The Picornavirus Precursor 3CD Has Different Conformational Dynamics Compared to 3Cpro and 3Dpol in Functionally Relevant Regions
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression and Isotopic Labeling
2.2. Protein Purification
2.3. NMR Sample Preparation
2.4. δ1-13. CH3 Resonance Assignments for 3C, 3D, and 3CD
2.5. Determination of Methyl Axis Order Parameters
2.6. CPMG Relaxation Dispersion
2.7. H and 13C CEST
3. Results
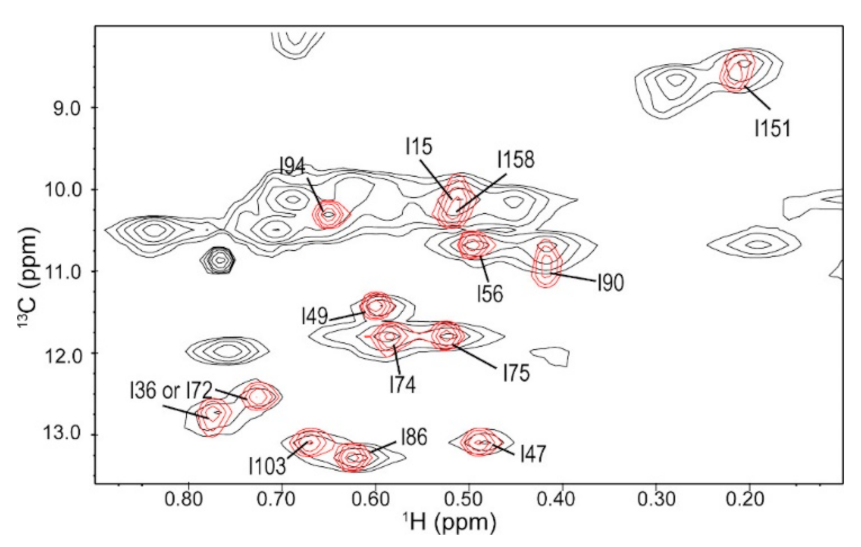
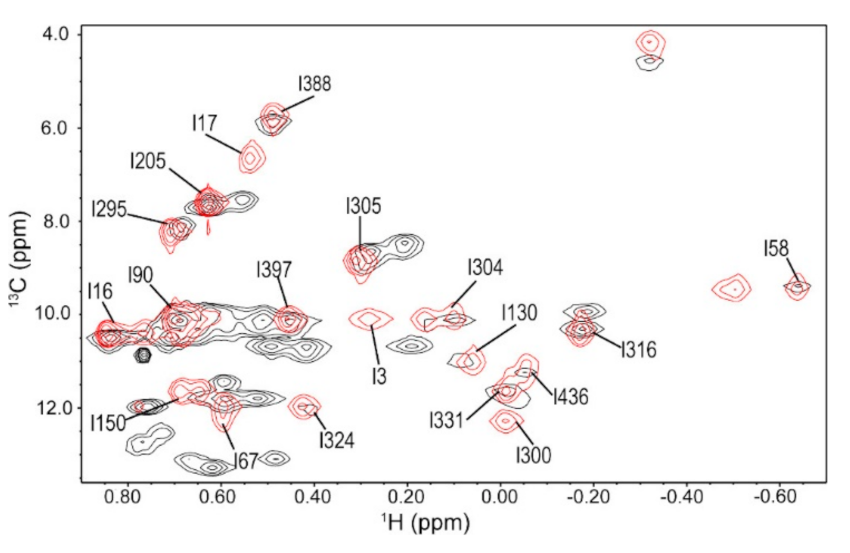
3.1. Ile Methyl Resonance Assignments for 3C
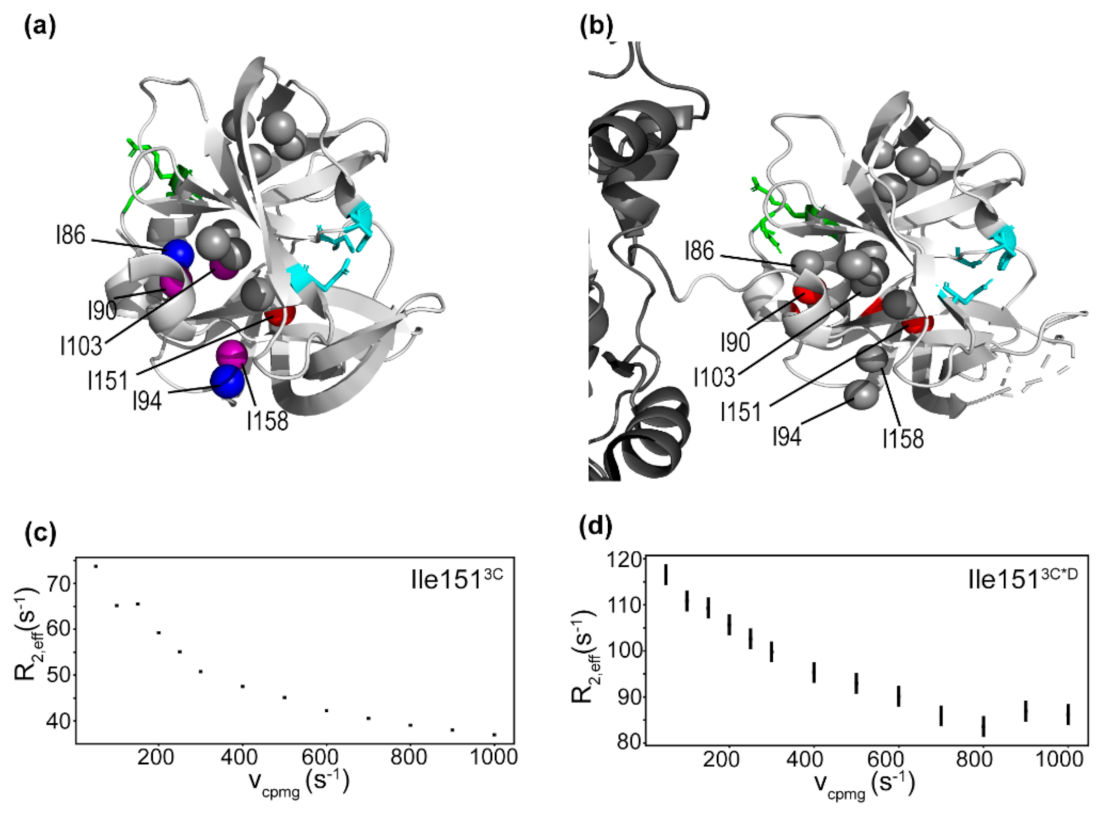
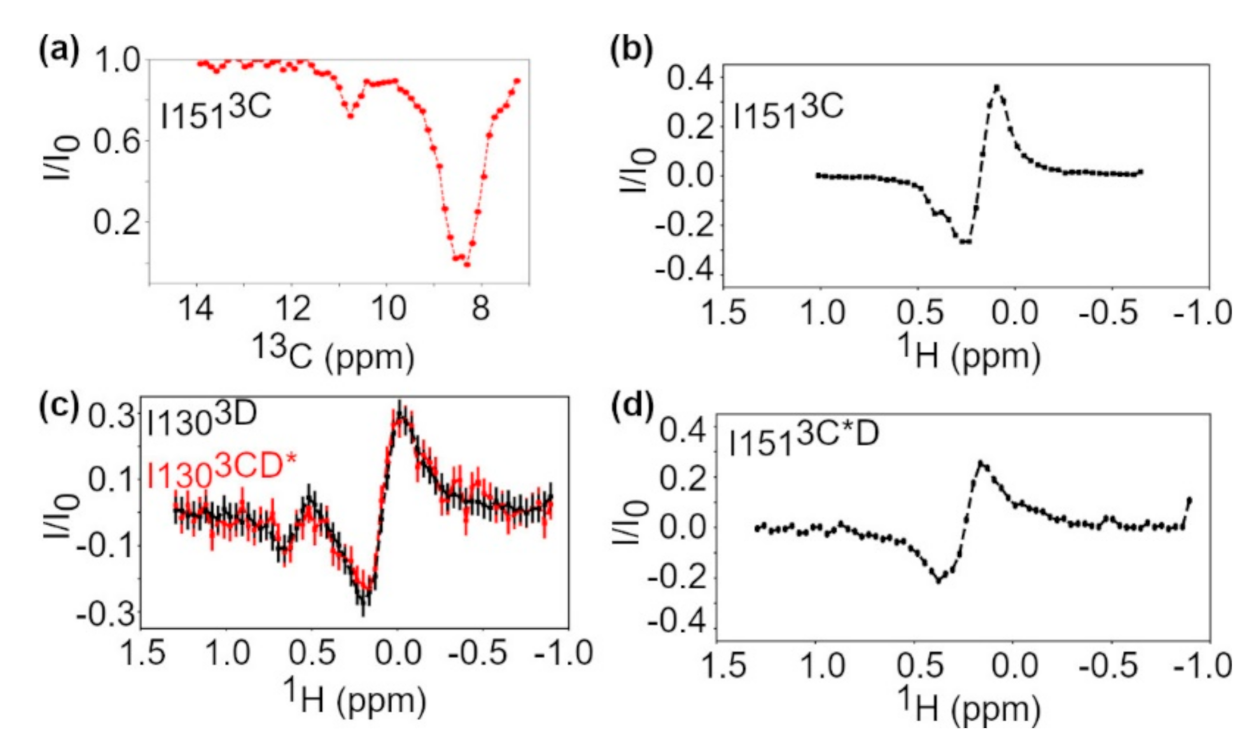
3.2. Conformational Dynamics of 3C on the Micro-to-Millisecond Timescale
3.3. Conformational Dynamics of 3C on the Millisecond-Second Timescale
3.4. Conformational Dynamics of 3C on the Picosecond-Nanosecond Timescale
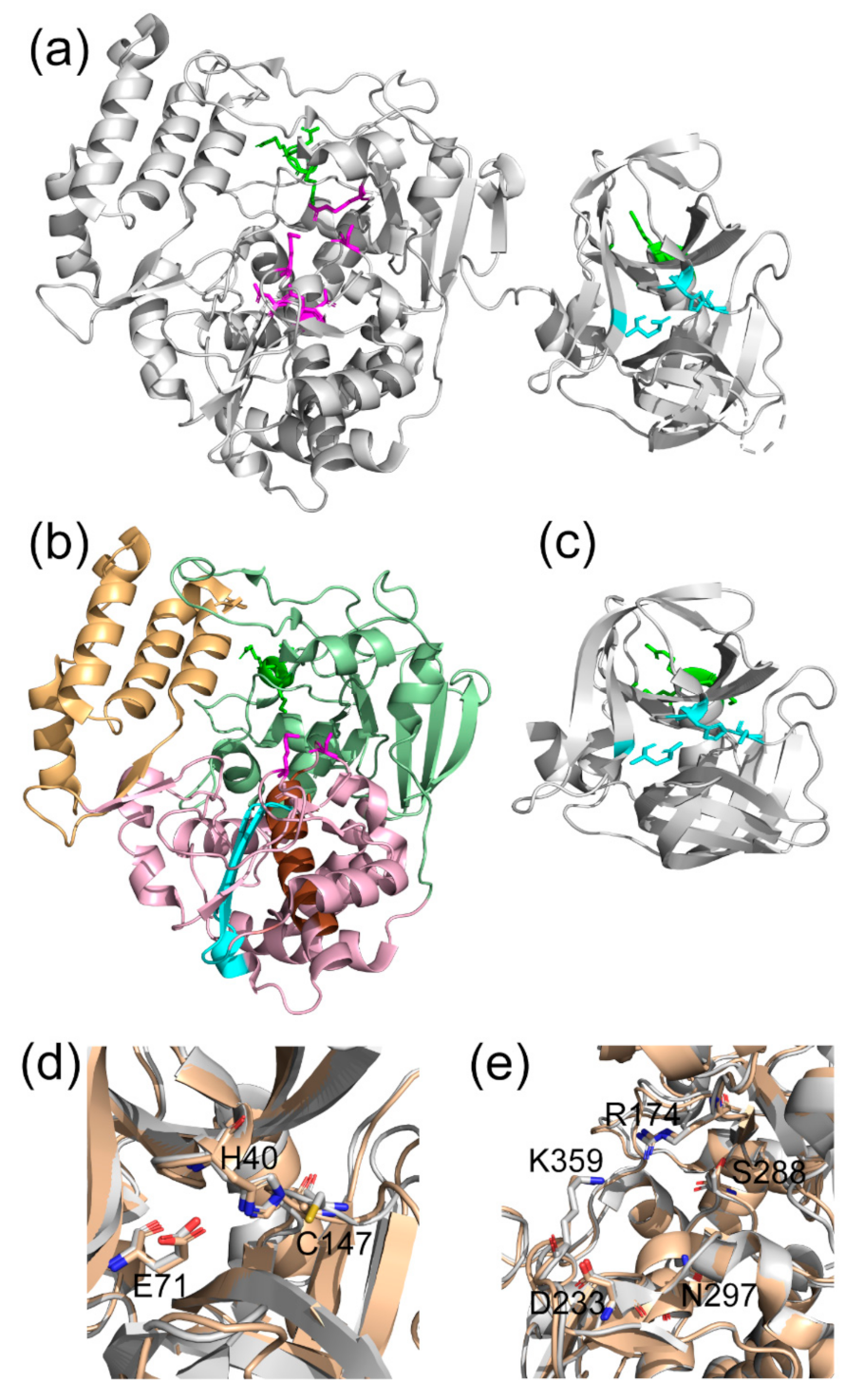
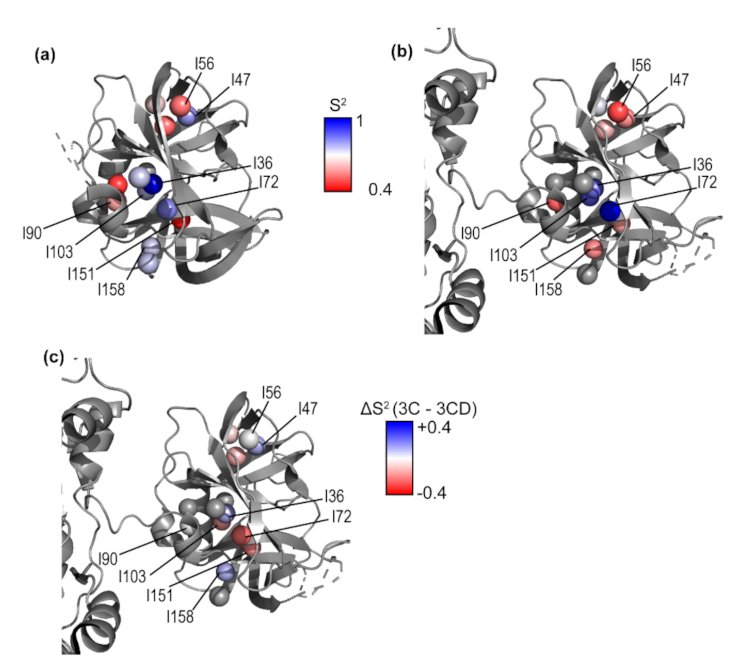
3.5. Conformational Dynamics of 3CD: 3C Domain
3.6. Ile Methyl Resonance Assignments for 3D
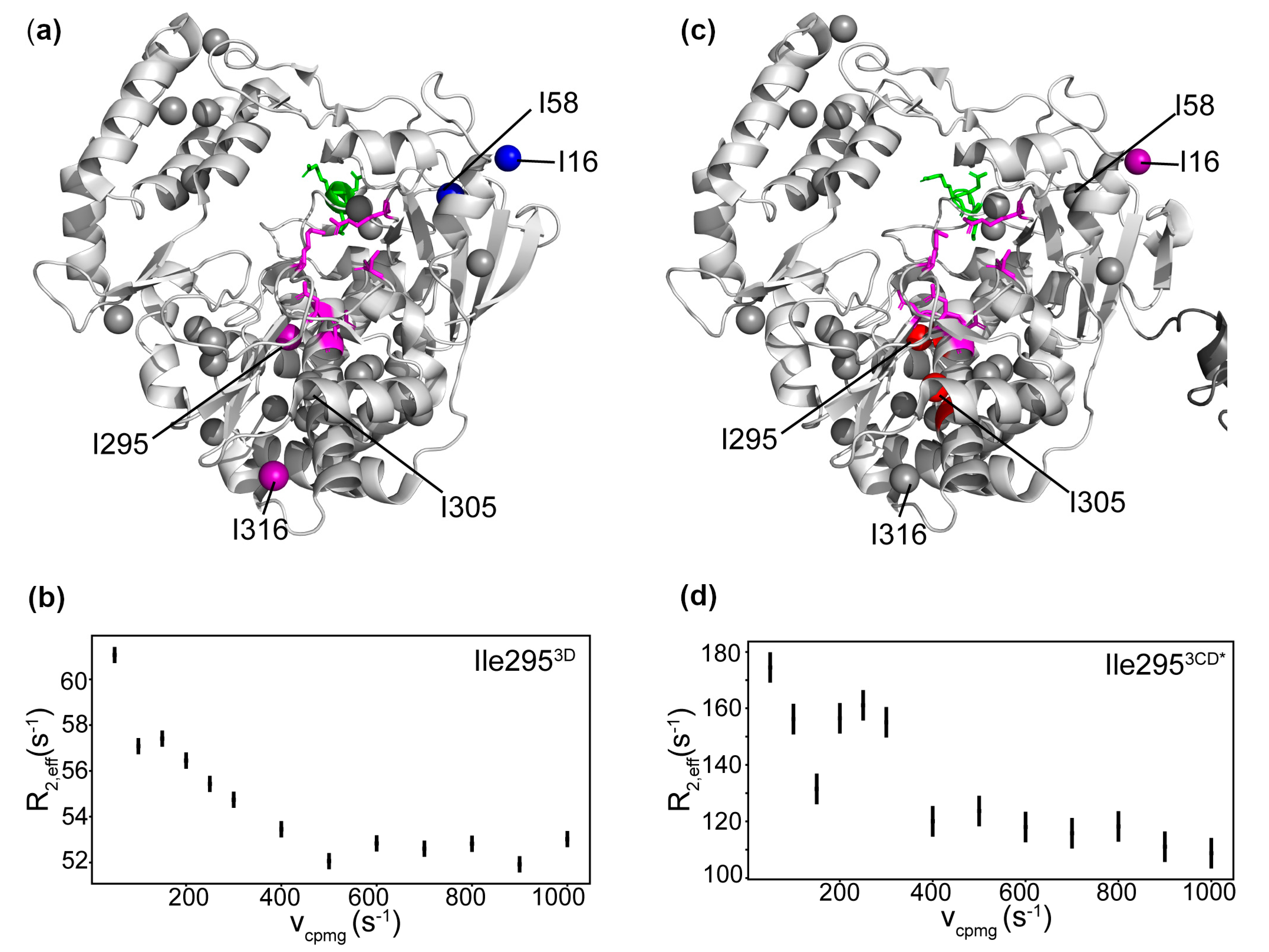
3.7. Conformational Dynamics of 3D on the Microsecond to Second Timescale
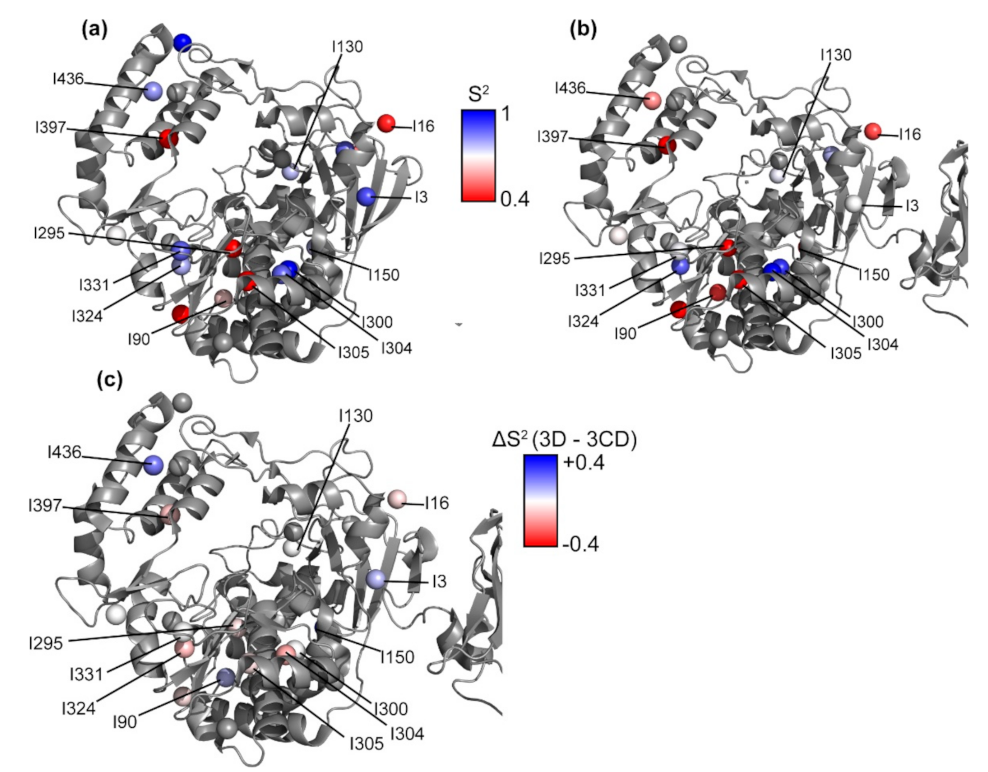
3.8. Conformational Dynamics of 3D on the Picosecond to Nanosecond Timescale
3.9. Conformational Dynamics of 3CD: 3D Domain
3.10. Domain Independence in 3CD
4. Discussion
4.1. Comparison of 3C and 3CD Conformational Dynamics
4.2. Comparison of 3D and 3CD Conformational Dynamics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spall, V.E.; Shanks, M.; Lomonossoff, G.P. Polyprotein Processing as a Strategy for Gene Expression in RNA Viruses. Semin. Virol. 1997, 8, 15–23. [Google Scholar] [CrossRef]
- Belshaw, R.; Pybus, O.G.; Rambaut, A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007, 17, 1496–1504. [Google Scholar] [CrossRef]
- Holmes, E.C. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 2003, 11, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Cheng Kao, C. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology 2013, 446, 123–132. [Google Scholar] [CrossRef]
- Cameron, C.E.; Oh, H.S.; Moustafa, I.M. Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future Microbiol. 2017, 5, 867–881. [Google Scholar] [CrossRef]
- Hellen, C.U.T.; Kraeusslich, H.G.; Wimmer, E. Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry 1989, 28, 9881–9890. [Google Scholar] [CrossRef]
- Emmott, E.; de Rougemont, A.; Hosmillo, M.; Lu, J.; Fitzmaurice, T.; Haas, J.; Goodfellow, I. Polyprotein processing and intermolecular interactions within the viral replication complex spatially and temporally control norovirus protease activity. J. Biol. Chem. 2019, 294, 4259–4271. [Google Scholar] [CrossRef]
- Marcotte, L.L.; Wass, A.B.; Gohara, D.W.; Pathak, H.B.; Arnold, J.J.; Filman, D.J.; Cameron, C.E.; Hogle, J.M. Crystal Structure of Poliovirus 3CD Protein: Virally Encoded Protease and Precursor to the RNA-Dependent RNA Polymerase. J. Virol. 2007, 81, 3583–3596. [Google Scholar] [CrossRef]
- Parsley, T.B.; Cornell, C.T.; Semler, B.L. Modulation of the RNA Binding and Protein Processing Activities of Poliovirus Polypeptide 3CD by the Viral RNA Polymerase Domain. J. Biol. Chem. 1999, 274, 12867–12876. [Google Scholar] [CrossRef] [PubMed]
- Ypma-Wong, M.F.; Dewalt, P.G.; Johnson, V.H.; Lamb, J.G.; Semler, B.L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the p1 capsid precursor. Virology 1988, 166, 265–270. [Google Scholar] [CrossRef]
- Belov, G.A.; Habbersett, C.; Franco, D.; Ehrenfeld, E. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J. Virol. 2007, 81, 9259–9267. [Google Scholar] [CrossRef]
- Banerjee, S.; Aponte-Diaz, D.; Yeager, C.; Sharma, S.D.; Ning, G.; Oh, H.S.; Han, Q.; Umeda, M.; Hara, Y.; Wang, R.Y.L.; et al. Hijacking of multiple phospholipid biosynthetic pathways and induction of membrane biogenesis by a picornaviral 3CD protein. PLoS Pathog. 2018, 14, e1007086. [Google Scholar] [CrossRef]
- Oh, H.S.; Banerjee, S.; Aponte-Diaz, D.; Sharma, S.D.; Aligo, J.; Lodeiro, M.F.; Ning, G.; Sharma, R.; Arnold, J.J.; Cameron, C.E. Multiple poliovirus-induced organelles suggested by comparison of spatiotemporal dynamics of membranous structures and phosphoinositides. PLoS Pathog. 2018, 14, e1007036. [Google Scholar] [CrossRef] [PubMed]
- Andino, R.; Rieckhof, G.E.; Achacoso, P.L.; Baltimore, D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993, 12, 3587–3598. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Interactions of Viral Protein 3CD and Poly(rC) Binding Protein with the 5′ Untranslated Region of the Poliovirus Genome. J. Virol. 2000, 74, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Spear, A.; Ogram, S.A.; Morasco, B.J.; Smerage, L.E.; Flanegan, J.B. Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex. Virology 2015, 485, 492–501. [Google Scholar] [CrossRef]
- Ypma-Wong, M.F.; Semler, B.L. Processing determinants required for in vitro cleavage of the poliovirus P1 precursor to capsid proteins. J. Virol. 1987, 61, 3181–3189. [Google Scholar] [CrossRef]
- Moustafa, I.M.; Gohara, D.W.; Uchida, A.; Yennawar, N.; Cameron, C.E. Conformational Ensemble of the Poliovirus 3CD Precursor Observed by MD Simulations and Confirmed by SAXS: A Strategy to Expand the Viral Proteome? Viruses 2015, 7, 5962–5986. [Google Scholar] [CrossRef]
- Mosimann, S.C.; Cherney, M.M.; Sia, S.; Plotch, S.; James, M.N.G. Refined X-ray crystallographic structure of the poliovirus 3C gene product11Edited By, D. Rees. J. Mol. Biol. 1997, 273, 1032–1047. [Google Scholar] [CrossRef] [PubMed]
- Amero, C.D.; Arnold, J.J.; Moustafa, I.M.M.; Cameron, C.E.E.; Foster, M.P. Identification of the oriI-Binding Site of Poliovirus 3C Protein by Nuclear Magnetic Resonance Spectroscopy. J. Virol. 2017, 82, 4363–4370. [Google Scholar] [CrossRef]
- Shengjuler, D.; Chan, Y.M.; Sun, S.; Moustafa, I.M.; Li, Z.-L.; Gohara, D.W.; Buck, M.; Cremer, P.S.; Boehr, D.D.; Cameron, C.E. The RNA-Binding Site of Poliovirus 3C Protein Doubles as a Phosphoinositide-Binding Domain. Structure 2017, 25, 1875–1886.e7. [Google Scholar] [CrossRef]
- Jore, J.; De Geus, B.; Jackson, R.J.; Pouwels, P.H.; Enger-Valk, B.E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J. Gen. Virol. 1988, 69, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Semler, B.L.; Johnson, V.H.; Dewalt, P.G.; Ypma-Wong, M.F. Site-specific mutagenesis of cDNA clones expressing a poliovirus proteinase. J. Cell. Biochem. 1987, 33, 39–51. [Google Scholar] [CrossRef]
- Černý, J.; Bolfíková, B.Č.; Valdés, J.J.; Grubhoffer, L.; Růžek, D. Evolution of Tertiary Structure of Viral RNA Dependent Polymerases. PLoS ONE 2014, 9, e96070. [Google Scholar] [CrossRef] [PubMed]
- Hobson, S.D.; Rosenblum, E.S.; Richards, O.C.; Richmond, K.; Kirkegaard, K.; Schultz, S.C. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001, 20, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, M.A.; Richards, O.C.; Amin, C.; Ehrenfeld, E. Enzymatic activity of poliovirus RNA polymerase synthesized in Escherichia coli from viral cDNA. Virology 1988, 164, 301–308. [Google Scholar] [CrossRef]
- Plotch, S.J.; Palant, O.; Gluzman, Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J. Virol. 1989, 63, 216–225. [Google Scholar] [CrossRef]
- Gohara, D.W.; Ha, C.S.; Kumar, S.; Ghosh, B.; Arnold, J.J.; Wisniewski, T.J.; Cameron, C.E. Production of “Authentic” Poliovirus RNA-Dependent RNA Polymerase (3Dpol) by Ubiquitin–Protease-Mediated Cleavage in Escherichia coli. Protein Expr. Purif. 1999, 17, 128–138. [Google Scholar] [CrossRef]
- Thompson, A.A.; Peersen, O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004, 23, 3462–3471. [Google Scholar] [CrossRef]
- Harris, K.S.; Reddigari, S.R.; Nicklin, M.J.; Hämmerle, T.; Wimmer, E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J. Virol. 1992, 66, 7481–7489. [Google Scholar] [CrossRef]
- Kim, T.W.; Brieba, L.G.; Ellenberger, T.; Kool, E.T. Functional Evidence for a Small and Rigid Active Site in a High Fidelity DNA Polymerase Probing T7 DNA polymerase with variably sized base pairs. J. Biol. Chem. 2006, 281, 2289–2295. [Google Scholar] [CrossRef]
- Belliot, G.; Sosnovtsev, S.V.; Chang, K.-O.; Babu, V.; Uche, U.; Arnold, J.J.; Cameron, C.E.; Green, K.Y. Norovirus Proteinase-Polymerase and Polymerase Are Both Active Forms of RNA-Dependent RNA Polymerase. J. Virol. 2005, 79, 2393–2403. [Google Scholar] [CrossRef]
- Verlinden, Y.; Cuconati, A.; Wimmer, E.; Rombau, B. The viral protein 3CD induces an equilibrium between the viral protein and RNA synthesis in a cell-free system for poliovirus replication. Arch. Virol. 2002, 147, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Blair, W.S.; Li, X.; Semler, B.L. 3CD Cleavage of the Poliovirus P1 Precursor: A Model for Complex Proteinase/Substrate Interactions. Regul. Gene Expr. Anim. Viruses 1993, 225–244. [Google Scholar] [CrossRef]
- Piana, S.; Carloni, P.; Parrinello, M. Role of Conformational Fluctuations in the Enzymatic Reaction of HIV-1 Protease. J. Mol. Biol. 2002, 319, 567–583. [Google Scholar] [CrossRef]
- Hedstrom, L.; Szilagyi, L.; Rutter, W.J. Converting trypsin to chymotrypsin: The role of surface loops. Science 1992, 255, 1249–1253. [Google Scholar] [CrossRef]
- Hedstrom, L.; Farr-Jones, S.; Kettner, C.A.; Rutter, W.J. Converting Trypsin to Chymotrypsin: Ground-State Binding Does Not Determine Substrate Specificity. Biochemistry 1994, 33, 8764–8769. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.W.; Agard, D.A. Enzyme specificity under dynamic control: A normal mode analysis of α-lytic protease 11Edited by P. E. Wright. J. Mol. Biol. 1999, 286, 267–278. [Google Scholar] [CrossRef]
- Ma, W.; Tang, C.; Lai, L. Specificity of Trypsin and Chymotrypsin: Loop-Motion-Controlled Dynamic Correlation as a Determinant. Biophys. J. 2005, 89, 1183–1193. [Google Scholar] [CrossRef]
- Moustafa, I.M.; Korboukh, V.K.; Arnold, J.J.; Smidansky, E.D.; Marcotte, L.L.; Gohara, D.W.; Yang, X.; Sánchez-Farrán, M.A.; Filman, D.; Maranas, J.K.; et al. Structural Dynamics as a Contributor to Error-prone Replication by an RNA-dependent RNA Polymerase. J. Biol. Chem. 2014, 289, 36229–36248. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.; Lee, C.A.; Moustafa, I.M.; Smidansky, E.D.; Lum, D.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Vaccine-derived Mutation in Motif D of Poliovirus RNA-dependent RNA Polymerase Lowers Nucleotide Incorporation Fidelity. J. Biol. Chem. 2013, 288, 32753–32765. [Google Scholar] [CrossRef]
- Shi, J.; Perryman, J.M.; Yang, X.; Liu, X.; Musser, D.M.; Boehr, A.K.; Moustafa, I.M.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Rational Control of Poliovirus RNA-Dependent RNA Polymerase Fidelity by Modulating Motif-D Loop Conformational Dynamics. Biochemistry 2019, 58, 3735–3743. [Google Scholar] [CrossRef]
- Ren, Z.; Ghose, R. Slow Conformational Dynamics in the Cystoviral RNA-Directed RNA Polymerase P2: Influence of Substrate Nucleotides and Template RNA. Biochemistry 2011, 50, 1875–1884. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alphonse, S.; Ghose, R. Methyl NMR spectroscopy: Measurement of dynamics in viral RNA-directed RNA polymerases. Methods 2018, 148, 100–114. [Google Scholar] [CrossRef]
- Damm, K.L.; Carlson, H.A. Exploring experimental sources of multiple protein conformations in structure-based drug design. J. Am. Chem. Soc. 2007, 129, 8225–8235. [Google Scholar] [CrossRef] [PubMed]
- Schames, J.R.; Henchman, R.H.; Siegel, J.S.; Sotriffer, C.A.; Ni, H.; McCammon, J.A. Discovery of a Novel Binding Trench in HIV Integrase. J. Med. Chem. 2004, 47, 1879–1881. [Google Scholar] [CrossRef]
- Cozzini, P.; Kellogg, G.E.; Spyrakis, F.; Abraham, D.J.; Costantino, G.; Emerson, A.; Fanelli, F.; Gohlke, H.; Kuhn, L.A.; Morris, G.M.; et al. Target flexibility: An emerging consideration in drug discovery and design. J. Med. Chem. 2008, 51, 6237–6255. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, G.; Dastidar, S.G.; Madhumalar, A.; Verma, C.S. Role of protein flexibility in the discovery of new drugs. Drug Dev. Res. 2011, 72, 26–35. [Google Scholar] [CrossRef]
- Antunes, D.A.; Devaurs, D.; Kavraki, L.E. Understanding the challenges of protein flexibility in drug design. Expert Opin. Drug Discov. 2015, 10, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Vallurupalli, P.; Sekhar, A.; Yuwen, T.; Kay, L.E. Probing conformational dynamics in biomolecules via chemical exchange saturation transfer: A primer. J. Biomol. NMR 2017, 67, 243–271. [Google Scholar] [CrossRef] [PubMed]
- Ishima, R. CPMG Relaxation Dispersion. In Protein Dynamics: Methods and Protocols; Livesay, D.R., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; pp. 29–49. ISBN 978-1-62703-658-0. [Google Scholar]
- Tugarinov, V.; Sprangers, R.; Kay, L.E. Probing Side-Chain Dynamics in the Proteasome by Relaxation Violated Coherence Transfer NMR Spectroscopy. J. Am. Chem. Soc. 2007, 129, 1743–1750. [Google Scholar] [CrossRef]
- Kovermann, M.; Rogne, P.; Wolf-Watz, M. Protein dynamics and function from solution state NMR spectroscopy. Q. Rev. Biophys. 2016, 49. [Google Scholar] [CrossRef]
- Mittermaier, A.K.; Kay, L.E. Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 2009, 34, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Caro, J.A.; Harpole, K.W.; Kasinath, V.; Lim, J.; Granja, J.; Valentine, K.G.; Sharp, K.A.; Wand, A.J. Entropy in molecular recognition by proteins. Proc. Natl. Acad. Sci. USA 2017, 114, 6563–6568. [Google Scholar] [CrossRef]
- Capdevila, D.A.; Braymer, J.J.; Edmonds, K.A.; Wu, H.; Giedroc, D.P. Entropy redistribution controls allostery in a metalloregulatory protein. Proc. Natl. Acad. Sci. USA 2017, 114, 4424–4429. [Google Scholar] [CrossRef]
- Kleckner, I.R.; Foster, M.P. An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta 2011, 1814, 942–968. [Google Scholar] [CrossRef] [PubMed]
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Azatian, S.B.; Kaur, N.; Latham, M.P. Increasing the buffering capacity of minimal media leads to higher protein yield. J. Biomol. NMR 2019, 73, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Welch, J.L.; Arnold, J.J.; Boehr, D.D. Long-Range Interaction Networks in the Function and Fidelity of Poliovirus RNA-Dependent RNA Polymerase Studied by Nuclear Magnetic Resonance. Biochemistry 2010, 49, 9361–9371. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Maciejewski, M.W.; Schuyler, A.D.; Gryk, M.R.; Moraru, I.I.; Romero, P.R.; Ulrich, E.L.; Eghbalnia, H.R.; Livny, M.; Delaglio, F.; Hoch, J.C. NMRbox: A Resource for Biomolecular NMR Computation. Biophys. J. 2017, 112, 1529–1534. [Google Scholar] [CrossRef]
- Tugarinov, V.; Kay, L.E. Ile, Leu, and Val Methyl Assignments of the 723-Residue Malate Synthase G Using a New Labeling Strategy and Novel NMR Methods. J. Am. Chem. Soc. 2003, 125, 13868–13878. [Google Scholar] [CrossRef]
- Sun, H.; Kay, L.E.; Tugarinov, V. An Optimized Relaxation-Based Coherence Transfer NMR Experiment for the Measurement of Side-Chain Order in Methyl-Protonated, Highly Deuterated Proteins. J. Phys. Chem. B 2011, 115, 14878–14884. [Google Scholar] [CrossRef]
- Hoffmann, F.; Xue, M.; Schäfer, L.V.; Mulder, F.A.A. Narrowing the gap between experimental and computational determination of methyl group dynamics in proteins. Phys. Chem. Chem. Phys. 2018, 20, 24577–24590. [Google Scholar] [CrossRef]
- Bernado, P.; Garcia de la Torre, J.; Pons, M. Interpretation of 15N NMR relaxation data of globular proteins using hydrodynamic calculations with HYDRONMR. J. Biomol. NMR 2002, 23, 139–150. [Google Scholar] [CrossRef]
- Tugarinov, V.; Kay, L.E. Relaxation rates of degenerate 1H transitions in methyl groups of proteins as reporters of side-chain dynamics. J. Am. Chem. Soc. 2006, 128, 7299–7308. [Google Scholar] [CrossRef] [PubMed]
- Tjandra, N.; Bax, A.; Feller, S.E.; Pastor, R.W. Rotational Diffusion Anisotropy of Human Ubiquitin from 15N NMR Relaxation. J. Am. Chem. Soc. 1995, 117, 12562–12566. [Google Scholar] [CrossRef]
- Woessner, D.E. Nuclear spin relaxation in ellipsoids undergoing rotational brownian motion. J. Chem. Phys. 1962, 37, 647–654. [Google Scholar] [CrossRef]
- Korzhnev, D.M.; Kloiber, K.; Kanelis, V.; Tugarinov, V.; Kay, L.E. Probing Slow Dynamics in High Molecular Weight Proteins by Methyl-TROSY NMR Spectroscopy: Application to a 723-Residue Enzyme. J. Am. Chem. Soc. 2004, 126, 3964–3973. [Google Scholar] [CrossRef] [PubMed]
- Kleckner, I.R.; Foster, M.P. GUARDD: User-friendly MATLAB software for rigorous analysis of CPMG RD NMR data. J. Biomol. NMR 2012, 52, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Yuwen, T.; Sekhar, A.; Kay, L.E. Separating Dipolar and Chemical Exchange Magnetization Transfer Processes in 1H-CEST. Angew. Chem. Int. Ed. 2017, 56, 6122–6125. [Google Scholar] [CrossRef]
- Bouvignies, G.; Kay, L.E. A 2D 13C-CEST experiment for studying slowly exchanging protein systems using methyl probes: An application to protein folding. J. Biomol. NMR 2012, 53, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Ollerenshaw, J.E.; Tugarinov, V.; Kay, L.E. Methyl TROSY: Explanation and experimental verification. Magn. Reson. Chem. 2003, 41, 843–852. [Google Scholar] [CrossRef]
- Farber, P.J.; Mittermaier, A. Relaxation dispersion NMR spectroscopy for the study of protein allostery. Biophys. Rev. 2015, 7, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.M. Functional Regulation and Protein Structural Dynamics of Poliovirus 3C Protease; The Pennsylvania State University: State College, PA, USA, 2015. [Google Scholar]
- Hass, M.A.S.; Hansen, D.F.; Christensen, H.E.M.; Led, J.J.; Kay, L.E. Characterization of conformational exchange of a histidine side chain: Protonation, rotamerization, and tautomerization of His61 in plastocyanin from Anabaena variabilis. J. Am. Chem. Soc. 2008. [Google Scholar] [CrossRef]
- Vallurupalli, P.; Bouvignies, G.; Kay, L.E. Studying “Invisible” Excited Protein States in Slow Exchange with a Major State Conformation. J. Am. Chem. Soc. 2012, 134, 8148–8161. [Google Scholar] [CrossRef]
- Lipari, G.; Szabo, A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982, 104, 4546–4559. [Google Scholar] [CrossRef]
- Frederick, K.K.; Sharp, K.A.; Warischalk, N.; Wand, A.J. Re-Evaluation of the Model-Free Analysis of Fast Internal Motion in Proteins Using NMR Relaxation. J. Phys. Chem. B 2008, 112, 12095–12103. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, D.; Hilty, C.; Wider, G.; Wüthrich, K. Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 2006, 178, 72–76. [Google Scholar] [CrossRef]
- Dewalt, P.G.; Semler, B.L. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J. Virol. 1987, 61, 2162–2170. [Google Scholar] [CrossRef]
- Shengjuler, D. Targeting Viral Proteins to Sites of Genome Replication: Discovery and Characterization of Viral Phosphoinositide-Binding Domains; Pennsylvania State University: Philadelphia, PA, USA, 2017. [Google Scholar]
- Peersen, O.B. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. 2017, 234, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.D.; Meier, K.; Ishima, R.; Gronenborn, A.M. NMR studies on domain diffusion and alignment in modular GB1 repeats. Biophys. J. 2010, 99, 2636–2646. [Google Scholar] [CrossRef]
- Jacobs, D.M.; Saxena, K.; Vogtherr, M.; Bernadó, P.; Pons, M.; Fiebig, K.M. Peptide Binding Induces Large Scale Changes in Inter-domain Mobility in Human Pin1. J. Biol. Chem. 2003, 278, 26174–26182. [Google Scholar] [CrossRef] [PubMed]
- Bernadó, P.; Fernandes, M.X.; Jacobs, D.M.; Fiebig, K.; García de la Torre, J.; Pons, M. Interpretation of NMR relaxation properties of Pin1, a two-domain protein, based on Brownian dynamic simulations. J. Biomol. NMR 2004, 29, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Halle, B. Self-association of lysozyme as seen by magnetic relaxation dispersion. J. Phys. Chem. B 2003, 107, 7914–7922. [Google Scholar] [CrossRef]
- Krushelnitsky, A. Intermolecular electrostatic interactions and Brownian tumbling in protein solutions. Phys. Chem. Chem. Phys. 2006, 8, 2117–2128. [Google Scholar] [CrossRef]
- Vugmeyster, L.; Raleigh, D.P.; Palmer, A.G.; Vugmeister, B.E. Beyond the decoupling approximation in the model free approach for the interpretation of NMR relaxation of macromolecules in solution. J. Am. Chem. Soc. 2003, 125, 8400–8404. [Google Scholar] [CrossRef]
- Chan, Y.M.; Moustafa, I.M.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Long-Range Communication between Different Functional Sites in the Picornaviral 3C Protein. Structure 2016, 24, 509–517. [Google Scholar] [CrossRef][Green Version]
- Gong, P.; Kortus, M.G.; Nix, J.C.; Davis, R.E.; Peersen, O.B. Structures of Coxsackievirus, Rhinovirus, and Poliovirus Polymerase Elongation Complexes Solved by Engineering RNA Mediated Crystal Contacts. PLoS ONE 2013, 8, e60272. [Google Scholar] [CrossRef] [PubMed]
- Kortus, M.G.; Kempf, B.J.; Haworth, K.G.; Barton, D.J.; Peersen, O.B. A Template RNA Entry Channel in the Fingers Domain of the Poliovirus Polymerase. J. Mol. Biol. 2012, 417, 263–278. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winston, D.S.; Boehr, D.D. The Picornavirus Precursor 3CD Has Different Conformational Dynamics Compared to 3Cpro and 3Dpol in Functionally Relevant Regions. Viruses 2021, 13, 442. https://doi.org/10.3390/v13030442
Winston DS, Boehr DD. The Picornavirus Precursor 3CD Has Different Conformational Dynamics Compared to 3Cpro and 3Dpol in Functionally Relevant Regions. Viruses. 2021; 13(3):442. https://doi.org/10.3390/v13030442
Chicago/Turabian StyleWinston, Dennis S., and David D. Boehr. 2021. "The Picornavirus Precursor 3CD Has Different Conformational Dynamics Compared to 3Cpro and 3Dpol in Functionally Relevant Regions" Viruses 13, no. 3: 442. https://doi.org/10.3390/v13030442
APA StyleWinston, D. S., & Boehr, D. D. (2021). The Picornavirus Precursor 3CD Has Different Conformational Dynamics Compared to 3Cpro and 3Dpol in Functionally Relevant Regions. Viruses, 13(3), 442. https://doi.org/10.3390/v13030442






