Abstract
Studies of conditionally lethal mutants can help delineate the structure-function relationships of biomolecules. Temperature-sensitive (ts) mammalian reovirus (MRV) mutants were isolated and characterized many years ago. Two of the most well-defined MRV ts mutants are tsC447, which contains mutations in the S2 gene encoding viral core protein σ2, and tsG453, which contains mutations in the S4 gene encoding major outer-capsid protein σ3. Because many MRV ts mutants, including both tsC447 and tsG453, encode multiple amino acid substitutions, the specific amino acid substitutions responsible for the ts phenotype are unknown. We used reverse genetics to recover recombinant reoviruses containing the single amino acid polymorphisms present in ts mutants tsC447 and tsG453 and assessed the recombinant viruses for temperature-sensitivity by efficiency-of-plating assays. Of the three amino acid substitutions in the tsG453 S4 gene, Asn16-Lys was solely responsible for the tsG453 ts phenotype. Additionally, the mutant tsC447 Ala188-Val mutation did not induce a temperature-sensitive phenotype. This study is the first to employ reverse genetics to identify the dominant amino acid substitutions responsible for the tsC447 and tsG453 mutations and relate these substitutions to respective phenotypes. Further studies of other MRV ts mutants are warranted to define the sequence polymorphisms responsible for temperature sensitivity.
1. Introduction
Mammalian reoviruses (MRVs) are nonenveloped, double-stranded RNA viruses that serve as prototypes of the family Reoviridae [1,2]. The MRV genome consists of 10 segments, three large (L1, L2, L3), three medium (M1, M2, M3), and four small (S1, S2, S3, S4) [3]. Other members of this family include rotavirus, which causes viral gastroenteritis in children [4,5,6] and animals, and orbiviruses, which include pathogens of cattle [7,8]. Three main MRV serotypes have been categorized by antibody-mediated neutralization and hemagglutination inhibition and are represented by the prototype strains type 1 Lang (T1L), type 2 Jones (T2J), and type 3 Dearing (T3D). MRV serotypes also are differentiated on the basis of host cell tropism, mechanisms of cell killing, modes of dissemination, and CNS disease [3]. Some MRV strains are reported to possess oncolytic properties against various cancers [9], which is largely mediated by activation of Ras signaling pathways [9,10,11]. T3D has undergone numerous clinical trials and possesses marked oncolytic effect against multiple tumors [12].
Conditionally lethal viral mutants have served as useful tools to study various stages of viral replication and assembly [13,14,15,16,17,18,19]. One of the most notable examples is the use of such mutants to elucidate bacteriophage T4 assembly [20]. These mutants have also been used in the design of attenuated viruses for vaccines [21,22]. Several groups generated sets of conditionally lethal temperature-sensitive (ts) MRV mutants in the 1960s and 1970s [23,24]. One of the more extensively studied panels of MRV ts mutants was isolated after chemical mutagenesis of wild type T3D by Dr. Bernard Fields and was characterized by his colleagues [24,25,26,27,28,29,30]. These mutants were identified by their capacity to form well-defined plaques at a lower “permissive” incubation temperature (generally 30–31 °C). They were also identified by limited replication defined by fewer plaques at a higher “non-permissive/restrictive” temperature (generally 39 °C), whereas wild type T3D forms comparable numbers of plaques at both temperatures. The difference in plaque forming capacity at the restrictive versus permissive temperature is known as efficiency of plating (EOP), and this value is usually <0.03 for many ts mutants [19]. To identify the genes responsible for temperature-sensitivity, mutant isolates were crossed with wild-type T1L, and progeny reassortant viruses were screened for EOP [28,29,31,32,33]. Genomic sequencing of some mutants identified specific amino acid residues altered in the ts strains [34,35,36,37]. For many of these mutants, the responsible genome segment encodes multiple amino acid substitutions. Therefore, the specific amino acids responsible for the ts phenotype are often undefined. For example, the tsC447 mutant, which fails to assemble core particles at the restrictive temperature, contains three mutations in the S2 gene that encodes major core scaffold protein σ2; Ala188→Val, Ala323→Val, and Asn383→Asp [35]. Passaging studies of this mutant, conducted at the restrictive temperature, led to the rescue of a small number of revertant viruses. Sequence determinations of these revertant S2 genes suggest that the Asn383→Asp alteration in tsC447 is responsible for the ts phenotype, because reversion at this site allowed wild-type levels of replication at the restrictive temperature [38]. Similarly, the tsG453 mutant, which only forms core-like particles at the non-permissive temperature [36], contains three mutations in the S4 gene that encodes major outer-capsid protein σ3; Asn16→Lys, Met141→Ile, and Glu229→Asp [36,37]. Mutant tsG453 σ3 does not associate with major outer-capsid protein µ1 [37], supporting a model in which inner core particles are incapable of obtaining an outer capsid and do not mature to virions at restrictive temperatures.
Kobayashi et al. [39] developed a reverse genetics system to introduce changes into specific MRV genome segments. Altered viruses are recovered following transfection of susceptible cells with 4 to 10 plasmids that encode cDNA for all 10 MRV RNA genome segments under transcriptional control of the bacteriophage T7 RNA polymerase promoter and are fused at the 3′ terminus with hepatitis delta virus ribozyme sequences [40]. We used this reverse genetics system to construct virus clones containing each of the individual tsC447 and tsG453 mutations to identify the specific amino acid residues responsible for the phenotype of each mutant. We recovered viral clones containing each of the individual tsG453 mutations, and EOP values of each indicated that the Asn16→Lys substitution was solely responsible for the tsG453 ts phenotype. We were unable to recover all of the tsC447 mutations individually, but the isolates examined suggest that Asn383→Asp is responsible for the tsC447 ts phenotype, in agreement with prior reversion analyses [38].
2. Materials and Methods
2.1. Cells
Murine L929 fibroblasts were maintained in Joklik-modified Eagle’s minimal essential medium (J-MEM) supplemented to contain 5% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U penicillin G/mL, 100 µg streptomycin/mL, and 0.25 µg amphotericin B/mL (Gibco/Life Technologies, Grand Island, New York). Baby hamster kidney cells engineered to stably express T7 RNA polymerase (BHK-T7) cells were maintained in Dulbecco’s modified Eagle medium (Gibco) supplemented to contain 5% FBS, 2 mM l-glutamine, 2% MEM non-essential amino acid solution (Gibco), and 1 mg/mL geneticin (Invitrogen, Waltham, MA, USA).
2.2. Plasmids
Seven plasmids were used that contained cDNAs corresponding to the 10 reovirus T3D gene segments cloned into pT7-cDNA, as described [41]. E. coli DH5α competent cells were transformed with T3D L1/M2, T3D L2/M3, T3D L3/S3, T3D M1, T3D S1, T3D S2, or T3D S4 plasmids. E. coli were amplified on Luria agar or in Luria broth supplemented with ampicillin, and plasmids were isolated using plasmid purification kits (Qiagen, Hilden, Germany).
2.3. Primer Design for ts Mutants
The QuikChange Primer Design tool (https://www.agilent.com/store/primerDesignProgram.jsp) was used to design primer sets to introduce each mutation in the ts S2 gene of tsC447 and the ts S4 gene of tsG453 into the wild-type T3D S2 and S4 gene plasmids, respectively. Corresponding primer sequences (listed 5′ to 3′) are:
- T3D S2 A188V F-CAATGTGTATGCAATCTCTACAAACGTGTGCCCGAAATA
- T3D S2 A188V R-TATTTCGGGCACACGTTTGTAGAGATTGCATACACATTG
- T3D S2 A323V F-CATGCAATTGGTTACCAACTCTACCAGTCCAGCCA
- T3D S2 A323V R-TGGCTGGACTGGTAGAGTTGGTAACCAATTGCATG
- T3D S2 N383D F-GGATGAGCCTGACTATATTGATCGTCTTCTCTCGCC
- T3D S2 N383D R-GGCGAGAGAAGACGATCAATATAGTCAGGCTCATCC
- T3D S2 N383S F-GGATGAGCCTGACTCTATTGATCGTCTTCTCTCGCC
- T3D S2 N383S R-GGCGAGAGAAGACGATCAATATAGTCAGGCTCATCC
- T3D S4 N16K F-CCTTCAAAAGCGTTCTTAATCAAGTCCACGACCTGAT
- T3D S4 N16K R-ATCAGGTCGTGGACTTGATTAAGAACGCTTTTGAAGG
- T3D S4 M141I F-CAACTTGAGTGTATTGATCTAAATATTGAATTTGGGTCAACCTGAAG
- T3D S4 M141I R-CTTCAGGTTGACCCAAATTCAATATTTAGATCAATACACTCAAGTTG
- T3D S4 E229D F-CCCTTCGATGGATCATGATCCAGCTCAGAGTAATC
- T3D S4 E229D R-GATTACTCTGAGCTGGATCATGATCCATCGAAGGG
2.4. Reovirus Plasmid Mutagenesis
PCR-based site-directed mutagenesis was used to introduce single or multiple mutations into the plasmids encoding wild-type genes. All PCR product sizes were confirmed by agarose gel electrophoresis. PCR products were digested with Dpn1 to remove template DNA and transformed into DH5α cells. Mutagenized and wild-type plasmid DNA was purified from DH5α and sequence-confirmed using Sanger sequencing and Serial Cloner 2.6.1 software.
2.5. Reovirus Reverse Genetics
BHK-T7 cells were passaged overnight in Dulbecco-modified Eagle medium (DMEM) medium supplemented to contain geneticin to maintain efficient expression of the T7 RNA polymerase, which mediates transcription of the reovirus cDNAs. Prior to transfection, cells were supplemented with geneticin-free DMEM medium and maintained at 37 °C. Transfection mixtures consisted of OptiMEM, TransIT-LTI, and 2.53 µg of each of the seven reverse genetics plasmids. Transfection mixtures were incubated at room temperature for 30 min and added dropwise to cells. Cells were incubated at 34.5 °C to allow rescue of the ts mutant virus. Positive control transfections were included that contained wild-type T3D S2 for tsC447 and wild-type T3D S4 for tsG453. Cells were observed for a maximum of 5 days for visible cytopathic effects. Cell lysates were prepared using two freeze-thaw cycles and stored at 4 °C for no more than a few days, or −80 °C for longer intervals. Fluorescent focus unit assays (FFUs) were conducted by inoculating L929 cells with lysates from transformed cells, followed by staining with polyclonal rabbit anti-reovirus antiserum (prepared by inoculating New Zealand white rabbits with reovirus strains T1L or T3D). Sera from T1L- and T3D-inoculated rabbits were mixed 1:1 (vol/vol), and nonspecific antibodies were depleted using cross-adsorption on methanol-fixed L929 cells. A fluorophore-conjugated, goat anti-rabbit secondary antibody was used to visualize reovirus infection.
2.6. Sanger Sequencing for Rescued Virus
L929 cells were inoculated with transformed cell lysates, and the resultant virus was plaque-purified and passaged in L929 cells at 34.5 °C (permissive temperature) to recover passage 1 (P1) and passage 2 (P2) viral stocks. Total RNA was purified from P2 cell lysates by phenol:chloroform extraction, and reovirus S2 or S4 RNA was converted to cDNA using primers specific for the termini of the S2 or S4 gene segments and the Qiagen OneStep RT-PCR kit according to the manufacturer’s instructions (Qiagen OneStep RT-PCR Handbook, 10/2012). Amplification primers were:
- T3D S2 F-GCTATTCGCTGGTCAGTTAT
- T3D S2 R-ATGAATGTGTGGTCAGTCGT
- T3D S4 F-CGTTGTCGCAATGGAGGTGTGCTTGC
- T3D S4 R-AGCCTGTCCCACGTCACACC
Thermal cycler conditions were maintained as follows: reverse transcription at 50 °C for 30 min; initial PCR activation step at 95 °C for 15 min; and 34 cycles of 3-step cycling that included denaturation at 94 °C for 60 s, annealing at 56 °C for 60 s, extension at 72 °C for 150 s, and final extension at 72 °C for 60 s. The sequence of amplified cDNA was determined by the Sanger method using amplification primers (listed above). Sequence data were analyzed using Sequence Scanner (Applied Biosystems, Foster City, California) and Serial Cloner.
2.7. Reovirus Efficiency of Plating (EOP) Assays
Reovirus EOP assays were conducted as described [19]. Briefly, sets of confluent monolayers of L929 cells in 6-well plates were inoculated with 10-fold serial dilutions of each P2 virus stock. After adsorption, inoculated cells were overlaid with a 1:1 mixture of 2% Difco-Bacto agar and completed plaque assay medium (2× Medium-199 supplemented to contain 6% FBS, 4mM l-glutamine, 200 U penicillin G/mL, 200 µg streptomycin/mL, and 0.5 µg amphotericin B/mL). Plates were incubated at 39 or 34.5 °C for a total of 5 or 8 days, respectively. Plates incubated at 34.5 °C were supplemented with additional agar:medium mixture on day 3 post-infection. At 5 or 8 days post-inoculation, monolayers were fixed with 2.5% formalin in PBS, and agar plugs were removed. Monolayers were re-fixed and stained with crystal violet. Plaques were photographed, and viral titers at different temperatures were compared to determine EOP.
2.8. 3-Dimensional Protein Analyses
The coordinates of the asymmetric unit of the reovirus core (PDB 1EJ6) and the reovirus μ1/σ3 heterohexamer (PDB 1JMU) were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) PDB protein databank (http://www.rcsb.org (accessed on 27 December 2020)) and exported to the UCSF Chimera program (version 1.13.1) to visualize reovirus protein structures. Relevant amino acids were identified and colored within the program.
3. Results
3.1. Reverse Genetics Rescue of Infectious Reovirus Clones Containing Individual and Sets of tsC447 and tsG453 Temperature-Sensitive Alterations
Plasmids containing each individual tsC447 S2 or tsG453 S4 mutation, or plasmids containing all three mutations of each ts mutant, were combined with plasmids containing the other nine reovirus genes and transfected into BHK-T7 cells. Rescue cell lysates were inoculated onto indicator cells and infection was monitored by fluorescent focus unit assay to confirm recovery of infectious reovirus (Figure 1). We recovered isolates containing each of the individual tsG453 polymorphisms (Asn16→Lys, Met141→Ile, and Glu229→Asp) and an isolate that contained all three parental tsG453 polymorphisms. We also recovered the S2 Ala188→Val single-mutant isolate. However, despite numerous attempts, we were unable to recover a plaque-forming virus containing either of the individual tsC447 Ala323-Val and Asn383-Asp polymorphisms.
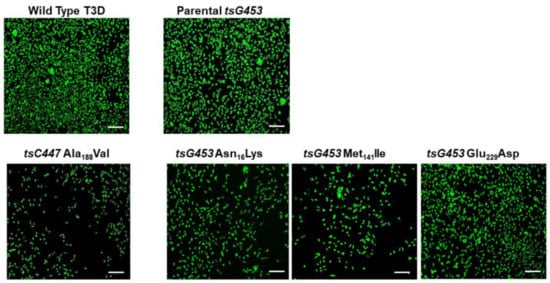
Figure 1.
Confirmation of recovery of wild-type T3D and five mutant isolates. Representative images of a fluorescent focus unit assay are shown. L929 cells were adsorbed with BHK-T7 cell lysates from reverse genetics experiments involving the indicated mutants. At 20 hpi, cells were fixed and stained with a rabbit polyclonal anti-reovirus antiserum and a fluorophore-conjugated, goat anti-rabbit secondary antibody to visualize reovirus infection. Scale bars are 200 µm.
Viral RNA was extracted from P2 stocks of each isolate, and sequences of purified cDNA were determined using the Sanger technique to confirm the presence of each mutation and the absence of any additional mutations. Sequencing confirmed the tsC447 Ala188-Val mutation and the tsG453 Asn16→Lys, Met141→Ile, and Glu229→Asp mutations in the expected isolates (Figure 2). Moreover, no additional mutations were observed in the altered genes of interest for these isolates.

Figure 2.
Sanger sequence confirmation of parental tsG453 and isolates containing each individual mutation. The top line depicts the wild-type T3D S4 nucleotide sequence and relevant σ3 amino acid residue. The second line depicts the corresponding sequence in the tsG453 mutant [36,37]. The bottom sequence line depicts the corresponding sequence in the indicated rescued isolate. Data for the T3D S2 gene (tsC447) are not shown because only a single recombinant isolate sequence was recovered.
3.2. Efficiency of Plating Values of Various Rescued Clones
We tested the temperature sensitivity of each rescued isolate to define the amino acid substitution(s) responsible for the respective mutant phenotypes. Because reovirus ts mutants replicate comparably at both 31 and 34.5 °C [19], we used 34.5 °C as the representative permissive temperature [19] and 39 °C as the non-permissive temperature. Therefore, we determined the capacity of each isolate to form plaques at each temperature and calculated the resulting EOP values. All isolates produced large, well-circumscribed plaques at 34.5 °C (Figure 3).
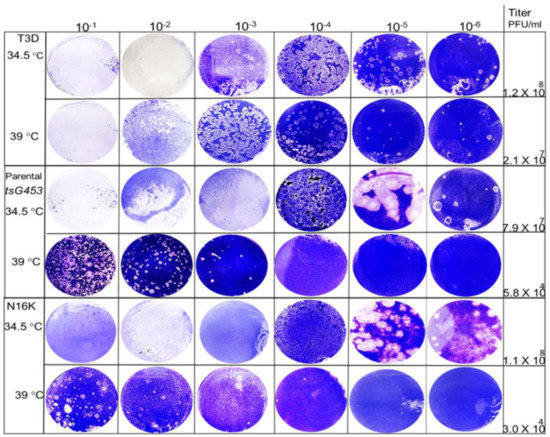
Figure 3.
Viral plaques produced by various rescued isolates at 34.5 and 39 °C. Ten-fold serial dilutions of P2 stocks for each isolate were adsorbed to mouse L929 monolayers and incubated for 5 days (39 °C) or 8 days (34.5 °C). Cell monolayers were fixed and stained with crystal violet.
At higher temperatures, plaques were also formed efficiently by the A188V, M141I, and E229D mutants at dilutions greater than 10−4. In contrast, the parental tsG453 and N16K mutants formed smaller-sized plaques at a higher temperature than those formed at 34.5 °C and only produced plaques at dilutions less than 10−3, indicating a potential ts defect (Figure 3). Indeed, the calculated 39 °C/34.5 °C EOP values indicate that only parental tsG453 and N16K had values significantly below 0.03 (Figure 4), indicating a ts phenotype.
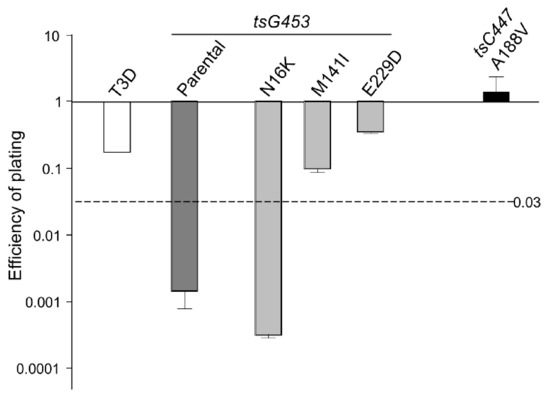
Figure 4.
Efficiency of plating (EOP) of rescued virus isolates. EOP values were calculated by dividing the titer produced by the viruses shown at the restrictive incubation temperature of 39 °C by the titer at the permissive temperature of 34.5 °C. n = 3; Error bars = SEM.
4. Discussion
The elucidation of macromolecular structure-function relationships has been aided by studies of conditionally lethal mutants. For example, many of the molecular steps in the assembly of bacteriophage T4 [20] and of mammalian orthoreoviruses [19,33,37] have been defined by analyses of such mutants. Two MRV ts mutants that have been studied extensively are the tsC447 mutant with lesions in the MRV S2 gene that encodes the σ2 core protein and the tsG453 mutant with lesions in the MRV S4 gene that encodes the σ3 outer-capsid protein [29]. The σ2 protein, present in 150 copies per virion [42], serves as a clamp to stabilize the λ1 core shell [43]. At the non-permissive temperature of 39 °C, the tsC447 mutant produces less RNA and fails to assemble core-like particles, yielding thin empty-shell structures [44]. The tsC447 S2 gene contains three polymorphisms compared to the wild-type T3D S2 sequence [35]. Identification of these polymorphism sites within the σ2 atomic structure demonstrates that the altered amino acids are located in different regions of the protein (Figure 5a). Both Ala188 and Asn383 are located near λ1; therefore, mutations at either site, or both, could explain the tsC447 phenotype. Successful recovery of the Ala188→Val isolate and its lack of temperature-sensitivity (Figure 4) indicate that the Ala188→Val alteration does not contribute to the ts phenotype of tsC447. Thus, either the Ala323→Val or Asn383→Asp, or both, mutations are responsible for the tsC447 ts phenotype. Ala323 is located at the periphery of the σ2 protein and, based on its position, is unlikely to interact with any other core proteins. It may interact with outer-capsid protein μ1, but this would likely not explain the failure of this mutant to produce a core particle, in apparent agreement with reversion analyses that indicated Asn383→Asp was solely responsible [38]. Unfortunately, the inability to rescue clones containing the individual Ala323→Val or Asn383→Asp mutations, despite repeated attempts, prevented us from further testing the roles of these individual amino acids in σ2 structure-function. It is possible that each of these individual polymorphisms, in the absence of the others, results in a lethal phenotype that prevented their rescue. Alternatively, RNA secondary structure negatively impedes translation of proteins by slowing or blocking the initiation and movement of ribosomes along the mRNA [45,46], and might explain our inability to rescue the individual Ala323→Val or Asn383→Asp mutations. However, RNA secondary structure predictions of the corresponding S2 genome segments of the corresponding Ala323→Val or Asn383→Asp mutants [47,48] did not reveal any noticeable differences in the predicted folding patterns compared to the parental T3D or tsC447 S2 genome folding patterns.
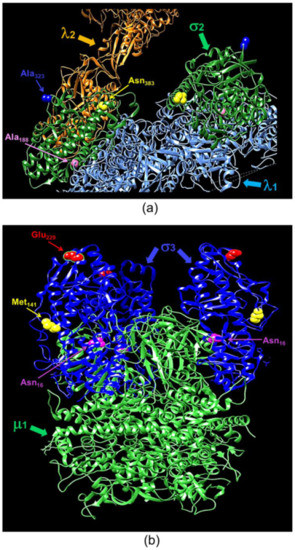
Figure 5.
Ribbon tracings of the reovirus σ2 and σ3 proteins with annotated point mutation sites. (a) A mammalian reoviruses (MRV) core with T1L σ2 depicted in green. Residues altered in tsC447 are indicated. Ala188 and Asn383 are located near λ1 (light blue), and A323 is located near λ2 (orange). (b) A heterohexamer assembly of T1L σ3 and μ1. Locations of tsG453 mutations are indicated on each of the three σ3 monomers (in blue). Asn16 is located near the σ3 interface with μ1 (green), Met141 is located adjacent to several unstructured loops on the lateral surface of σ3, and Glu229 is located on the apical surface of σ3. A lysine substitution at Asn16 is postulated to alter σ3 contacts with the neighboring μ1 protein. Ribbon tracing images were produced using PDB 1EJ6 (λ1-λ2-σ2) and PDB 1JMU (μ1-σ3) and UCSF Chimera.
The MRV σ3 protein, present in 600 copies in complex with the μ1 protein, forms the outermost shell of the virion [42,49]. It is the first protein removed by proteolysis during viral entry into cells [50,51,52,53] or in the intestinal lumen [54,55]. This protein has numerous functions during viral replication, including suppression of protein kinase R (PKR) activation [56,57]. The tsG453 mutant produces comparable levels of viral protein and RNA during infections at permissive and non-permissive temperatures [26,58], but this mutant produces only core-like particles [36]. The failure of this mutant to assemble intact virions has been attributed to lack of mutant σ3 protein association with the μ1 protein at restrictive temperatures [37]. Sequencing of the tsG453 mutant S4 gene identified three polymorphisms compared to wild-type T3D S4 [36,37]. Identification of these polymorphism sites within the σ3 atomic structure demonstrates that the altered amino acids are located in different regions of the protein (Figure 5b). Met141 and Glu229 are located on the periphery of the σ3 protein and, based on their positions, are unlikely to interact with any other viral proteins. Furthermore, EOP analyses (Figure 4) indicate that neither the M141I nor the E229D alterations contribute to the mutant phenotype. Of the isolates tested, only the parental rescued mutant with all three amino acid substitutions and the isolate containing the Asn16→Lys alteration had EOP values substantially below 0.03, indicating that this single amino acid residue is responsible for the ts phenotype of tsG453. Asn16 is located at an interface with μ1; therefore, it is possible that a change from asparagine to a substantially more basic, extended lysine residue could perturb the σ3 protein so that at the non-permissive temperature σ3 is unable to interact stably with μ1 [37]. The failure of the tsG453 mutant to assemble beyond a core-like particle at the non-permissive temperature (Figure 6), combined with the apparent incapacity of mutant σ3 to interact with μ1 [37], suggest that σ3/μ1 interactions and formation of the heterohexameric complex are prerequisites for assembly of the reovirus outer capsid.
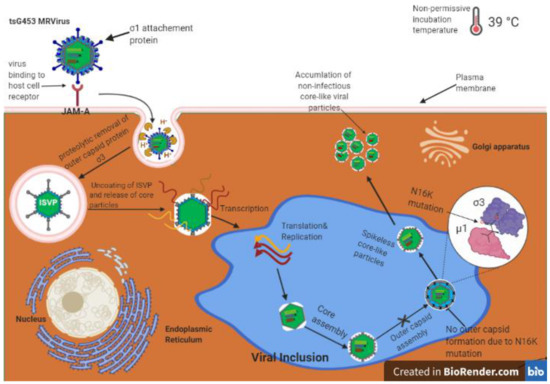
Figure 6.
Model for reovirus tsG453 mutant capsid assembly. Reovirus binds to the receptor junction adhesion molecule A (JAM-A; top left), undergoes internalization, and outer-capsid proteins are removed to yield transcriptionally active core particles (center). Viral mRNAs are translated to produce viral proteins and also serve as templates for replication of progeny (−)-sense RNA. Viral proteins encapsidate viral RNA to produce progeny particles in viral inclusions (blue area in center). At the non-permissive temperature of 39 °C, progeny core-like particles are produced. However, the σ3-N16K mutation prevents σ3 from forming complexes with outer-capsid protein µ1. Therefore, core-like particles accumulate in cells, and viral replication is inhibited.
5. Conclusions
In conclusion, this study is the first to employ reverse genetics to precisely define the amino acid polymorphisms responsible for the ts phenotype in at least one of the Fields’ panel of MRV ts mutants. Several ts isolates remain for which there is no information about the amino acid alternations that confer the ts phenotype. In addition, further studies using the mutants studied here should be conducted to determine what effects, if any, alter the atomic structures of the relevant proteins, thereby better delineating the functions of these proteins.
Author Contributions
Conceptualization, K.K.M.G., D.M.S., T.S.D., and K.M.C.; methodology, K.K.M.G. and D.M.S.; validation, K.K.M.G. and D.M.S.; formal analysis, K.K.M.G., D.M.S., T.S.D., and K.M.C.; investigation, K.K.M.G. and D.M.S.; writing—original draft preparation, K.K.M.G.; writing—review and editing, K.K.M.G., D.M.S., T.S.D., and K.M.C; visualization, K.K.M.G. and K.M.C.; supervision, T.S.D. and K.M.C.; project administration, K.M.C.; funding acquisition, T.S.D. and K.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Natural Sciences and Engineering Research Council Discovery Grant to K.M.C., by a Mitacs Globalink Research Award to K.K.M.G, and U.S. Public Health Service award R01 AI038296 to D.M.S. and T.S.D.
Data Availability Statement
All data present within manuscript.
Acknowledgments
We thank members of the Dermody Lab for useful suggestions during the conduct of this research and members of the Coombs Lab for review of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Dermody, T.S.; Parker, J.S.L.; Sherry, B. Orthoreoviruses. In Fields Virology, 6th ed.; Knipe, D., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Abad, A.T.; Danthi, P. Recognition of reovirus rnas by the innate immune system. Viruses (Basel) 2020, 12, 667. [Google Scholar] [CrossRef] [PubMed]
- Boehme, K.W.; Lai, C.M.; Dermody, T.S. Mechanisms of reovirus bloodstream dissemination. Adv. Virus Res. 2013, 87, 1–35. [Google Scholar] [PubMed]
- Estes, M.K.; Kapikian, A.Z. Rotaviruses. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 5, pp. 1917–1974. [Google Scholar]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D.; WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Karampatsas, K.; Osborne, L.; Seat, M.L.; Tong, C.Y.W.; Prendergast, A.J. Clinical characteristics and complications of rotavirus gastroenteritis in children in east london: A retrospective case-control study. PLoS ONE 2018, 13, e0194009. [Google Scholar] [CrossRef]
- Roy, P. Orbiviruses. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippencott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 5, pp. 1975–1997. [Google Scholar]
- Coetzee, P.; Stokstad, M.; Venter, E.H.; Myrmel, M.; Van Vuuren, M. Bluetongue: A historical and epidemiological perspective with the emphasis on south africa. Virol. J. 2012, 9. [Google Scholar] [CrossRef]
- Gong, J.; Sachdev, E.; Mita, A.C.; Mita, M.M. Clinical development of reovirus for cancer therapy: An oncolytic virus with immune-mediated antitumor activity. World J. Methodol. 2016, 6, 25–42. [Google Scholar] [CrossRef]
- Strong, J.E.; Coffey, M.C.; Tang, D.; Sabinin, P.; Lee, P.W. The molecular basis of viral oncolysis: Usurpation of the ras signaling pathway by reovirus. EMBO J. 1998, 17, 3351–3362. [Google Scholar] [CrossRef]
- Shmulevitz, M.; Marcato, P.; Lee, P.W. Unshackling the links between reovirus oncolysis, ras signaling, translational control and cancer. Oncogene 2005, 24, 7720–7728. [Google Scholar] [CrossRef]
- Galanis, E.; Markovic, S.N.; Suman, V.J.; Nuovo, G.J.; Vile, R.G.; Kottke, T.J.; Nevala, W.K.; Thompson, M.A.; Lewis, J.E.; Rumilla, K.M.; et al. Phase ii trial of intravenous administration of reolysin (r) (reovirus serotype-3-dearing strain) in patients with metastatic melanoma. Mol. Ther. 2012, 20, 1998–2003. [Google Scholar] [CrossRef]
- Chen, H.F.; Ramachandra, M.; Padmanabhan, R. Biochemical characterization of a temperature-sensitive adenovirus DNA polymerase. Virology 1994, 205, 364–370. [Google Scholar] [CrossRef]
- Millns, A.K.; Carpenter, M.S.; DeLange, A.M. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology 1994, 198, 504–513. [Google Scholar] [CrossRef]
- Schwartzberg, P.L.; Roth, M.J.; Tanese, N.; Goff, S.P. Analysis of a temperature-sensitive mutation affecting the integration protein of moloney murine leukemia virus. Virology 1993, 192, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Dzianott, A.; Ahlquist, P.; Bujarski, J.J. Mutations in the helicase-like domain of protein 1a alter the sites of rna-rna recombination in brome mosaic virus. J. Virol. 1995, 69, 2547–2556. [Google Scholar] [CrossRef]
- Shikova, E.; Lin, Y.C.; Saha, K.; Brooks, B.R.; Wong, P.K.Y. Correlation of specific virus-astrocyte interactions and cytopathic effects induced by ts1, a neurovirulent mutant of moloney murine leukemia virus. J. Virol. 1993, 67, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Wiskerchen, M.; Muesing, M.A. Human-immunodeficiency-virus type-1 integrase—effects of mutations on viral ability to integrate, direct viral gene-expression from unintegrated viral-DNA templates, and sustain viral propagation in primary-cells. J. Virol. 1995, 69, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M. Temperature-sensitive mutants of reovirus. Curr. Top. Microbiol. Immunol. 1998, 233, 69–107. [Google Scholar] [PubMed]
- Black, L.W.; Showe, M.K.; Steven, A.C. Morphogenesis of the t4 head. In Molecular Biology of Bacteriophage T4; Karam, J.D., Drake, J.W., Kreuzer, K.N., Mosig, G., Hall, D., Eiserling, F.A., Black, L.W., Kutter, E., Spicer, E., Carlson, K., et al., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 218–258. [Google Scholar]
- Tolley, K.P.; Marriott, A.C.; Simpson, A.; Plows, D.J.; Matthews, D.A.; Longhurst, S.J.; Evans, J.E.; Johnson, J.L.; Cane, P.A.; Randolph, V.B.; et al. Identification of mutations contributing to the reduced virulence of a modified strain of respiratory syncytial virus. Vaccine 1996, 14, 1637–1646. [Google Scholar] [CrossRef]
- Murphy, B.R.; Richman, D.D.; Spring, S.B.; Chanock, R.M. Use of temperature-sensitive mutants of influenza-a virus as live virus-vaccine strains—evaluation in laboratory-animals, adults and children. Postgrad. Med. J. 1976, 52, 381–388. [Google Scholar] [CrossRef][Green Version]
- Ikegami, N.; Gomatos, P.J. Temperature-sensitive conditional-lethal mutants of reovirus 3: I. Isolation and characterization. Virology 1968, 36, 447–458. [Google Scholar] [CrossRef]
- Fields, B.N.; Joklik, W.K. Isolation and preliminary genetic and biochemical characterization of temperature-sensitive mutants of reovirus. Virology 1969, 37, 335–342. [Google Scholar] [CrossRef]
- Fields, B.N. Temperature-sensitive mutants of reovirus type 3 features of genetic recombination. Virology 1971, 46, 142–148. [Google Scholar] [CrossRef]
- Fields, B.N.; Laskov, R.; Scharff, M.D. Temperature-sensitive mutants of reovirus type 3: Studies on the synthesis of viral peptides. Virology 1972, 50, 209–215. [Google Scholar] [CrossRef]
- Ramig, R.F.; Fields, B.N. Method for rapidly screening revertants of reovirus temperature-sensitive mutants for extragenic suppression. Virology 1977, 81, 170–173. [Google Scholar]
- Ramig, R.F.; Mustoe, T.A.; Sharpe, A.H.; Fields, B.N. A genetic map of reovirus. Ii. Assignment of the double-stranded rna-negative mutant groups c, d, and e to genome segments. Virology 1978, 85, 531–534. [Google Scholar] [CrossRef]
- Mustoe, T.A.; Ramig, R.F.; Sharpe, A.H.; Fields, B.N. Genetic map of reovirus.3. Assignment of double-stranded rna-positive mutant group-a, group-b, and group-g to genome segments. Virology 1978, 85, 545–556. [Google Scholar] [CrossRef]
- Chakraborty, P.R.; Ahmed, R.; Fields, B.N. Genetics of reovirus: The relationship of interference to complementation and reassortment of temperature-sensitive mutants at nonpermissive temperature. Virology 1979, 94, 119–127. [Google Scholar] [CrossRef]
- Hazelton, P.R.; Coombs, K.M. The reovirus mutant tsa279 has temperature-sensitive lesions in the M2 and L2 genes: The M2 gene is associated with decreased viral protein production and blockade in transmembrane transport. Virology 1995, 207, 46–58. [Google Scholar] [CrossRef]
- Coombs, K.M. Identification and characterization of a double-stranded rna- reovirus temperature-sensitive mutant defective in minor core protein mu2. J. Virol. 1996, 70, 4237–4245. [Google Scholar] [CrossRef] [PubMed]
- Hazelton, P.R.; Coombs, K.M. The reovirus mutant tsa279 L2 gene is associated with generation of a spikeless core particle: Implications for capsid assembly. J. Virol. 1999, 73, 2298–2308. [Google Scholar] [CrossRef]
- Wiener, J.R.; McLaughlin, T.; Joklik, W.K. The sequences of the S2 genome segments of reovirus serotype 3 and of the dsrna-negative mutant ts447. Virology 1989, 170, 340–341. [Google Scholar] [CrossRef]
- Dermody, T.S.; Schiff, L.A.; Nibert, M.L.; Coombs, K.M.; Fields, B.N. The S2 gene nucleotide sequences of prototype strains of the three reovirus serotypes: Characterization of reovirus core protein sigma 2. J. Virol. 1991, 65, 5721–5731. [Google Scholar] [CrossRef] [PubMed]
- Danis, C.; Garzon, S.; Lemay, G. Further characterization of the ts453 mutant of mammalian orthoreovirus serotype 3 and nucleotide sequence of the mutated s4 gene. Virology 1992, 190, 494–498. [Google Scholar] [CrossRef]
- Shing, M.; Coombs, K.M. Assembly of the reovirus outer capsid requires mu 1/sigma 3 interactions which are prevented by misfolded sigma 3 protein in temperature-sensitive mutant tsg453. Virus Res. 1996, 46, 19–29. [Google Scholar] [CrossRef]
- Coombs, K.M.; Mak, S.C.; Petrycky-Cox, L.D. Studies of the major reovirus core protein sigma 2: Reversion of the assembly-defective mutant tsc447 is an intragenic process and involves back mutation of asp-383 to asn. J. Virol. 1994, 68, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Antar, A.A.; Boehme, K.W.; Danthi, P.; Eby, E.A.; Guglielmi, K.M.; Holm, G.H.; Johnson, E.M.; Maginnis, M.S.; Naik, S.; et al. A plasmid-based reverse genetics system for animal double-stranded rna viruses. Cell Host Microbe 2007, 1, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ooms, L.S.; Ikizler, M.; Chappell, J.D.; Dermody, T.S. An improved reverse genetics system for mammalian orthoreoviruses. Virology 2010, 398, 194–200. [Google Scholar] [CrossRef]
- Boehme, K.W.; Ikizler, M.; Kobayashi, T.; Dermody, T.S. Reverse genetics for mammalian reovirus. Methods Companion Methods Enzymol. 2011, 55, 109–113. [Google Scholar] [CrossRef]
- Dryden, K.A.; Wang, G.; Yeager, M.; Nibert, M.L.; Coombs, K.M.; Furlong, D.B.; Fields, B.N.; Baker, T.S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: Analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1993, 122, 1023–1041. [Google Scholar] [CrossRef]
- Reinisch, K.M.; Nibert, M.L.; Harrison, S.C. Structure of the reovirus core at 3.6 a resolution. Nature 2000, 404, 960–967. [Google Scholar] [CrossRef]
- Matsuhisa, T.; Joklik, W.K. Temperature-sensitive mutants of reovirus. V. Studies on the nature of the temperature-sensitive lesion of the group c mutant ts447. Virology 1974, 60, 380–389. [Google Scholar] [CrossRef]
- Gaspar, P.; Moura, G.; Santos, M.A.S.; Oliveira, J.L. Mrna secondary structure optimization using a correlated stem-loop prediction. Nucleic Acids Res. 2013, 41, e73. [Google Scholar] [CrossRef]
- Mao, Y.H.; Liu, H.L.; Liu, Y.L.; Tao, S.H. Deciphering the rules by which dynamics of mrna secondary structure affect translation efficiency in saccharomyces cerevisiae. Nucleic Acids Res. 2014, 42, 4813–4822. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuboock, R.; Hofacker, I.L. The vienna rna websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Siederdissen, C.H.Z.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Viennarna package 2.0. Algorithms Mol. Biol. 2011, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M. Stoichiometry of reovirus structural proteins in virus, isvp, and core particles. Virology 1998, 243, 218–228. [Google Scholar] [CrossRef]
- Ebert, D.H.; Deussing, J.; Peters, C.; Dermody, T.S. Cathepsin l and cathepsin b mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002, 277, 24609–24617. [Google Scholar] [CrossRef] [PubMed]
- Jane-Valbuena, J.; Breun, L.A.; Schiff, L.A.; Nibert, M.L. Sites and determinants of early cleavages in the proteolytic processing pathway of reovirus surface protein sigma3. J. Virol. 2002, 76, 5184–5197. [Google Scholar] [CrossRef]
- Mendez, I.I.; She, Y.M.; Ens, W.; Coombs, K.M. Digestion pattern of reovirus outer capsid protein sigma3 determined by mass spectrometry. Virology 2003, 311, 289–304. [Google Scholar] [CrossRef]
- Golden, J.W.; Bahe, J.A.; Lucas, W.T.; Nibert, M.L.; Schiff, L.A. Cathepsin s supports acid-independent infection by some reoviruses. J. Biol. Chem. 2004, 279, 8547–8557. [Google Scholar] [CrossRef]
- Bodkin, D.K.; Nibert, M.L.; Fields, B.N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 1989, 63, 4676–4681. [Google Scholar] [CrossRef]
- Bass, D.M.; Bodkin, D.; Dambrauskas, R.; Trier, J.S.; Fields, B.N.; Wolf, J.L. Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J. Virol. 1990, 64, 1830–1833. [Google Scholar] [CrossRef] [PubMed]
- Imani, F.; Jacobs, B.L. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 sigma 3 protein. Proc. Natl. Acad. Sci. USA 1988, 85, 7887–7891. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Schmechel, S.C.; Williams, B.R.; Silverman, R.H.; Schiff, L.A. Involvement of the interferon-regulated antiviral proteins pkr and rnase l in reovirus-induced shutoff of cellular translation. J. Virol. 2005, 79, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.K.; Fields, B.N. Temperature-sensitive mutants of reovirus type 3: Studies on the synthesis of viral rna. Virology 1972, 50, 799–809. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).