H7N7 Avian Influenza Virus Mutation from Low to High Pathogenicity on a Layer Chicken Farm in the UK
Abstract
1. Introduction
2. Materials and Methods
2.1. Epidemiological Investigation and Sample Collection
2.2. Pathology and Immunohistochemistry (IHC)
2.3. RNA Extraction and Real-Time Reverse Transcription-PCR (RRT-PCR)
2.4. Sequencing and Phylogenetic Analysis
2.5. Virus Isolation
2.6. Hemagglutination Inhibition (HI) and Neuraminidase Inhibition (NI) Tests
2.7. Intravenous Pathogenicity Index (IVPI)
2.8. Statistical Analyses
3. Results
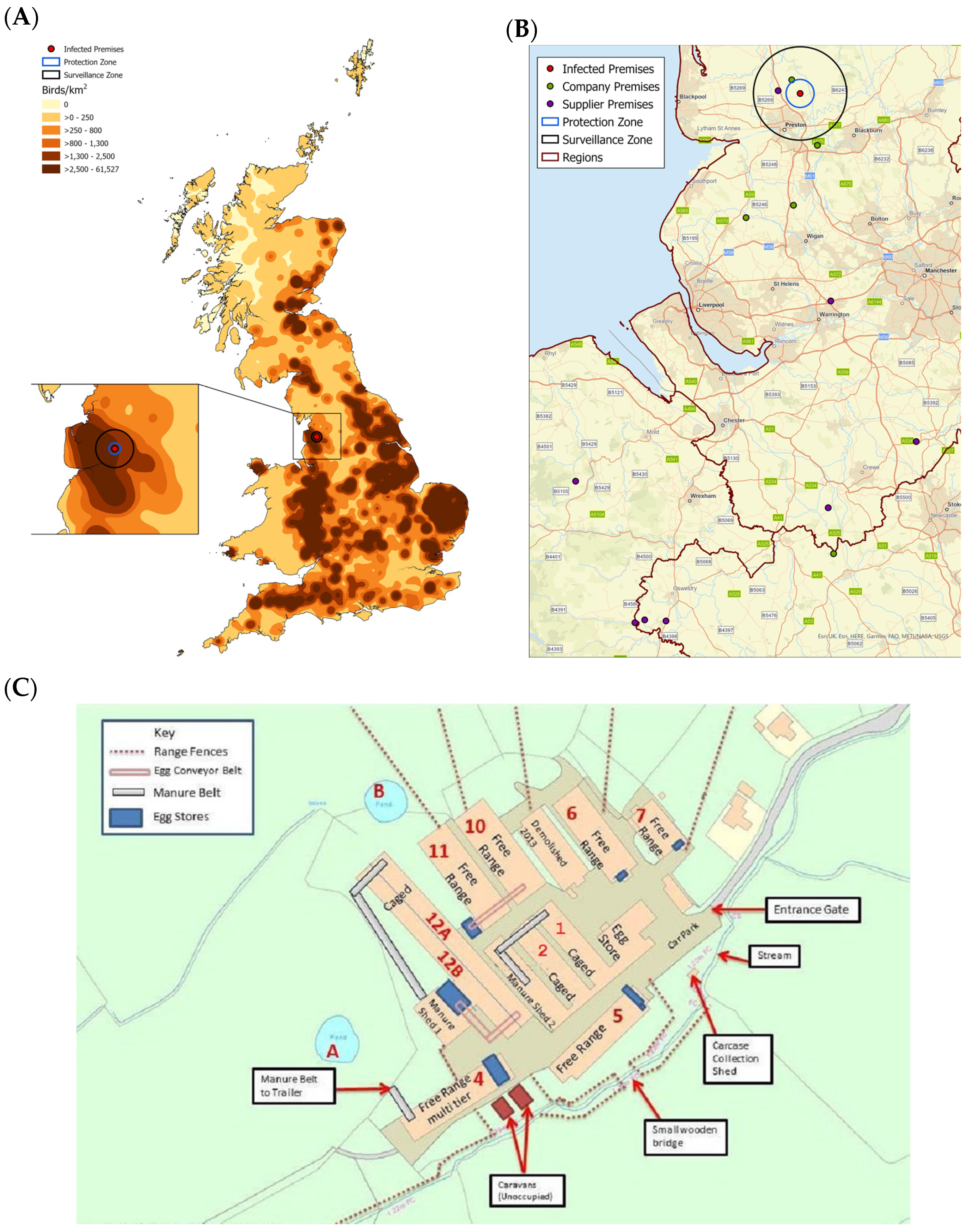
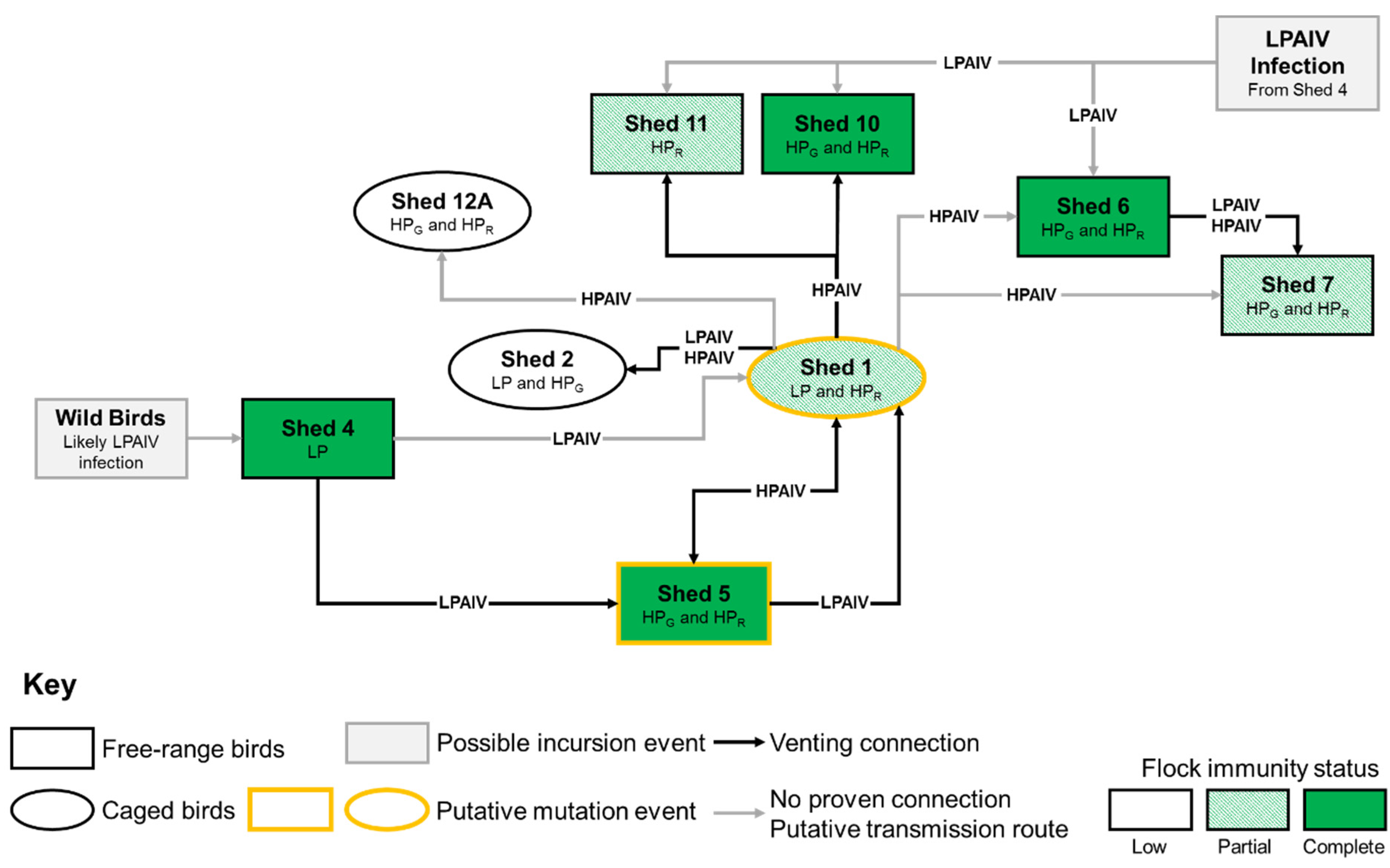
3.1. Case Description
3.2. Clinical Background and Submission of Samples
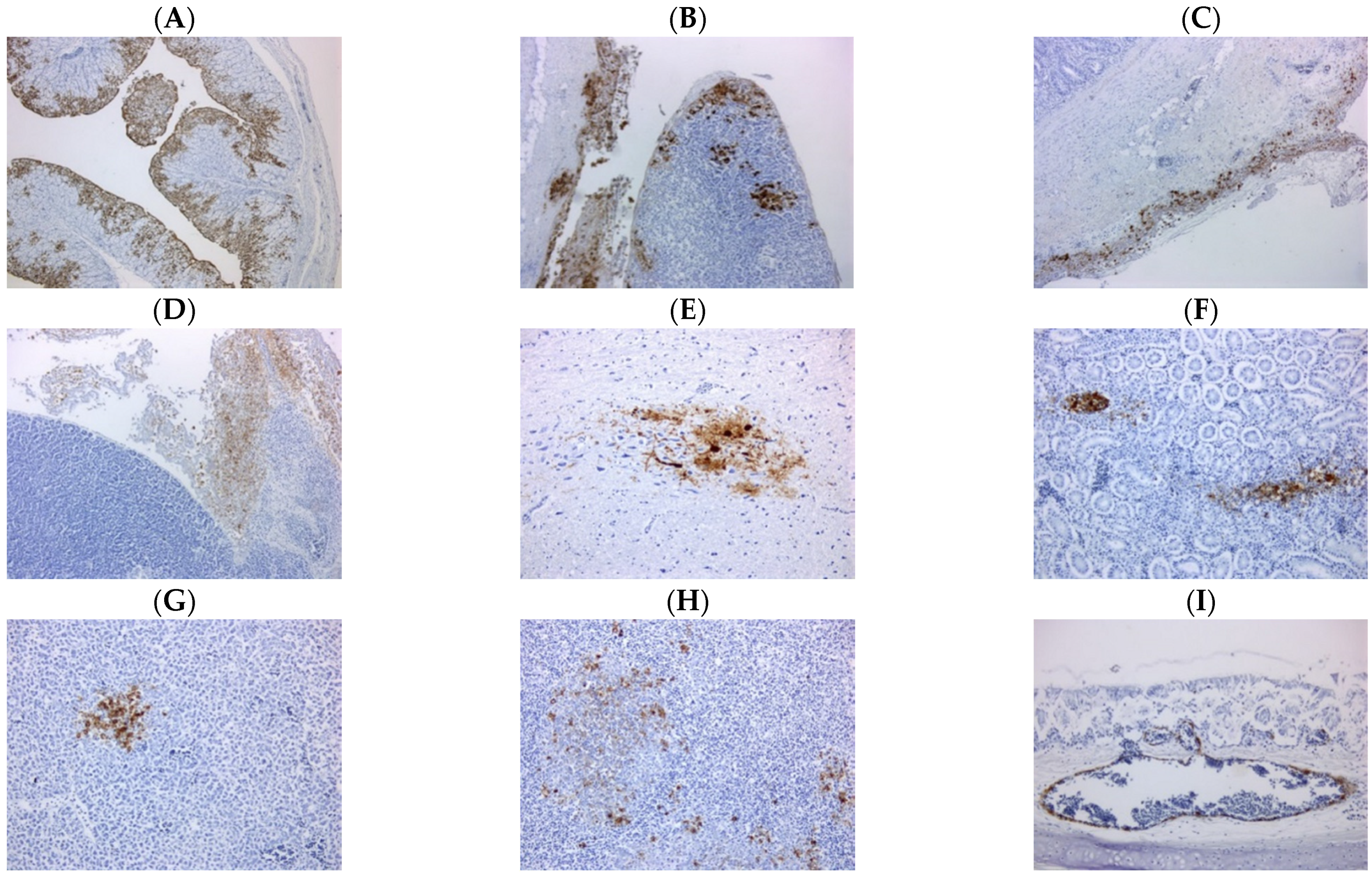
3.3. Pathology
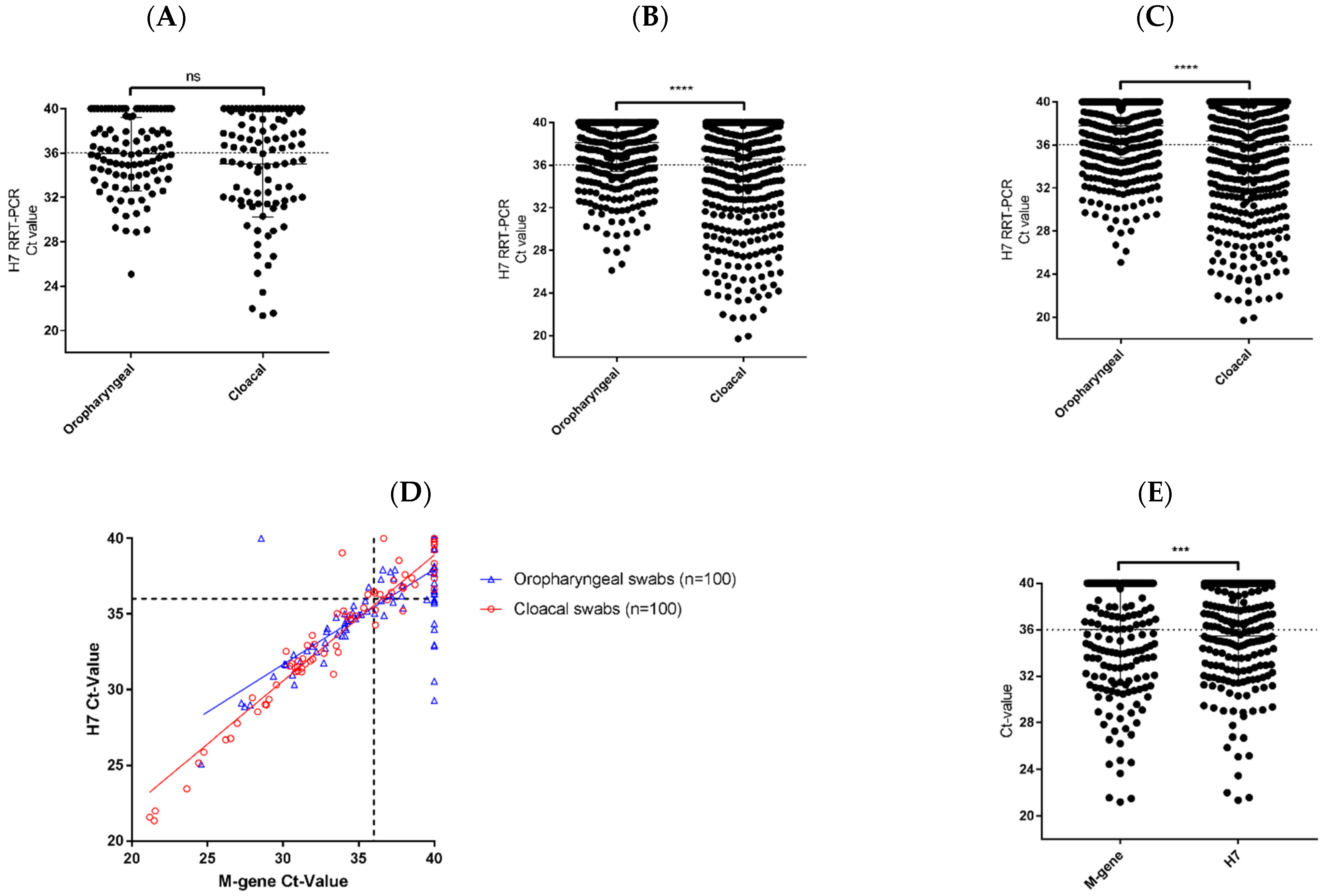
3.4. RRT-PCR Investigations
3.5. Sequencing of the HA CS Motif
3.6. Virus Isolation, Conventional Typing and IVPI
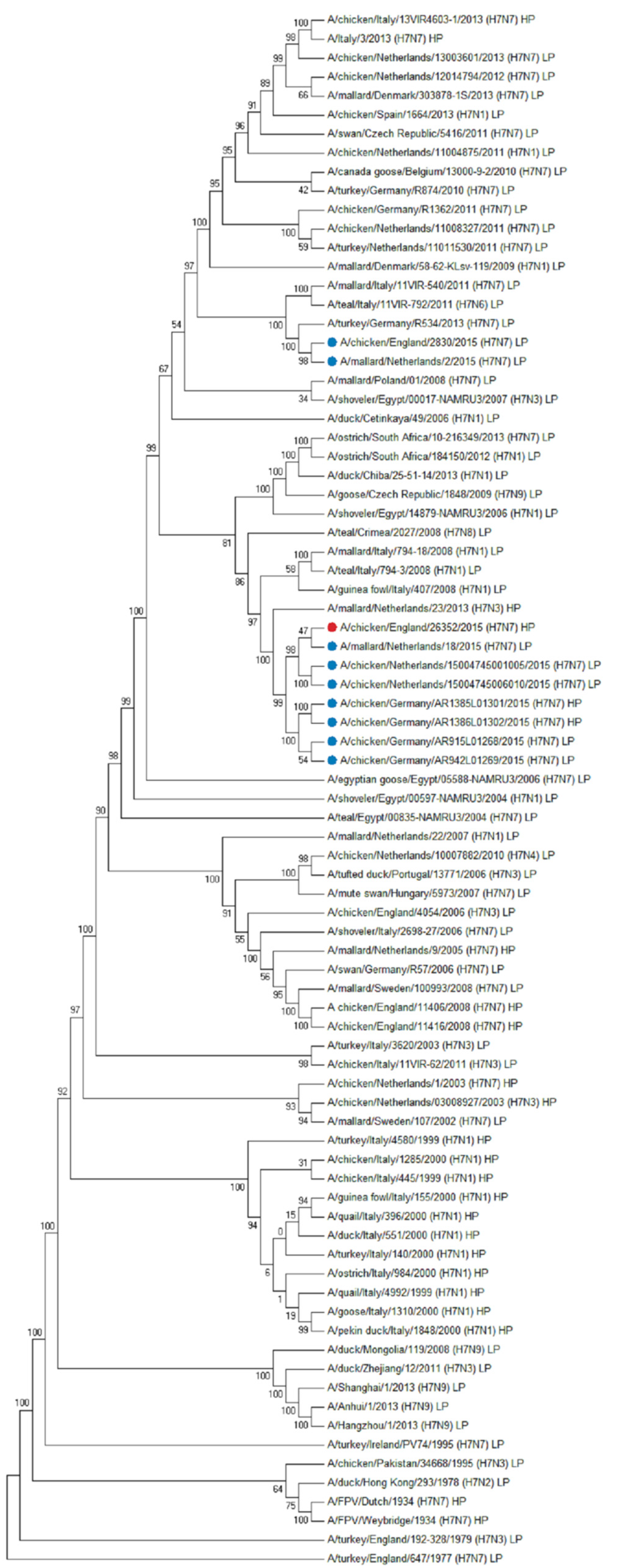
3.7. WGS and Phylogenetic Analysis
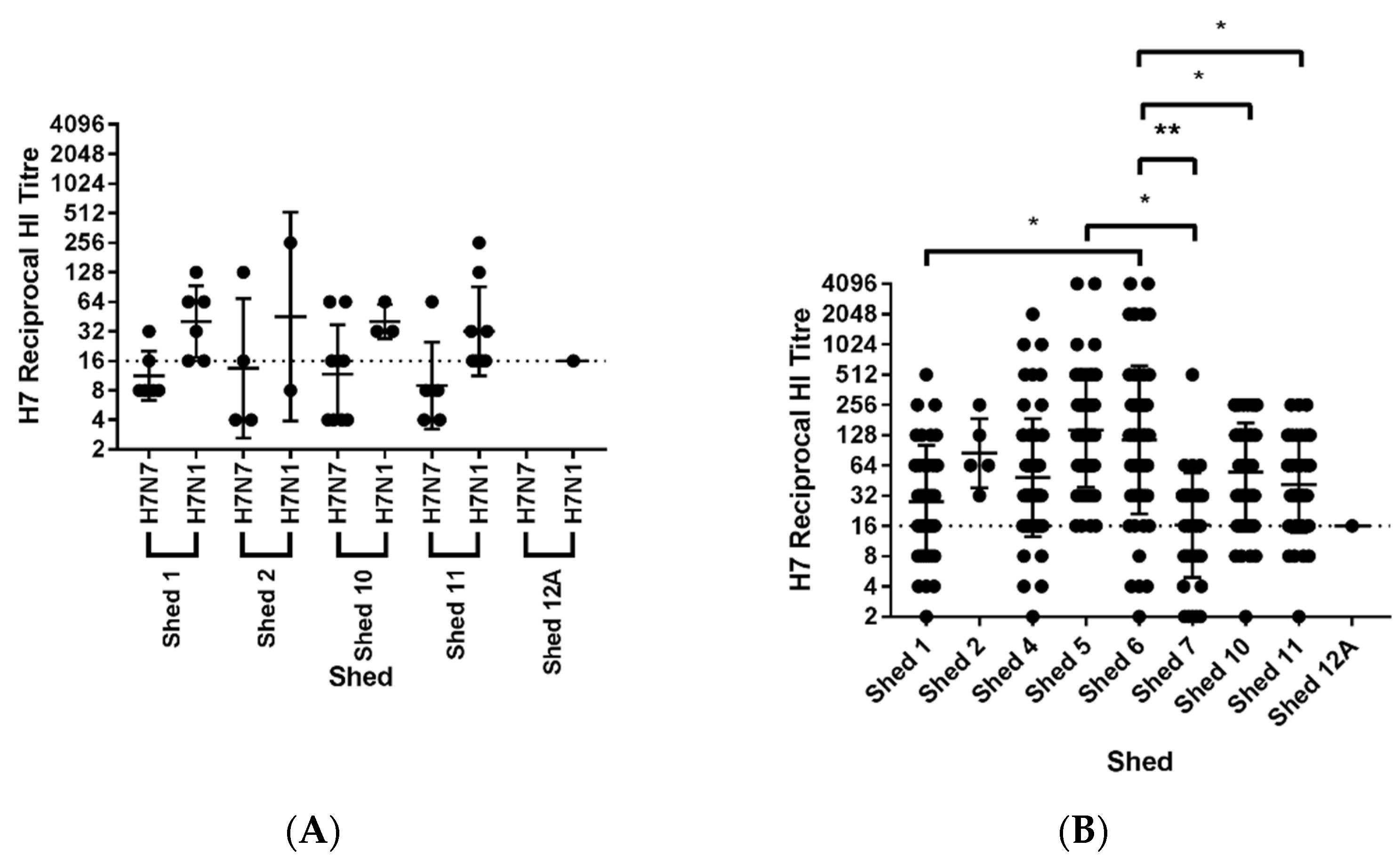
3.8. Serological Investigation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alexander, D.J.; Brown, I.H. History of Highly Pathogenic Avian Influenza. Rev. Sci. Tech. 2009, 28, 19–38. [Google Scholar] [CrossRef]
- Nunez, A.; Brookes, S.M.; Reid, S.M.; Rueda, G.C.; Hicks, D.J.; Seekings, J.M.; Spencer, Y.I.; Brown, I.H. Highly Pathogenic Avian Influenza H5N8 Clade 2.3.4.4 Virus: Equivocal Pathogenicity and Implications for Surveillance Following Natural Infection in Breeder Ducks in the United Kingdom. Transbound. Emerg. Dis. 2016, 63, 5–9. [Google Scholar] [CrossRef]
- Reid, S.M.; Núnez, A.; Seekings, A.H.; Thomas, S.S.; Slomka, M.J.; Mahmood, S.; Clark, J.R.; Banks, J.; Brookes, S.M.; Brown, I.H. Two Single Incursions of H7N7 and H5N1 Low Pathogenicity Avian Influenza in U.K. Broiler Breeders During 2015 and 2016. Avian Dis. 2019, 63, 181–192. [Google Scholar] [CrossRef]
- Seekings, A.H.; Slomka, M.J.; Russell, C.; Howard, W.A.; Choudhury, B.; Nunez, A.; Londt, B.Z.; Cox, W.; Ceeraz, V.; Thoren, P.; et al. Direct evidence of H7N7 avian influenza virus mutation from low to high virulence on a single poultry premises during an outbreak in free range chickens in the UK, 2008. Infect. Genet. Evol. 2018, 64, 13–31. [Google Scholar] [CrossRef]
- APHA. Epidemiology Report: Pathogenic Avian Influenza H7N7 (AIV2015/02; Formerly AIV SO2015/0001), in Table-Egg Laying Hens; Defra: London, UK, 2015.
- Manvell, R.J.; Londt, B.Z.; Ceeraz, V.; Cox, W.J.; Essen, S.; Banks, J.; Slomka, M.J.; Pavlidis, T.; Irvine, R.M.; Wilesmith, J.W.; et al. Low pathogenic avian influenza in domestic fowl in Norfolk, England, March and April, 2006. Vet. Rec. 2008, 162, 278–280. [Google Scholar] [CrossRef]
- Slomka, M.J.; Irvine, R.M.; Pavlidis, T.; Banks, J.; Brown, I.H. Role of real-time RT-PCR platform technology in the diagnosis and management of notifiable avian influenza outbreaks: Experiences in Great Britain. Avian Dis. 2010, 54, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Dietze, K.; Graaf, A.; Homeier-Bachmann, T.; Grund, C.; Forth, L.; Pohlmann, A.; Jeske, C.; Wintermann, M.; Beer, M.; Conraths, F.J.; et al. From low to high pathogenicity-Characterization of H7N7 avian influenza viruses in two epidemiologically linked outbreaks. Transbound. Emerg. Dis. 2018, 65, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Lebarbenchon, C.; Stallknecht, D.E. Host shifts and molecular evolution of H7 avian influenza virus hemagglutinin. Virol. J. 2011, 8, 328. [Google Scholar] [CrossRef] [PubMed]
- European Directive. Council Directive 2005/94/EC of 20 December 2005 on Community measures for the control of avian influenza and repealing Directive 92/40/EEC. Off. J. Eur. Union 2006, 10, 16–64. [Google Scholar]
- Slomka, M.J.; Pavlidis, T.; Coward, V.J.; Voermans, J.; Koch, G.; Hanna, A.; Banks, J.; Brown, I.H. Validated RealTime reverse transcriptase PCR methods for the diagnosis and pathotyping of Eurasian H7 avian influenza viruses. Influenza Respir. Viruses 2009, 3, 151–164. [Google Scholar] [CrossRef]
- Nagy, A.; Vostinakova, V.; Pirchanova, Z.; Cernikova, L.; Dirbakova, Z.; Mojzis, M.; Jirincova, H.; Havlickova, M.; Dan, A.; Ursu, K.; et al. Development and evaluation of a one-step real-time RT-PCR assay for universal detection of influenza A viruses from avian and mammal species. Arch. Virol. 2010, 155, 665–673. [Google Scholar] [CrossRef]
- Slomka, M.J.; Pavlidis, T.; Banks, J.; Shell, W.; McNally, A.; Essen, S.; Brown, I.H. Validated H5 Eurasian real-time reverse transcriptase-polymerase chain reaction and its application in H5N1 outbreaks in 2005–2006. Avian Dis. 2007, 51, 373–377. [Google Scholar] [CrossRef]
- Fuller, C.M.; Brodd, L.; Irvine, R.M.; Alexander, D.J.; Aldous, E.W. Development of an L gene real-time reverse-transcription PCR assay for the detection of avian paramyxovirus type 1 RNA in clinical samples. Arch. Virol. 2010, 155, 817–823. [Google Scholar] [CrossRef]
- Slomka, M.J.; Coward, V.J.; Banks, J.; Londt, B.Z.; Brown, I.H.; Voermans, J.; Koch, G.; Handberg, K.J.; Jorgensen, P.H.; Cherbonnel-Pansart, M.; et al. Identification of sensitive and specific avian influenza polymerase chain reaction methods through blind ring trials organized in the European Union. Avian Dis. 2007, 51, 227–234. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- European Commission. European Commission Decision 2006/437/EC of 4 August 2006 approving Diagnostic Manual for avian influenza as provided for in Council Directive 2005/94/EC. Off. J. Eur. Union L. 2006, 237, 1–27. [Google Scholar]
- OIE. Terrestrial Manual: Avian Influenza (Infection with Avian Influenza Viruses); OIE: Paris, France, 2019. [Google Scholar]
- European Commission. European Commission Decision 2010/367/EU of 25 June 2010 on the implementation by Member States of surveillance programmes for avian influenza in poultry and wild birds. Off. J. Eur. Union 2010, 50, 22–32. [Google Scholar]
- CDC. H5N1 Genetic Changes Inventor: A Tool for Influenza Surveillance and Preparedness; CDC: Atlanta, GA, USA, 2020.
- Fereidouni, S.R.; Starick, E.; Beer, M.; Wilking, H.; Kalthoff, D.; Grund, C.; Hauslaigner, R.; Breithaupt, A.; Lange, E.; Harder, T.C. Highly pathogenic avian influenza virus infection of mallards with homo- and heterosubtypic immunity induced by low pathogenic avian influenza viruses. PLoS ONE 2009, 4, 6706. [Google Scholar] [CrossRef]
- Vergara-Alert, J.; Moreno, A.; Zabala, J.G.; Bertran, K.; Costa, T.P.; Cordon, I.; Rivas, R.; Majo, N.; Busquets, N.; Cordioli, P.; et al. Exposure to a low pathogenic A/H7N2 virus in chickens protects against highly pathogenic A/H7N1 virus but not against subsequent infection with A/H5N1. PLoS ONE 2013, 8, 8692. [Google Scholar] [CrossRef]
- Capua, I.; Mutinelli, F.; Marangon, S.; Alexander, D.J. H7N1 avian influenza in Italy (1999 to 2000) in intensively reared chickens and turkeys. Avian Pathol. 2000, 29, 537–543. [Google Scholar] [CrossRef]
- Bergervoet, S.A.; Pritz-Verschuren, S.B.E.; Gonzales, J.L.; Bossers, A.; Poen, M.J.; Dutta, J.; Khan, Z.; Kriti, D.; van Bakel, H.; Bouwstra, R.; et al. Circulation of low pathogenic avian influenza (LPAI) viruses in wild birds and poultry in the Netherlands, 2006-2016. Sci. Rep. 2019, 9, 3681. [Google Scholar] [CrossRef]
- OIE. Terrestrial Animal Health Code; OIE: Paris, France, 2019. [Google Scholar]
- WAHIS: Low Pathogenicity Avian Influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=14012 (accessed on 1 June 2020).
- Abdelwhab, E.M.; Veits, J.; Mettenleiter, T.C. Prevalence and control of H7 avian influenza viruses in birds and humans. Epidemiol. Infect. 2014, 142, 896–920. [Google Scholar] [CrossRef]
- Probst, C.; Gethmann, J.M.; Petermann, H.J.; Neudecker, J.; Jacobsen, K.; Conraths, F.J. Low pathogenic avian influenza H7N7 in domestic poultry in Germany in 2011. Vet. Rec. 2012, 171, 624. [Google Scholar] [CrossRef]
- WAHIS: Low Pathogenicity Avian Influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=8009 (accessed on 1 June 2020).
- WAHIS: Low Pathogenicity Avian Influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=10633 (accessed on 1 June 2020).
- WAHIS: Low Pathogenicity Avian influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=13464 (accessed on 1 June 2020).
- WAHIS: Low Pathogenicity Avian Influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=10580 (accessed on 1 June 2020).
- WAHIS: Low Pathogenicity Avian Influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=10739 (accessed on 1 June 2020).
- Wahis: Low Pathogenicity Avian Influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=12194 (accessed on 1 June 2020).
- WAHIS: Low Pathogenicity Avian Influenza (poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=13128 (accessed on 1 June 2020).
- WAHIS: Low Pathogenicity Avian Influenza (Poultry): Summary of Immediate Notifications and Follow-Ups. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=13177 (accessed on 1 June 2020).
- ECDC. ECDC Rapid Risk Assessment: Human Infection by Low Pathogenic Avian Influenza A(H7) Viruses; ECDC: Solna, Sweden, 2015.
- Gibbens, N.; Brown, I.H.; Irvine, R.M. Testing for exclusion of notifiable avian disease. Vet. Rec. 2014, 174, 534–535. [Google Scholar] [CrossRef]
- Garten, W.; Klenk, H.D. Understanding influenza virus pathogenicity. Trends Microbiol. 1999, 7, 99–100. [Google Scholar] [CrossRef]
- Kawaoka, Y.; Webster, R.G. Molecular mechanism of acquisition of virulence in influenza virus in nature. Microb. Pathog. 1988, 5, 311–318. [Google Scholar] [CrossRef]
- Bulach, D.; Halpin, R.; Spiro, D.; Pomeroy, L.; Janies, D.; Boyle, D.B. Molecular analysis of H7 avian influenza viruses from Australia and New Zealand: Genetic diversity and relationships from 1976 to 2007. J. Virol. 2010, 84, 9957–9966. [Google Scholar] [CrossRef] [PubMed]
- Elbers, A.R.; Fabri, T.H.; de Vries, T.S.; de Wit, J.J.; Pijpers, A.; Koch, G. The highly pathogenic avian influenza A (H7N7) virus epidemic in The Netherlands in 2003--lessons learned from the first five outbreaks. Avian Dis. 2004, 48, 691–705. [Google Scholar] [CrossRef]
- Graaf, A.; Ulrich, R.; Maksimov, P.; Scheibner, D.; Koethe, S.; Abdelwhab, E.M.; Mettenleiter, T.C.; Beer, M.; Harder, T. A viral race for primacy: Co-infection of a natural pair of low and highly pathogenic H7N7 avian influenza viruses in chickens and embryonated chicken eggs. Emerg. Microbes Infect. 2018, 7, 204. [Google Scholar] [CrossRef]
| Set 1 | Set 2 | Set 3 | |
|---|---|---|---|
| Date of submission | 7 July 2015 | 9 July 2015 | 11–13 July 2015 |
| Sample type and number | 11 carcasses | 20 OP swabs, per shed 20 C swabs, per shed 20 clotted bloods, per shed | 60 OP swabs, per shed 60 C swabs, per shed 60 clotted bloods, per shed 2 carcasses, per shed |
| Origin of samples | Sheds 10 (N = 5) and 12A (N = 6) | Sheds 1, 2, 10, 11, and 12A | Sheds 1, 2, 4, 5, 6, 7, 10, 11, and 12A |
| Shed | Type | Percentage of RRT-PCR Positive Samples at Whole Bird Level (%) (N = 20) | Cleavage Site Motif Detected | AIV Molecular Pathotype | |||
|---|---|---|---|---|---|---|---|
| Influenza A | AOAV-1 | H5 | H7 | ||||
| 1 | Caged | 50 | 0 | 0 | 50 | PEIPRHRKRRGLF | HPR |
| 2 | Caged | 70 | 0 | 0 | 75 | PEIPRHRKGRGLF | HPG |
| 10 | Free-range | 70 | 0 | 0 | 75 | PEIPRHRKGRGLF | HPG |
| 11 | Free-range | 65 | 0 | 0 | 90 | PEIPRHRKGRGLF PEIPRHRKRRGLF | HPG/HPR |
| 12A | Caged | 50 | 0 | 0 | 65 | PEIPRHRKRRGLF | HPR |
| Shed | Type | Percentage of RRT-PCR Positive Samples at Whole Bird Level by H7 RRT-PCR (%) (N = 60) | Cleavage Site Motif Detected | AIV Molecular Pathotype |
|---|---|---|---|---|
| 1 | Caged | 47 | PEIPKGRGLF PEIPRHRKRRGLF | LP/HPR |
| 2 | Caged | 83 | PEIPKGRGLF PEIPRHRKRRGLF | LP/HPR |
| 4 * | Free-range | 0 | PEIPKGRGLF | LP |
| 5 | Free-range | 32 | PEIPRHRKGRGLF PEIPRHRKRRGLF | HPG/HPR |
| 6 | Free-range | 3 | PEIPRHRKGRGLF PEIPRHRKRRGLF | HPG/HPR |
| 7 | Free-range | 2 | PEIPRHRKGRGLF PEIPRHRKRRGLF | HPG/HPR |
| 10 | Free-range | 52 | PEIPRHRKGRGLF PEIPRHRKRRGLF | HPG/HPR |
| 11 | Free-range | 90 | PEIPRHRKRRGLF | HPR |
| 12A | Caged | 38 | PEIPRHRKGRGLF PEIPRHRKRRGLF | HPG/HPR |
| Shed | Type | Tissue RRT-PCR Results (2 Carcasses per Shed) | ||
|---|---|---|---|---|
| Set 1 | Set 2 | Set 3 | ||
| 7 July 2015 | 9 July 2015 | 11–13 July 2015 | ||
| 1 | Caged | - | HPR | LP/HPR |
| 2 | Caged | - | HPG | LP/HPR |
| 4 | Free-range | - | - | LP |
| 5 | Free-range | - | - | HPG/HPR |
| 6 | Free-range | - | - | HPG/HPR |
| 7 | Free-range | - | - | HPG/HPR |
| 10 | Free-range | HPG | HPG | HPG/HPR |
| 11 | Free-range | HPR | HPG/HPR | HPR |
| 12A | Caged | - | HPR | HPG/HPR |
| Shed | Type | Percentage Antibody Positive Samples by HI at Whole Bird Level (%) | ||
|---|---|---|---|---|
| AOAV-1 | H5 1 | H7 2 | ||
| 1 (N = 14) | Caged | 0 | 0 | 43 |
| 2 (N = 17) | Caged | 0 | 0 | 12 |
| 10 (N = 18) | Free-range | 0 | 0 | 33 |
| 11 (N = 20) | Free-range | 0 | 0 | 45 |
| 12A (N = 15) | Caged | 0 | 0 | 7 |
| Shed | Type | Percentage H7N1 Antibody Positive Samples by HI at Whole Bird Level (%) |
|---|---|---|
| 1 (N = 54) | Caged | 54 |
| 2 (N = 60) | Caged | 8 |
| 4 (N = 60) | Free-range | 90 |
| 5 (N = 60) | Free-range | 100 |
| 6 (N = 58) | Free-range | 91 |
| 7 (N = 59) | Free-range | 41 |
| 10 (N = 60) | Free-range | 82 |
| 11 (N = 60) | Free-range | 60 |
| 12A (N = 60) | Caged | 2 |
| Shed | Type | Birds within Each Category (%) | AIV Molecular Pathotype | |||
|---|---|---|---|---|---|---|
| Category I | Category II | Category III | Category IV | |||
| H7 RRT-PCR Negative | H7 RRT-PCR Positive | H7 RRT-PCR Positive | H7 RRT-PCR Negative | |||
| H7 AIV Seronegative | H7 AIV Seronegative | H7 AIV Seropositive | H7 AIV Seropositive | |||
| 1 | Caged | 21.6 | 31.1 | 13.5 | 33.8 | LP/HPR |
| 2 | Caged | 15.0 | 76.3 | 5.0 | 3.8 | LP/HPG/HPR |
| 4 | Free-range | 10.0 | 0.0 | 0.0 | 90.0 | LP |
| 5 | Free-range | 0.0 | 0.0 | 31.7 | 68.3 | HPG/HPR |
| 6 | Free-range | 8.6 | 0.0 | 1.7 | 89.7 | HPG/HPR |
| 7 | Free-range | 59.3 | 0.0 | 1.7 | 39.0 | HPG/HPR |
| 10 | Free-range | 1.3 | 28.2 | 28.2 | 42.3 | HPG/HPR |
| 11 | Free-range | 1.3 | 42.5 | 47.5 | 8.8 | HPG/HPR |
| 12A | Caged | 52.5 | 45.0 | 0.0 | 2.5 | HPG/HPR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byrne, A.M.P.; Reid, S.M.; Seekings, A.H.; Núñez, A.; Obeso Prieto, A.B.; Ridout, S.; Warren, C.J.; Puranik, A.; Ceeraz, V.; Essen, S.; et al. H7N7 Avian Influenza Virus Mutation from Low to High Pathogenicity on a Layer Chicken Farm in the UK. Viruses 2021, 13, 259. https://doi.org/10.3390/v13020259
Byrne AMP, Reid SM, Seekings AH, Núñez A, Obeso Prieto AB, Ridout S, Warren CJ, Puranik A, Ceeraz V, Essen S, et al. H7N7 Avian Influenza Virus Mutation from Low to High Pathogenicity on a Layer Chicken Farm in the UK. Viruses. 2021; 13(2):259. https://doi.org/10.3390/v13020259
Chicago/Turabian StyleByrne, Alexander M. P., Scott M. Reid, Amanda H. Seekings, Alejandro Núñez, Ana B. Obeso Prieto, Susan Ridout, Caroline J. Warren, Anita Puranik, Vanessa Ceeraz, Stephen Essen, and et al. 2021. "H7N7 Avian Influenza Virus Mutation from Low to High Pathogenicity on a Layer Chicken Farm in the UK" Viruses 13, no. 2: 259. https://doi.org/10.3390/v13020259
APA StyleByrne, A. M. P., Reid, S. M., Seekings, A. H., Núñez, A., Obeso Prieto, A. B., Ridout, S., Warren, C. J., Puranik, A., Ceeraz, V., Essen, S., Slomka, M. J., Banks, J., Brown, I. H., & Brookes, S. M. (2021). H7N7 Avian Influenza Virus Mutation from Low to High Pathogenicity on a Layer Chicken Farm in the UK. Viruses, 13(2), 259. https://doi.org/10.3390/v13020259







