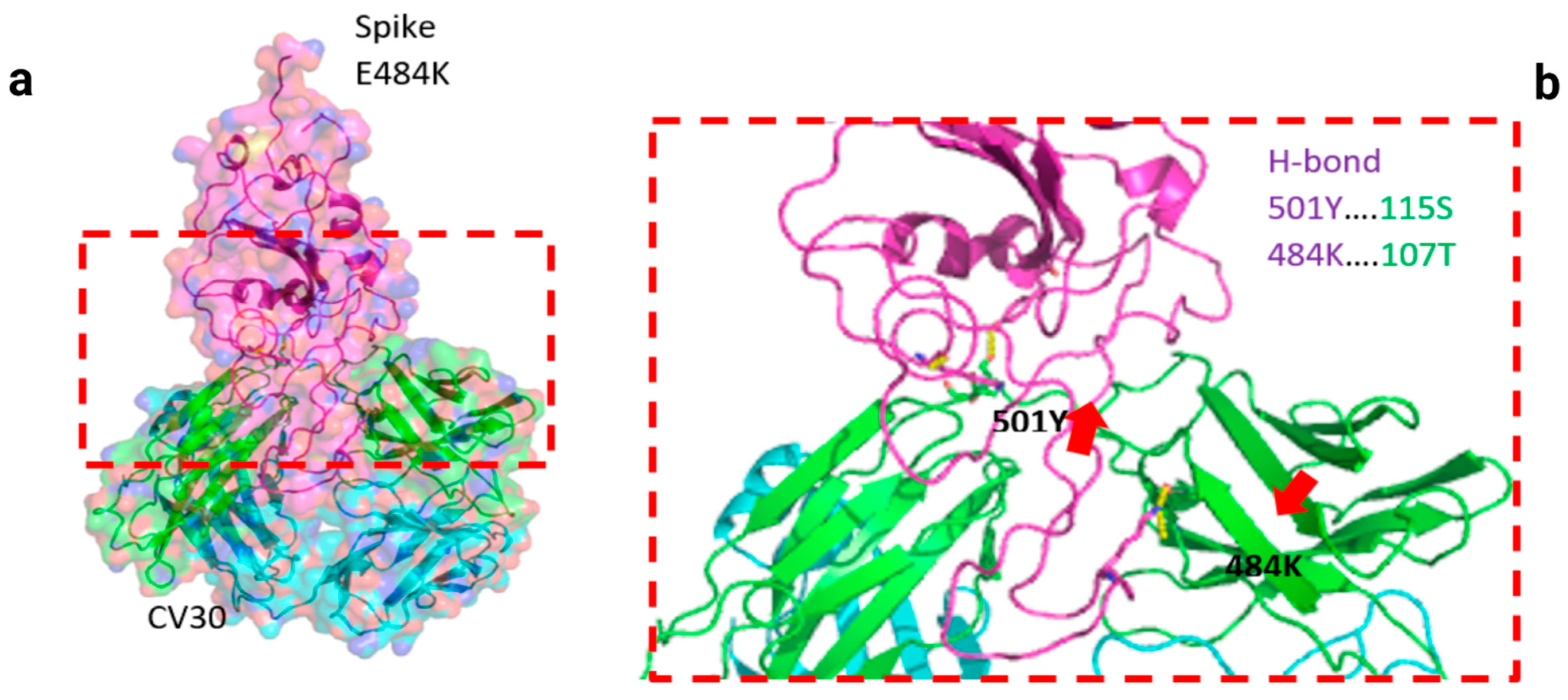
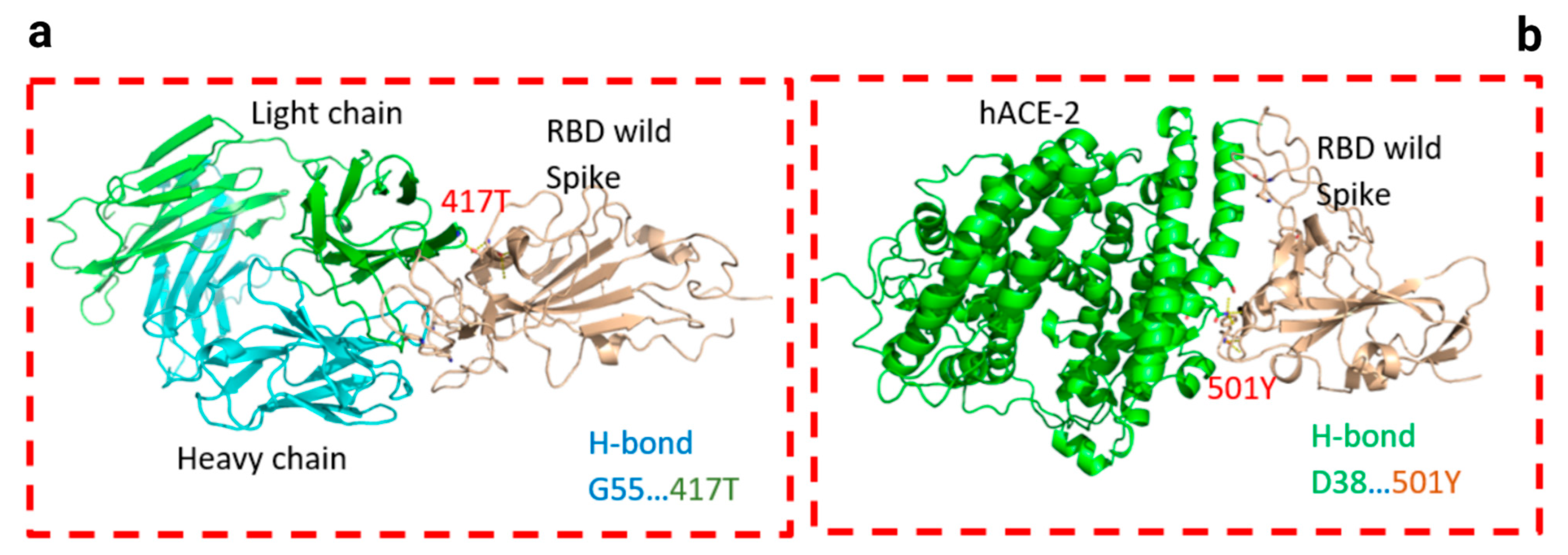
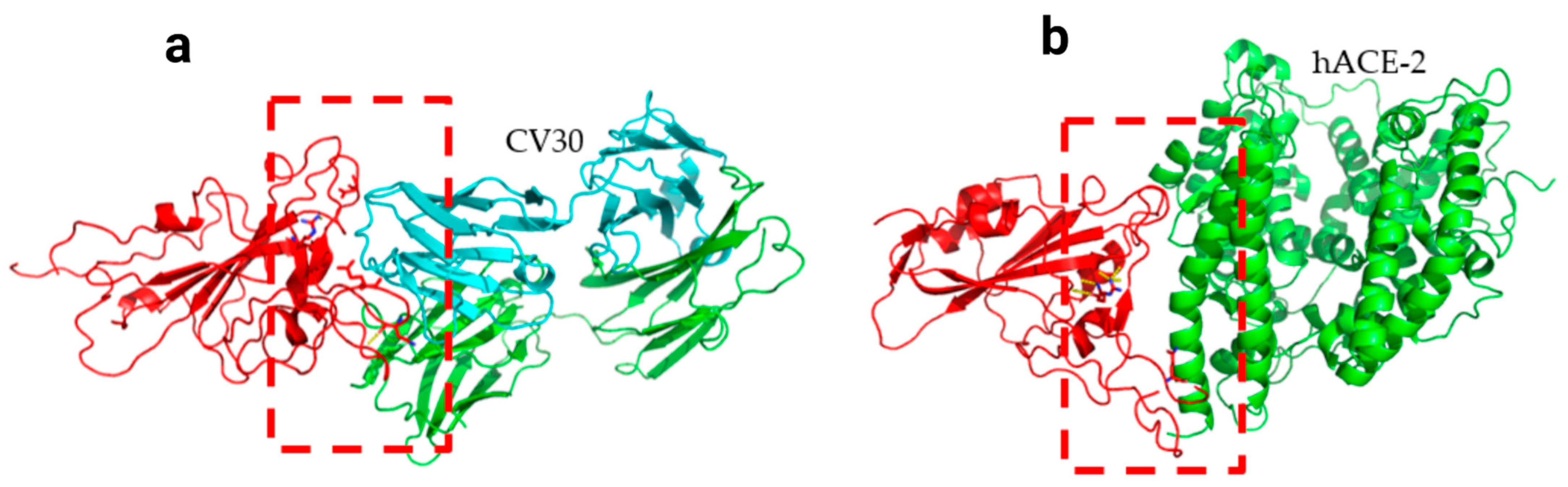
| CV30/Spike | hACE2/Spike | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank | * WT | B.1.1.7 | B.1.351 | P.1 | E484K | B.1.617 | * WT | B.1.1.7 | B.1.351 | E484K | B.1.617 |
| Docking Score | −371.61 | −337.93 | −260.34 | −270.72 | −263.21 | −269.54 | −310.19 | −353.32 | −266.76 | −323.78 | −341.31 |
| Ligand RMSD (Å2) | 2.32 | 142.44 | 324.79 | 385.41 | 74.78 | 384.23 | 3.27 | 0.79 | 93.91 | 0.56 | 0.36 |
| Score Affinity % | 100 | 90.93 | 70.05 | 72.85 | 70.82 | 72.53 | 100 | 113.9 | 85.99 | 104.38 | 110.03 |
| MOL1/MOL2 | WT/CV30 | B.1.1.7 /CV30 | B.1.351 /CV30 | 484/ CV30 | P.1/ CV30 | B.1.617/ CV30 |
|---|---|---|---|---|---|---|
| Buried area upon the complex formation (Å2) | 2010.8 | 2010.9 | 1952.8 | 1977.8 | 2061.7 | 1850.4 |
| Buried area upon the complex formation (%) | 6.51 | 6.47 | 5.69 | 8.19 | 6.68 | 6.01 |
| Interface area (Å2) | 1005.4 | 1005.45 | 976.4 | 988.9 | 1030.85 | 925.2 |
| Interface area MOL1 (%) | 8.69 | 8.69 | 6.52 | 7.74 | 8.91 | 8.09 |
| Interface area MOL2 (%) | 5.2 | 5.16 | 5.05 | 8.69 | 5.34 | 4.79 |
| POLAR Buried area upon the complex formation (Å2) | 1084 | 1084.1 | 956.2 | 1149.3 | 1042.4 | 930.5 |
| POLAR Interface (%) | 53.91 | 53.91 | 48.97 | 58.11 | 50.56 | 50.29 |
| POLAR Interface area (Å2) | 542 | 542.05 | 478.1 | 574.65 | 521.2 | 465.25 |
| NON POLAR Buried area upon the complex formation (Å2) | 927 | 927 | 996.6 | 828.5 | 1019.1 | 919.9 |
| NON POLAR Interface (%) | 46.1 | 46.1 | 51.03 | 41.89 | 49.43 | 49.71 |
| NON POLAR Interface area (Å2) | 463.5 | 463.5 | 498.3 | 414.25 | 509.55 | 459.95 |
| Residues at the interface_TOT (n) | 256 | 249 | 228 | 66 | 247 | 232 |
| Residues at the interface MOL1 | 32 | 32 | 30 | 33 | 28 | 27 |
| Residues at the interface MOL2 | 224 | 217 | 198 | 33 | 219 | 205 |
| Mol1/Mol2 | WT/ hACE2 | B.1.1.7 /hACE2 | B.1.351 /hACE2 | 484 /hACE2 | P.1/ hACE2 | B.1.617 /hACE2 |
|---|---|---|---|---|---|---|
| Buried area upon the complex formation (Å2) | 1902.5 | 1955.3 | 2568.1 | 1944.6 | 1894.6 | 1986.4 |
| Buried area upon the complex formation (%) | 5.46 | 5.63 | 6.48 | 5.6 | 5.45 | 5.77 |
| Interface area (Å2) | 951.25 | 977.65 | 1284.05 | 972.3 | 947.3 | 993.2 |
| Interface area MOL1 (%) | 9.54 | 9.78 | 8.57 | 9.75 | 9.52 | 9.93 |
| Interface area MOL2 (%) | 3.82 | 3.95 | 5.21 | 3.93 | 3.82 | 4.06 |
| POLAR Buried area upon the complex formation (Å2) | 1122.7 | 1103.8 | 1572.3 | 1109.7 | 1120 | 1111 |
| POLAR Interface (%) | 59.01 | 56.45 | 61.22 | 57.07 | 59.12 | 55.93 |
| POLAR Interface area (Å2) | 561.35 | 551.9 | 786.15 | 554.85 | 560 | 555.5 |
| NON POLAR Buried area upon the complex formation (Å2) | 779.9 | 851.6 | 995.8 | 835 | 774.6 | 875.5 |
| NON POLAR Interface (%) | 40.99 | 43.55 | 38.78 | 42.94 | 40.88 | 44.07 |
| NON POLAR Interface area (Å2) | 389.95 | 425.8 | 497.9 | 417.5 | 387.3 | 437.75 |
| Residues at the interface TOT (n) | 56 | 53 | 78 | 54 | 53 | 58 |
| Residues at the interface MOL1 | 27 | 27 | 35 | 26 | 26 | 29 |
| Residues at the interface MOL2 | 29 | 26 | 43 | 43 | 27 | 29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quinonez, E.; Vahed, M.; Hashemi Shahraki, A.; Mirsaeidi, M. Structural Analysis of the Novel Variants of SARS-CoV-2 and Forecasting in North America. Viruses 2021, 13, 930. https://doi.org/10.3390/v13050930
Quinonez E, Vahed M, Hashemi Shahraki A, Mirsaeidi M. Structural Analysis of the Novel Variants of SARS-CoV-2 and Forecasting in North America. Viruses. 2021; 13(5):930. https://doi.org/10.3390/v13050930
Chicago/Turabian StyleQuinonez, Elena, Majid Vahed, Abdolrazagh Hashemi Shahraki, and Mehdi Mirsaeidi. 2021. "Structural Analysis of the Novel Variants of SARS-CoV-2 and Forecasting in North America" Viruses 13, no. 5: 930. https://doi.org/10.3390/v13050930
APA StyleQuinonez, E., Vahed, M., Hashemi Shahraki, A., & Mirsaeidi, M. (2021). Structural Analysis of the Novel Variants of SARS-CoV-2 and Forecasting in North America. Viruses, 13(5), 930. https://doi.org/10.3390/v13050930








