Abstract
Plant RNA viruses encode essential viral proteins that depend on the host translation machinery for their expression. However, genomic RNAs of most plant RNA viruses lack the classical characteristics of eukaryotic cellular mRNAs, such as mono-cistron, 5′ cap structure, and 3′ polyadenylation. To adapt and utilize the eukaryotic translation machinery, plant RNA viruses have evolved a variety of translation strategies such as cap-independent translation, translation recoding on initiation and termination sites, and post-translation processes. This review focuses on advances in cap-independent translation and translation recoding in plant viruses.
1. Introduction
Plant viruses usually encode several viral proteins essential for the important processes in the viral life cycle, such as replication, translation, movement, and virus packaging. However, translation of viral proteins totally depends on the host translation machinery, which prefers RNA templates with the same characteristics as cellular mRNA. For DNA viruses, the genomic transcripts have the same characteristics as cellular mRNA since their viral genome enters the nucleus. Translation of the viral RNA for DNA viruses is accomplished using the canonical ribosome scanning model. For RNA viruses, their viral genome does not enter the nucleus and remains the intrinsic characteristic of the virus particle. The genomic RNA of many plant RNA viruses lacks the 5′ cap and/or 3′ poly(A) as cellular mRNA, which are essential factors ensuring the effective translation of cellular mRNA. In addition, the viral RNA of some RNA viruses is multi-cistronic, which is remarkably different from mono-cistronic cellular mRNA. It is suggested that the internal open reading frame (ORF) and ORFs located at the 3’ part may be expressed by several strategy. Although most plant RNA viruses present differences in terms of the 5′ end, 3′ end, and encoding characteristic from cellular mRNA, they accomplish the expression of viral proteins through many strategies, including the synthesis of subgenomic RNA at the transcriptional level, cap-independent translation, and translation recoding at the translational level [1,2,3]. This review focuses on advances in cap-independent translation and translation recoding in plant viruses.
2. Characteristics of 5′ and 3′ Ends of Viral RNAs in Plant RNA Viruses
To study the translation of viral proteins, the characteristics of the 5′ and 3′ ends of the viral RNA are the primary factors to be considered. If viral RNA has a capped 5′ end and polyadenylated 3′ end, it can express the viral protein through the canonical ribosome scanning model as cellular mRNA. In the ribosome scanning model of eukaryotic cellular mRNA, the 7-methylguanosine cap at the 5′ end of cellular mRNA is first bound by eIF4E, part of the eIF4F complex, which recruits the 43S preinitiation complex, including the 40S small ribosomal subunit, eIF2–GTP–Met-tRNAi ternary complex, along with eIF1, eIF1A, eIF3, and eIF5. The resulting 48S preinitiation complex is then scanned from 5′ to 3′ in an ATP-dependent manner until the charged initiator Met-tRNAi is base-paired with an AUG start codon surrounding by suitable context sequences. A number of initiation factors are then displaced to allow the joining of the 60S large ribosomal subunit to form the complete 80S ribosome to initiate translation. The 3′ poly (A) can be bound by PABP, which also binds eIF4G to cyclize the cellular mRNA, greatly enhancing translation by avoiding cellular mRNA decay and cyclic utilization of ribosomal subunits [4,5].
Here, all genera of plant RNA viruses are listed with characteristics of the 5′ and 3′ ends, as well as the potential translation recoding strategy (Table 1). Of the 106 assigned genera of plant RNA viruses [6,7], only 18 (17.0%) had genomic RNA with both 5′ cap and 3′ poly(A), while 24 (22.6%) had genomic RNA without 5′ cap and 3′ poly(A) (Table 1). In addition, 64 (60.4%) had genomic RNA with a 5′ cap (30 genera) or 3′ poly(A) (34 genera) (Table 1). For genera without the 5′ cap, seven in the family Secoviridae, two in the family Solemoviridae, 12 in the family Potyviridae of the order Patatavirales, and three in the family Luteoviridae had a viral genome-linked protein (VPg) at the 5′ end of the genomic RNA. For genera without 3′ poly(A), six in the family Bromoviridae, seven in the family Virgaviridae, and one in the family Tymoviridae had a 3′ tRNA-like structure (Table 1). There were 58 genera (54.7%) of plant RNA viruses that lack the 5′ cap (Table 1), which may translate the viral proteins through a cap-independent translation strategy to recruit translation factors in different manners.

Table 1.
Characteristic of 5′ and 3′ ends of plant RNA viruses and their potential translational recoding strategy.
3. Cap-Independent Translation in Plant RNA Viruses
Cap-independent translation in plant RNA viruses is often mediated by two types of cis-elements: internal ribosome entry site (IRES) and 3′ cap-independent translation enhancer (3′ CITE). IRES was first reported in animal RNA viruses such as poliovirus and Encephalomyocarditis virus (EMCV), and it is mainly located at the 5′ upstream region of corresponding open reading frames (ORFs) [102,103]. 3′CITE was first reported in plant RNA viruses, such as satellite tobacco necrosis virus (sTNV), and it is located at 3′ downstream of the corresponding ORFs [2,104,105]. In addition to RNA viruses, IRES and 3′CITE were reported in eukaryotic cellular mRNAs, which may play a role when cap recognition is suppressed under special conditions and act as modulators of enhanced stress resistance, metabolic processes, and development [105,106,107,108].
3.1. IRESes in Plant RNA Viruses
Since IRES was first reported in picornavirus RNAs [102,103], it has been reported in many animal and plant RNA viruses, as well as in host cellular mRNAs [109,110,111,112,113]. For animal RNA viruses, viral IRESes are classified into six classes on the basis of their structural characteristics and the requirements for various translation initiation factors and IRES trans-acting factors (ITAFs). Picornavirus IRESes are classified into five types (I, II, III, HCV-like, and AV-like), and dicistrovirus IRESes are classified into another type [114,115,116,117]. The IRES sequences in animal RNA viruses are long (at least 450 nt), and the corresponding RNA tertiary structures are very complex. Various translation initiation factors, as well as ITAFs, are required by these IRESes to play a role during translation initiation [117,118,119,120,121,122,123,124]. There seemed to be an inverse correlation between the degree of the stable structure of the IRES element and the number of factors. Most animal IRESes do not require eIF4E, which is one of the main targets of cellular translation regulation [125]. IRES activity in some animal RNA viruses is also synergistically enhanced by the long-distance RNA–RNA interaction between the 5’ and 3’ ends [126,127,128,129,130,131,132].
Studies on IRESes in plant RNA viruses have mainly focused on members of the family Potyviridae, such as tobacco etch virus (TEV), turnip mosaic virus (TuMV), potato virus Y (PVY), triticum mosaic virus (TriMV), and wheat yellow mosaic virus (WYMV) [111,113,133,134,135,136]. IRESes in plant RNA viruses have shorter sequence lengths (60–190 nt, excluding TriMV) and simpler structures than those in animal RNA viruses. The characteristics of genomic RNA in the family Potyviridae are similar to those in the family Picornaviridae. They all have VPg at the 5’ end and poly (A) at the 3’ end. They all encode a single polyprotein to produce functional proteins via the cleavage of proteinases. However, the 5’UTR of the family Potyviridae is relatively shorter than that of the family Picornaviridae (about 150 nt in Potyviridae and 600–1200 nt in Picornaviridae), and the structure of the 5’UTR in the family Potyviridae is simpler than that in the family Picornaviridae. In addition, the VPg of the family Potyviridae is relatively larger than that in the family Picornaviridae (20–23 kDa in Potyviridae, and 2–3 kDa in Picornaviridae) [111]. VPg can interact with eIF4E or eIFiso4E, which was associated with the regulation on translation of viral RNA and host mRNA in several potyviruses such as TEV, TuMv and PVA [137,138,139,140]. Eukaryotic eIF4E-mediated recessive resistance to plant viruses were reported, which is a direction of virus-resistant crop breeding [141]. In the genus Calicivirus, VPg- eIF4E interaction is required for translation [142]. However, But the VPg of members in the family Picornaviridae is dispensable in the process of translation regulation [143]. TheVPg in the family Picornaviridae can be released from viral RNA though “unlinkase” activity via TDP2 enzyme [144]. Due to the two state of VPg including dissociative state and RNA-linked state, the relationship between VPg and translation is complicated. The possible function and mechanism of VPg in translation are not discussed here. Here, reported IRESes in plant RNA viruses are shown and summarized below.
3.1.1. IRESes in Members of Family Potyviridae
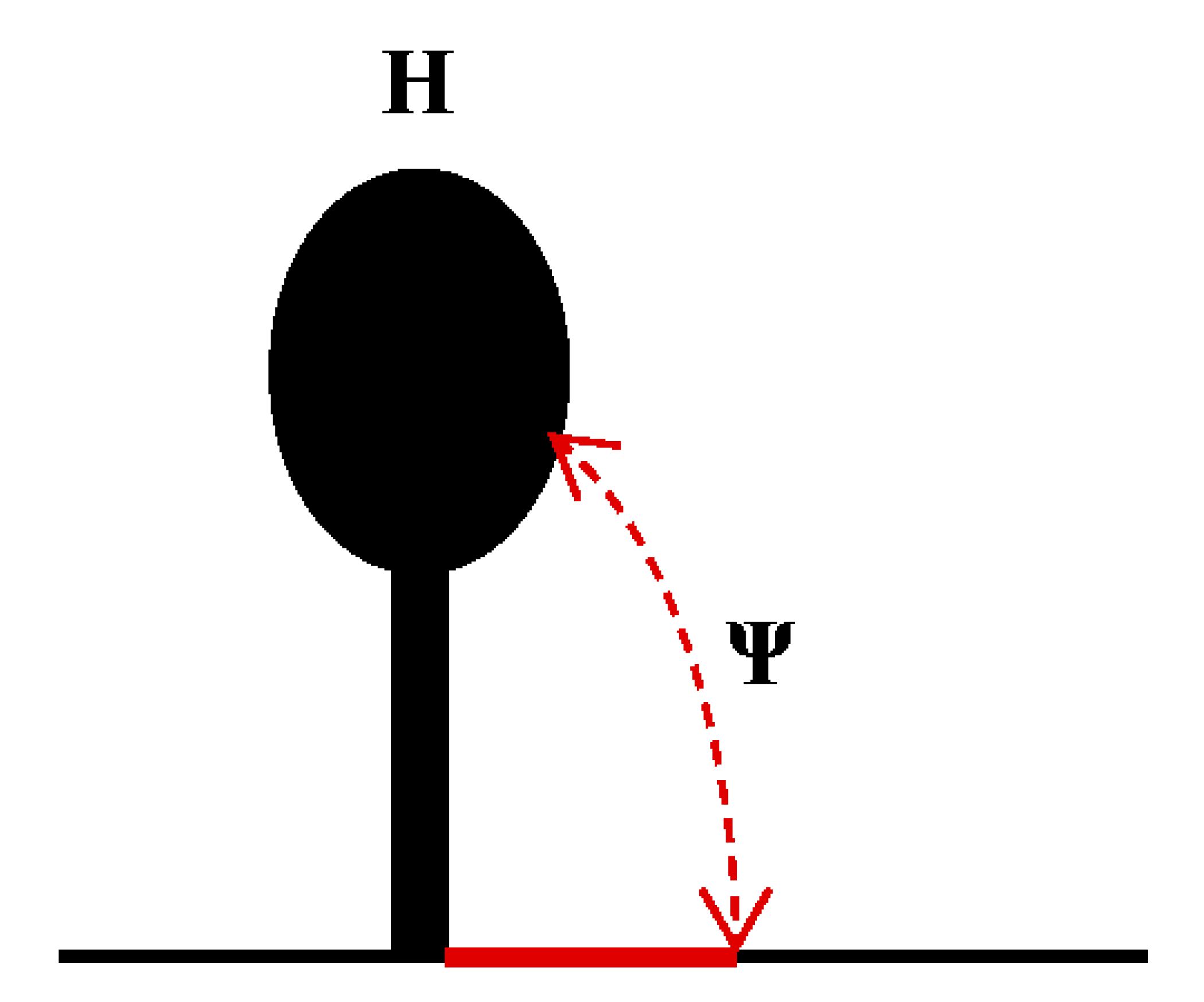
TEV is a model virus used to study the translation regulation of the family Potyviridae [145,146]. Early studies have shown that two regulatory elements (CIRE and CIRE-2) in the 5’UTR (144 nt) of TEV can improve the cap-independent translation efficiency by 8–21-fold [133,147], and poly (A) can synergistically improve the translation efficiency mediated by the 5’UTR [147,148]. The 5’ proximal domain (38–75 nt) in TEV can fold into a pseudoknot (Ψ), which is essential for cap-independent translation [149]. In addition, loop sequences (UACUUCU) in L3 can pair with the 1117–1123 nt of 18S rRNA. It is suggested that the base-pairing sequences between the 5′UTR and 18S RNA may directly recruit ribosomal subunits to enhance translation [149]. When the 5’UTR of TEV is placed in the intergenic region of the bi-cistronic reporter vector, it promotes the expression of the second ORF, indicating IRES activity [150]. When a stable secondary structure is placed before the 5’ end, the IRES efficiency is reduced by nearly 10-fold, indicating that maximal activity of the IRES requires an open 5’ end. [147]. The IRES activity from the 5’UTR of TEV is eIF4F-dependent rather than eIFiso4F-dependent due to the interaction between eIF4G and the 5’UTR of TEV, and this interaction facilitates cap-independent translation [138,150,151].
Insertion of the 5’UTR (184 nt) of PVY into the intergenic region of the bi-cistronic reporter vector can promote the expression of the second gene, which indicates that the 5’UTR of PVY has IRES activity [134]. On the basis of this prediction, the 5’UTR of PVY has two hairpins [152]. Deletion of the first hairpin increases translation, whereas deletion of the second hairpin slightly decreases translation [152]. It is suggested that the first hairpin has a negative effect on IRES activity because of the potential block on the scanning of ribosomes [152]. In addition, the 3’ terminal 55 nt region in the 5’UTR of PVY is crucial for cap-independent translation [152]. However, the detailed mechanism and required translation factors have not yet been determined.
The 5’UTR of TuMV (130 nt) can promote translation in vivo and in vitro, but the exact mechanism is still unclear [135]. When a stable hairpin is added before the TuMV 5’UTR, its translation level is reduced by 70%. The complementary sequences to the 5’UTR of TuMV inhibited cap-independent translation in a trans competition experiment, while the identical complementary sequences located at reporter gene increased translation [135]. It is suggested that both 5’UTR of TuMV and its complementary sequences can support the cap-independent translation.
TriMV is a newly discovered virus that infects wheat. Compared with other members of the genus Potyvirus, its 5’UTR with 739 nt is very long and its opening reading frame starts from the 13th initiation codon [153,154]. The 5’UTR of TriMV enhances cap-independent translation in vivo and in vitro [136]. IRES activity from the TriMV 5’UTR requires a hairpin structure at position 469–490 nt [136]. The 5’UTR of TriMV can directly interact with eIF4G and eIFiso4G, and the hairpin structure at position 469–490 nt is very important for this interaction between the 5’UTR and eIF4G [155]. In addition, cap-independent translation mediated by the 5’UTR also requires eIF4A instead of eIF4E [155].
In PVA, another member of the genus Potyvirus, the 5’UTR (161 nt) without remarkable structure characteristic played a key role in the translation of viral RNA stimulated by VPg and ribosomal protein P0 [156]. It is implied the possible synergistic function of VPg and 5’UTR on translation.
Recently, a novel IRES element was found in the 5′UTR (162 nt) of RNA1 of WYMV [103]. The core elements of IRES in WYMV RNA1 have two hairpins (H1 and H2) and an internal linker region (LR1). IRES activity from the 5′UTR can be synergistically enhanced via long-distance RNA–RNA interaction between C80U in the 5′UTR and A7574G in the 3′UTR [103]. Structural stability of the stem and nucleotide specificity of the upper loop in H1, along with the length of discontinuous stems and nucleotide specificity of the upper loop in H2 are the core cis-element for IRES activity from the 5′UTR [103]. The IRES of WYMV RNA1 5′UTR is eIF4E-dependent, and the target site of eIF4E is the top loop of H2, especially C114UUUCC [103]. In addition, the cytosines (C55, C66, C105, and C108) in the hairpins H1 and H2 and the guanines (G73, G79, and G85) in LR1 form discontinuous base pairing to maintain a dynamic equilibrium state. Dynamic base pairs between C55 and C66 in H1 and guanines (G73, G79, and G85) in LR1 have positive effects on IRES activity, while dynamic base pairs between C105 and C108 in H2 and guanines (G73, G79, and G85) in LR1 negatively regulate the IRES activity [103]. Dynamic base pairs among cytosines (C55, C66, C105, and C108) in H1/H2 and guanines (G73, G79, and G85) in LR1 maintain a tertiary equilibrium state to ensure that the IRES activity of the RNA1 5’UTR is at a suitable level, which is suggested to be the evolution target of WYMV RNA1 [103].
3.1.2. IRESes in Other Plant RNA Viruses
In addition to IRES found in species of the family Potyviridae, it has been reported in other types of plant RNA viruses. The upstream coat protein (CP) ORF in TCV contains an IRES, which can regulate the expression of CP protein [157]. The low-level expression of CP protein can be detected even if CP subgenomic RNA is not synthesized. The IRES located upstream of the CP ORF in TCV does not present structural characteristics, and the IRES activity is related to an unstructured A-rich sequence. Moreover, IRES activity depends on eIF4G instead of eIF4E [157]. IRES with A-rich sequences has also been found upstream of the CP and MP ORFs in crTMV [158]. Similar IRES elements have also been found in a variety of viruses of the family Tombusviridae, such as HCRSV and PFBV [159,160]. These IRES elements present unstructured characteristic [157,158,159,160]. An IRES has been found in the 5’UTR of RNA2 of the blackcurrant conversion virus (BRV), a Nepovirus. This IRES did not present a remarkable secondary structure, but it did contain multiple segments of an 8–10 nt sequence motif essential for IRES activity, which can complement the position of the 1113–1123 nt region of 18S rRNA [161]. Subsequently, similar regions complementary to 18S rRNA have been found in the 5’UTR of other species of Nepovirus. It is speculated that these IRESes may directly recruit 40S subunit, because 18s rRNA is part of the 40S subunit [161]. IRES has also been found in the genome of the potato leaf roll virus (PLRV). The IRES is completely located in the ORF. The core cis-elements include a conserved AUG codon and adjacent inverse symmetric motif (GGAGAGAGAGG) [162].
To date, IRESes of plant RNA viruses have presented multifarious structural characteristics and can be roughly divided into three types according to their structural characteristics (Table 2). Type I IRES is unstructured and generally contains a section of an A-rich sequence. The representative viruses are TCV/HCRSV/PFBV/BRV. Type II IRES is structural. All of these IRESes have one or several hairpins. According to the numbers and other characteristics of hairpin structures, they can be divided into the single hairpin type such as in TEV, the double hairpin type such as in PVY and TriMV, and the equilibrium state structure type such as in WYMV. In WYMV, there are two hairpins in IRES with an equilibrium-state structure, which is mediated by discontinuous C–G base pairing between the two hairpins: dynamic base pairs among cytosines (C55, C66, C105, and C108) in two hairpins and guanines (G73, G79, and G85) in the linker region. In addition to types I and II, there are several IRESes whose structural characteristics are unclear. The IRES in TuMV/PVA/PLRV has been classified as type III. Although there are three type of IRESes based on their structural characteristics, IRESes in plant RNA viruses appear to have simpler structure than that in animal RNA viruses. Similarly, IRESes of mammalian cellular mRNA are also less structured than that in animal RNA viruses [163,164]. In addition, IREses in yeast and fruit fly exhibit a weak secondary structure, which was correlated with high IRES activity [165]. With the exception of animal RNA viruses, RNAs from other resources including plant RNA viruses contain less structured IRESes. The less structured nature of IRESes in plant RNA viruses may be related to the shorter 5′UTR than that in animal RNA viruses. In addition, the 5′UTR length (a median length of approximately 53–218 nucleotides) of cellular mRNA appears to be shorter than that of IRESes (at least 450 nt) in animal RNA viruses [166]. A subset of eukaryotic IRESs exhibit very low secondary structure in the 5′UTR sequences immediately upstream of the initiation codon [165]. However, not all IRESes in plant RNA viruses exhibit a less structure characteristic. For instance, IRESes in WYMV presented a tertiary equilibrium-state structure, which was an alternative complex structure of IRES [103]. It has been suggested that the length of IRES sequences is not the sole determinant on the complexity of tertiary structure. The determinant of structure complexity of IRES can be a future research direction, in addition to the dialectic relationship between IRES structure and IRES activity.

Table 2.
Summary of IRESes in plant RNA viruses.
In addition to the different structural characteristics, IRESes in plant RNA viruses present different mechanisms to recruit translation factors or ribosomes. Some IRESes such as TCV and TEV can bind eIF4G, while others such as WYMV can bind eIF4E. In addition, some IRESes, such as BRV, TEV and TriMV, can directly bind 18S rRNA. Although the genomic RNA of these plant RNA viruses does not contain the 5′cap, they can recruit the translation initiation complex by binding to specific components, such as eIF4E, eIF4G, and/or 18S rRNA. To date, IRESes of plant RNA viruses have appeared to function without the assistance of ITAF, while some IRESes in animal RNA viruses and cellular mRNA required ITAF [118,119,120,121,122,123,124,164,167,168]. Whether some potential ITAFs modulate the activity of IRESes in plant RNA viruses can be a future research direction. According to the sequence or structural characteristics of these IRESes in plant RNA viruses, a potential control strategy for the plant RNA viruses was tested. In studies of TuMV, TriMV and WYMV, complementary oligonucleotide with core cis-elements shows remarkable inhibition on translation [113,135], which implied that complementary oligonucleotides could be efficient agents against plant disease through the inhibition of translation of viral proteins. In addition, other types of small molecules, such as specific nucleotides and peptides, can be used as inhibitors of viral disease if they can block the essential interactions of IRESes and host translation factors or ribosomes. These types of molecules have been reported in studies of animal RNA viruses [169,170,171,172,173,174], which implies that this strategy can be applied to the control of diseases caused by plant RNA viruses. Small molecules blocking the core region in IRESes or the essential interaction between IRESes and host translation factors or ribosomes are potential agents for the management of viruses. In addition to resolving the detailed characteristic of core cis-elements in IRESes, the precise interaction sites between IRESes and host translation factors or ribosomes can be a future research direction, which will provide insight into the design of small-molecule blocking agents.
The primary function of IRES is the regulation of cap-independent translation, and different types of IRESes have been identified in some plant RNA viruses, which recruit translational initiation factors in different manners (Table 2). In addition to regulating cap-independent translation, the IRES of WYMV can play a positive role in regulating the translation of RNA with 5′ cap [113]. It implied a potential interaction between IRES and the 5′ cap in translation. Similarly, the 5′UTR (14 nt and 21 nt) of RNA3 and RNA10 in RBSDV presents IRES activity and can enhance the translation of RNA with a 5′cap [175]. The mechanism of the potential synergistic function between the 5′ cap and IRES requires further identification in future.
3.2. 3′CITE in Plant RNA Viruses
The 3’CITE was firstly reported in sTNV [104] and subsequently discovered in a large number of positive-strand RNA plant viruses [2,176,177]. 3’CITEs have also been found in both eukaryotic cells and animal RNA viruses [112,178,179]. In general, 3’CITEs can recruit diverse translation initiation factors or directly recruit and bind to the ribosome subunit, and the translation initiation complex is subsequently brought to the 5’ end of the RNA through long-distance RNA–RNA interaction to initiate translation [2]. Detailed characteristics of 3’CITEs have mainly been determined for plant RNA viruses. According to their recruitment on different host translation initiation factors, RNA structures, and circularization mechanisms, these 3’CITEs can be classified into seven classes: TED, BTE, PTE, TSS, ISS, YSS, and CXTE (Table 3).

Table 3.
Summary of 3′ CITEes in plant RNA viruses.
3.2.1. Translation Enhancer Domain (TED)
TED was discovered in the 3′UTR (619 nt) of sTNV, a parasitic subviral agent, and it enhances translation in vitro and in vivo [104]. Its structure is predicted to form a long hairpin containing several internal bulges and a 6 nt apical loop, which has no strong uninterrupted helices [104,180,198,199]. TED can recruit eIF4F or eIFiso4F, typically preferring eIF4F, which is essential for translation functions [200,201]. The apical loop of TED in sTNV contains sequences complemented by the apical loop of the 5′UTR, while mutation to disrupt the potential base pairing slightly reduces translation [180]. The detailed mechanism of how the ribosome complex recruited by TED is brought to the 5′UTR remains unclear.
Similar TED elements have been experimentally verified for other related viruses. Three carmoviruses including pelargonium line pattern virus (PLPV), pelargonium chlorotic ring pattern virus, and pelargonium ring spot virus have been shown to contain a TED-like element, which has sequences in its apical loops putatively forming a kissing-loop interaction with a 5′ proximal hairpin [187]. The core sequences (YGCCA; Y is a pyrimidine) in the apical loop of the TED-like element are conserved, which mediate the long-distance kissing-loop interaction with the 5′ proximal sequences. When the predicted long-range base pairing with the TED-like element in the PLPV is disrupted, the translation efficiency is reduced to less than 10% of the wt levels. It has been revealed that maintenance of the 5′–3′ gRNA communication is imperative for efficient translation mediated by the TED-like element [98].
3.2.2. Barley Yellow Dwarf Virus (BYDV)-Like Element (BTE)
BTE was first reported in BYDV and subsequently found in all species of the genus Luteovirus, as well as in some species of the genera Necrovirus, Umbravirus, and Dianthovirus [182,202]. BTE contains a highly conserved 17 nt sequence (GGAUCCUGGNRNACAGG, the underlined base pair; N is any base and R is a purine) and a stable stem-loop SL-IIII pairing with the 5′UTR (140 nt) [183,203]. This long-distance RNA–RNA interaction between BTE and the 5′ ends of the gRNAs and/or sgRNAs is necessary for efficient translation [183]. eIF3 can bind to both the UTRs of BYDV to stabilize the 3′UTR–5′UTR interaction and facilitate the transfer of the translation machinery from the 3′BTE to the 5′UTR [204]. BTE of BYDV bound eIF4G with unusually high affinity and recruited translation machinery in an eIF4G-dependent manner [205]. The three-dimensional structure of BTE of BYDV was determined via crystallization and preliminary X-ray diffraction analysis [206].
BTE-like elements have also been reported in tobacco bushy top virus (TBTV) [207]. A study on the BTE of TBTV identified the structural evolution of BTE, which is mediated by the mutation of nucleotides outside of the BTE regions at the 3′ end. It is suggested that other regions at the 3′ end regulate translation by affecting the structure of the BTE region [207]. In addition, we found that the 5′ terminal region of the TBTV genome has a local molecule regulating the formation or deformation of long-distance RNA–RNA interactions between the 5′UTR (10 nt) and BTE (G. Geng and X. Yuan, unpublished data). In addition, a recent study revealed that opium poppy mosaic virus (OPMV) has a BTE at the 3′ end, which also contains another 3′CITE termed TSS [208]. The BTE, not the TSS in OPMV, contributes to the translation of the reporter constructs [208]. In addition to the conserved 17 nt sequences, BTE in species of the genus Umbravirus and seven additional BTEs from species of the family Tombusviridae and Luteoviridae have additional structural and sequence similarities, including the distance between SL1 and SL2, conserved sequences located downstream of SL2 and SL3 [208].
3.2.3. Panicum Mosaic Virus-Like Translation Element (PTE)
PTE is present in several species of the genera Carmovirus and Panicovirus, pea enation mosaic virus RNA 2 (PEMV2) of the genus Umbravirus, and pothos latent virus of the genus Aureusvirus [187]. The PTE consists of a three-way branched helix with a large G-rich bulge (G domain) in the main stem and two helical branches at the branch point with a short C- or pyrimidine-rich bulge (C domain) [186]. The 5′ side hairpin of PTEs, excluding the PTE in PEMV2, has an apical loop complementary to the apical loop of a hairpin at or near the 5′ end of the viral RNA. In PEMV2, the translation complex recruited by the PTE is brought to the 5′ end through the 5′–3′ interaction mediated by kl-TSS. Meanwhile, the PTE of PEMV2 has been shown to bind eIF4E with high affinity [187,188]. MCMV also contains a 3′CITE mainly similar to PTE, termed MTE, which can interact with eIF4E with high affinity [209]. However, MTE lacks a strong pseudoknot, unlike most PTEs, and stimulates cap-independent translation with less efficiency than most PTEs [209].
3.2.4. T-Shaped Structure (TSS)
TSS was first discovered in the turnip crinkle virus (TCV) of the genus Carmovirus [189]. The TSS contains a unique set of three hairpins and two pseudoknots that fold into a structure similar to that of tRNAs, as predicted by molecular modeling and confirmed by small-angle X-ray scattering (SAXS)/NMR, which was the first resolved 3D structure of a 3′CITE [190]. The TSS of TCV binds to the ribosome 60S subunit, and this binding is not only important for TSS activity, but also for circularization of the RNA template [189]. In addition, TSS is a scaffold that forms a highly interactive structure at the 3′ end of TCV, which undergoes a widespread conformation shift upon binding to RNA-dependent RNA polymerase [210,211].
A similar TSS structure has been proposed for the related cardamine chlorotic fleck virus of the genus Carmovirus. The 3′UTR of PEMV2 of the genus Umbravirus contains two functional TSSs. One is termed kl-TSS, located 9 nt upstream of the PTE, and the other is termed 3′TSS, located near the 3′ end of the genomic RNA, which is predicted to fold into structures similar to tRNAs [191,192]. In addition, TSS was also discovered in TBTV of the genus Umbravirus (X. Yuan, unpublished data) and also contained BTE, another type of 3′CITE [207].
3.2.5. I-Shaped Structure (ISS)
ISS, the shortest 3′CITE, has been found in the different genera of the family Tombusviridae such as maize necrotic spot virus of the genus Tombusvirus and melon necrotic spot virus (MNSV) of the genus Carmovirus [194,212]. The ISS consists of a stem-loop structure (approximately 60 nt) with a four-base helix and flanking bulged sequences, which is similar to the RNA structure of TED. However, the sequences and motifs between the ISS and TED are fundamentally different. Two different types of ISS were previously identified in different MNSV isolates: MNSV-Ma5 and MNSV-264. Although there is considerable sequence divergence between the 3′CITEs of the MNSV isolates, all have the shape of an “I”. ISS binds the translation initiation factor eIF4F and engages in an RNA–RNA kissing-loop interaction with a hairpin loop located at the 5′ end of the genomic RNA [194,195,212].
3.2.6. Y-Shaped Structure (YSS)
Nearly all members of the genus Tombusvirus are predicted to have a conserved YSS at the 3′ end [2,99,196]. The YSS consists of three long helices (SL-A, SL-B, and SL-C) protruding from a central hub and folded into structures similar to the shape of a “Y”. Mutations in three extended helices altering the structure of the stem or bulge formations reduce translation mediated by the YSS of TBSV [196]. The YSS of TBSV engages a 5′–3′ RNA–RNA interaction to facilitate cap- and poly(A)-independent translation [99]. YSS has also been discovered in carnation Italian ring spot virus and pelargonium leaf curl virus, whose activity for efficient translation requires eIF4F or eIFiso4F [213].
3.2.7. Cucurbit Aphid-Borne Yellows Virus (CABYV) Xinjiang-Like Translation Element (CXTE)
A small 3′CITE termed CXTE was identified from the 3′UTR of the MNSV-N isolate, which can overcome eIF4E-mediated resistance due to the insertion of 55 nt sequences from CABYV [197]. These 55 nt sequences form a new 3′-CITE called CXTE. In the MNSV-N isolate, CXTE is responsible for recruiting translation machinery, and ISS is responsible for the formation of the 5′–3′ interaction [2,197]. Cap-independent translation mediated by CXTE may occur in an eIF4E-independent manner. This is the first report of 3′CITE transferring between different families by recombination in nature [197].
To date, the detailed characteristics of 3′CITE have been mainly identified in plant RNA viruses. Different 3′CITEs present different structural characteristics and regulate translation in a different manner recruiting host translation machinery (Table 3). Some plant RNA viruses such as species of Umbravirus contain multiple 3′CITEs. BTE, not TSS, in OPMV plays a role in cap-independent translation [208]. However, two types of 3′CITEs (CXTE and ISS) synergistically play a role in translation in the case of MNSV-N [197]. PTE and kl-TSS also synergistically play a role in translation in the case of PEMV 2 [192]. In addition to plant RNA viruses, there are many possible 3′CITEs in animal RNA viruses and eukaryotic cellular mRNAs [112,178,179], whose characterization is the virgin land for cap-independent translation and even translation regulation. This implies the universal existence of 3′CITE in different types of RNA, even in different organisms. Characteristics and mechanisms of 3′CITE in plant RNA viruses, animal RNA viruses, and eukaryotic cellular mRNA need to be identified, which will provide new insight into the translational regulation and evolution of different types of RNA. In addition, the 3′ end of genomic RNA in plant RNA viruses has also been reported to contain cis-elements regulating viral replication, which is another important biological process in viral life cycle. 3′CITE and replication-associated cis-elements located at the 3′end of virus genome may induce reciprocal actions or effects due to the local tertiary structure [210,211], which can be a future research direction for the deeper characterization of 3′CITE. In addition, switching between translation mediated by 3′CITE and replication mediated by corresponding cis-elements may also play an essential role in the viral life cycle, because the 3′ end could undergo a structural change upon binding to RdRp, the core component of viral replication [210].
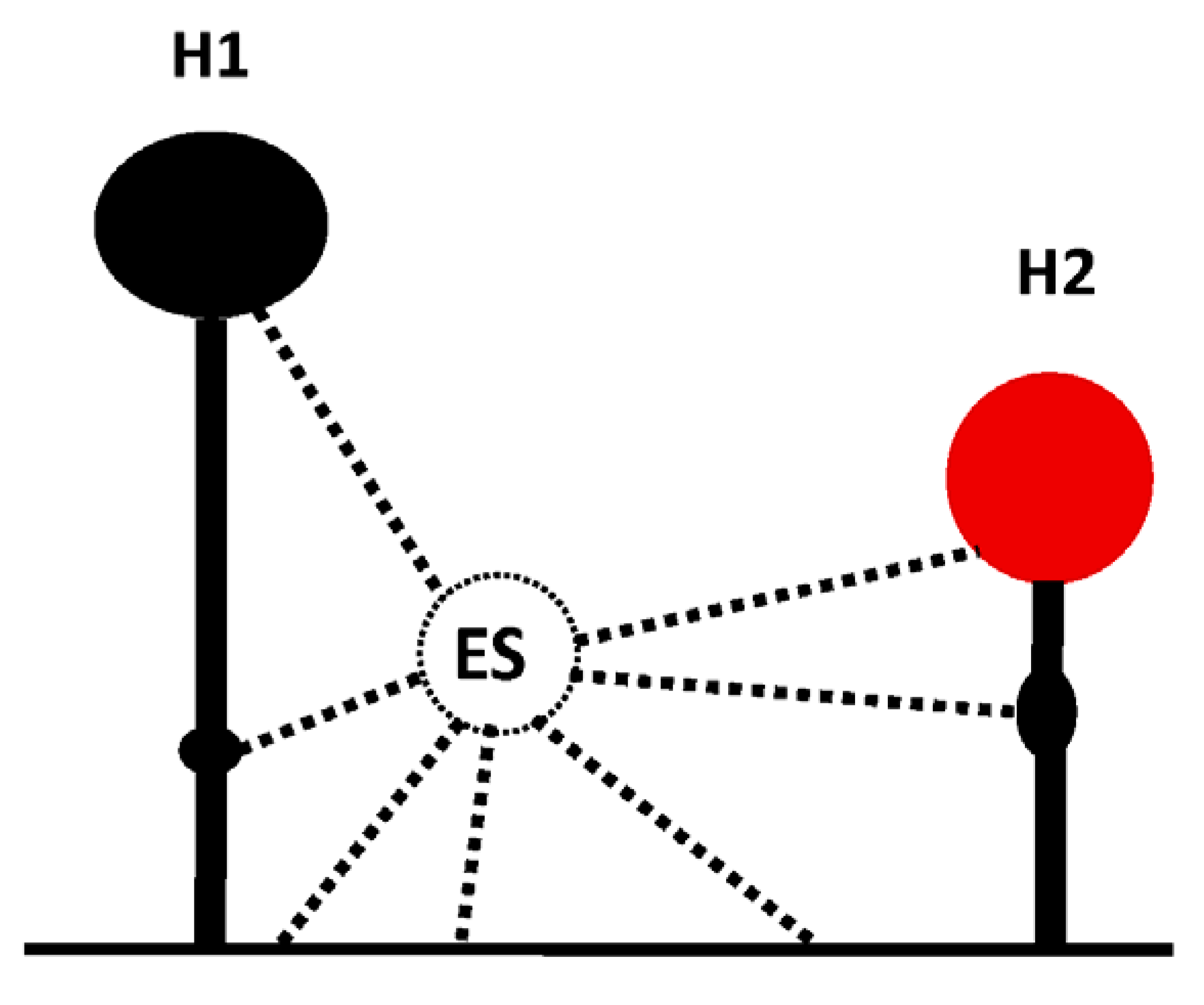
IRES and 3′CITE are two types of cis-elements that mediate the cap-independent translation of viral RNA without the 5′ cap. According to information from previous studies and this review, IRES and 3′CITE in plant RNA viruses appear to present different structural characteristics. IRESes in plant RNA viruses present one of three characteristics: unstructured, structured, or unclear structure (Table 2). All 3′CITEs present seven types of remarkable structural characteristics (Table 3). Although the structure presents variety with few similarities, some conserved characteristics can be identified related to translation initiation factors or ribosome RNA bound by IRES or 3′CITE. According to the reported data, IRES or 3′CITE can bind to at least one of three components including eIF4E, eIF4G, or 18S RNA, which are important components for the translation initiation complex. It is suggested that both IRES and 3′CITE can recruit the translation initiation complex through three different pathways: type I, II, or III (Figure 1). Through the interaction with eIF4E, eIF4G, and/or 18S RNA, IRES or 3′CITE in plant RNA viruses can recruit the host translation initiation complex to ensure the translation initiation of viral RNA (Figure 1). In addition, cap-independent translation is also synergistically enhanced by viral RNA civilization mediated by the long-distance interaction between IRES or 3′CITE and cis-elements located at another end of the viral genome (Figure 1). cis-Elements in IRES and/or 3′CITE involved in the interaction with host translation initiation complex or the civilization of viral RNAs are the target for the design the small-molecule agents for the management of virus diseases.
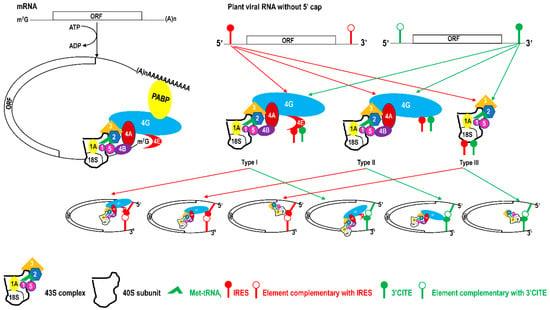
Figure 1.
Schematics of translation initiation in cellular mRNA and plant viral RNA without 5′cap. All eukaryotic initiation factors (eIFs) are indicated by number as 1, 1A, 2, 3, 4A, 4B, 4E, 4G, and 5. PABP, poly(A)-binding protein.
4. Translation Recoding
Cap-independent translation is responsible for the expression of viral proteins located at the 5′ proximal region in viral genomic RNAs or subgenomic RNAs without the 5′ cap. However, some plant RNA viruses are multi-cistronic, and viral proteins located at the internal or 3′ part of genomic RNAs are expressed through other strategies at the transcriptional or translational level. At the transcriptional level, some plant RNA viruses can produce subgenomic RNAs, in which some viral proteins located at the internal or 3′ part of the genomic RNAs are changed to 5′ proximal [1]. At the translational level, these proteins unlocated at the 5′ proximal in genomic or subgenomic RNAs can be expressed through translational recoding, which is an alternative method of identifying the message of the initiation or stop codon of the ORF. Translation recoding includes several types: leaky scanning or non-AUG initiation standing for the recoding of the initiation codon, ribosomal read-through or ribosomal frameshift standing for the recoding of the stop codon, and translational bypassing standing for special expression of the peptide from a discontinuous frame [3,214,215].
4.1. Leaky Scanning
Leaky scanning was first discovered in the genus Orthobunyavirus, in which the ORF of NSs was located completely inside the ORF of N. The NSs are expressed via leaky scanning at the AUG of the ORF of the N protein [216]. Leaky scanning is a mechanism of the translation initiation complex skipping the first initiation codon AUG of the corresponding ORF and initiating translation at the downstream initiation codon AUG due to the nonoptimal context surrounding the first initiation codon AUG [217]. The optical context of the initiation codon AUG is (A/G)CCAUGG in mammalian systems and ACAAUGG in the plant systems, in which purine (A/G) at the −3 position and G at the +4 position are the strongest indicators of translation initiation in animals, plants, and fungi [218,219,220]. This process allows the expression of multiple C-terminally coincident isoforms of a single protein (in-frame alternative initiation sites), distinct proteins encoded by different overlapping ORFs (alternative initiation sites in different reading frames), or even distinct proteins encoded by nonoverlapping continuous ORFs [221].
According to the genome organization of plant RNA viruses, leaky scanning can occur at eight genera in the family Alphaflexiviridae, three genera in the family Betaflexivridae, one genus in the family Benyviridae, one genus in the family Kitaviridae, three genera in the family Luteoviridae, two genera in the family Reoviridae, three genera in the family Secoviridae, two genera in the family Solemoviridae, six genera in the family Virgaviridae, nine genera in the family Tombusviridae, and three genera in the family Tymoviridae (Table 1). Leaky scanning is a universal translation recoding strategy for plant RNA viruses. The 17K protein in BYDV-PAV sgRNA1 is produced by leaky scanning from the start codon (UGAAUGA) of CP ORF, in which the leaky ratio was about 50% [222]. The p39 protein is the leaky scanning product of p23 in the peanut clump virus, in which the ratio of leaky scanning on p23 was 20–30% [223]. βc protein is translated using a leaky scanning mechanism from the start codon ofβd ORF in RNAβ of barley strip mosaic virus [224]. Rice tungro bacilliform virus (RTBV) can express the internal ORFs II and III by leaky scanning [225]. In potato virus X, the 8K ORF is translated by leaky ribosome scanning through the 12K ORF [226]. In the PLPV, the expression of p9.7 is the leaky scanning product from p7 with the ratio of 10% of p7. In addition, p37 is produced through the leaky scanning over the start codon of p7 and non-AUG start codon of p9.7, and the ratio of p37 to p7 was about 50% [227]. Translation of ORF 2a in sobemoviruses is also dependent on the leaky scanning mechanism [228].
4.2. Non-AUG Initiation
The leaky scanning mechanism implies the importance of the context of initiation codon AUG, which can also be confirmed by another translation recoding mechanism termed non-AUG initiation. No AUG initiation was first discovered in the sendai virus in 1988, and translation can be initiated from the ACG codon [229]. In the non-AUG initiation strategy, some codons such as CUG, GUG, ACG, AUU, AUA, AUC, and UUG have been confirmed to initiate translation at a 2–30% level, and CUG surrounded by an optimal context is the most efficient non-AUG initiation codon [230]. Initiation at a non-AUG codon normally requires a strict context such as an A or G at −3 and a G at +4. In addition, a stem loop located at approximately 14 nt downstream of the initiation codon could enhance non-AUG initiation [231,232]. Non-AUG initiation has been discovered in many plant viruses, such as RTBV, species of the family Tombuviridae, and species of the family Panicovirus [225,227,233,234].
4.3. Ribosomal Frameshift
During the special translation process, ribosomes show an abnormal shift of non-three codons on the RNA template, which changes the reading frame of the ORF. This phenomenon is termed ribosomal frameshift [235]. The ribosome can slip one or two nucleotides (−1 or −2 frameshifts) to the 5′ end or one or two nucleotides (+1 or +2 frameshifts) to the 3′ end [25,236,237,238], in which the programmed −1 ribosomal frameshift is the type with detailed characteristics [239]. Programmed −1 ribosomal frameshift was first described by the expression of the Gag–Pol polyprotein of the rous sarcoma alpharetrovirus, which is a chimeric product of overlapping gag and pol ORFs [240,241]. The −1 ribosomal frameshift is involved in several levels of cis-elements: a “slippery site” composed of seven nucleotides with the characteristic XXXYYYZ (X is any base, Y is A or U, and Z is not G) [241,242,243]; a downstream stimulatory structure, typically a stem loop or pseudoknot [244,245,246]; a suitable 5–9 nt between the slippery site and downstream stimulatory structure; an element at the 3′ end forming a long-distance RNA–RNA interaction with the downstream stimulatory structure [247,248,249].
Ribosomal frameshifting is an important gene expression strategy in plant RNA viruses, which can occur in one genus of the family Amalgaviridae, one genus of the family Aspiviridae, four genera of the family Closteroviridae, four genera of the family Luteoviridae, and two genera of the family Solemoviridae (Table 1). The ORF2a–ORF2b protein in PLRV, CP-12K protein in potato virus M, and the Pipo protein in TuMV are expressed via a −1 ribosomal frameshift [250,251]. PEMV2 expresses its RNA polymerase using a −1 ribosomal frameshift, which is regulated through multiple cis-acting elements [252]. The viral RdRp of species of the genus Closterovirus such as CTV and BYV is possibly translated through a +1 frameshift [253]. Viruses of the family Luteoviridae express ORF 2 via a −1 ribosomal frameshift from ORF 1, thereby giving an ORF 1/2 fusion protein [254]. p98 (RdRP) in TBTV is expressed via a −1 ribosomal frameshift [255], which is regulated by downstream kissing-linker and multiple pairs of long-distance RNA–RNA interactions between downstream regions of slippery sequences and the 3′ end (Yu and Yuan, unpublished data).
In addition to the above translational ribosomal frameshift, there is a transcriptional frameshift due to RNA editing on the RNA template to change the sequences of the ORF. The expression of P3N-PIPO in PVY and TuMV, as well as of P1N-PISPO in sweet potato feather mottle virus, has been confirmed through the transcriptional frameshift mechanism [251,256,257,258,259]. The transcriptional frameshift mechanism has also been found in prokaryotes, eukaryotes, and chloroplasts [260,261,262,263,264].
4.4. Ribosomal Read-Through
During the special translation process, the ribosome can fail to terminate translation at the stop codon and pass through the stop codon to produce a C-terminal extended peptide at a proportion of 0.3–5%. This phenomenon during translation is termed ribosomal read-through [265,266]. Ribosomal read-through was first discovered in TMV, in which read-through of an amber stop codon produced a 183K protein, which requires the existence of two naturally tyrosine-specific suppressor tRNAs [267,268,269]. Ribosomal read-through may be involved in several cis-elements including suppressor tRNA and a local stimulatory structure, such as a stem loop or pseudoknot, downstream of the stop codon, which is an element at the proximal 3′ end engaging in long-distance RNA–RNA interaction with the local stimulatory structure [270,271,272,273,274,275].
In plant RNA viruses, ribosomal read-through can occur in the genus Benyvirus in the family Benyviridae, three genera in the family Luteoviridae, seven genera in the family Virgaviridae, and 14 genera in the family Tombusviridae (Table 1) and is used to express two types of proteins: including replicase and CP [271]. The RNA replicase of TMV is translated by read-through [276]. In the genus Tobamovirus, the read-through replicase p182 is sufficient for viral replication and transcription [277]. The CP of BYDV is expressed by readthrough [278]. Tobacco necrosis virus-D expresses its polymerase via read-through [279]. The family Luteoviridae encodes two forms of CP: the major component, CP, and read-through protein (CP readthrough domain (RTD)) [280]. The readthrough (RT) product of CP is involved in vector transmission through specific interactions between CP-readthrough and vector proteins [270,281,282]. The ratio of CP-RT to CP is regulated such that the surface of the virion contains a suitable CP-RT to facilitate vector transmission. Replicases in the family Tombusviridae and Virgaviridae are expressed via read-through; the ratio of read-through is about 5–10% and is regulated by several cis-elements [283]. During genome replication, plant RNA viruses produce dsRNA, which can induce gene-silencing cleavage. These plant RNA viruses express replicase at a suitable level via read-through to control the speed of replication of the genome.
Although different types of translation recoding in plant RNA viruses present different mechanisms involving different cis-elements and various viral or host trans-factors, the expression of translation recoding products at the suitable level is the common point for different RNA viruses. The suitable ratio of translation recoding is essential for fitness of corresponding RNA viruses, suggesting that small molecules interfering with the suitable ratio of translation recoding in plant RNA viruses can be effective agents for the management of virus diseases. The design of this type of small molecule relies on the detailed characterization of cis-elements or trans-factors involved in the different translation recoding processes, which is also a future research direction in translation recoding.
5. Conclusions
IRESes and 3’CITEes in plant RNA viruses presented different structure characteristic in different viruses and recruited host translation machinery through eIF4E, eIF4G or 18s rRNA. However, detailed information about interaction between cap-independent translation elements and translation machinery was rough, which require further identification in future. Both ribosomal frameshift and ribosomal read through in plant RNA viruses were involved in several levels of cis-elements, but detailed characterization on structure of these core cis-elements was few and rough. In addition, potential relationship between the ratio of translation recoding and virus fitness was unclear. During translation regulation of plant RNA viruses, different RNA cis-element may undergo structure shift on the interaction with other cis- or- trans- element and/or factors. However, the study on the structure shift of cis-elements was few. All these information gap will be future direction.
Translation of viral proteins is a vital process during the life cycle of viruses. Comprehensive and persistent identification of different translation strategies in plant RNA viruses will provide new insights into translation regulation and new mechanisms in virus evolution, which will result in new strategies, target sites, and agents for the management of viral diseases of plants.
Author Contributions
Data curation, G.G., D.W., C.Y. and X.Y.; writing-original draft preparation, G.G., D.W., C.Y. and X.Y.; writing-review and editing, G.G., Z.L., Y.W., M.Z., C.Y. and X.Y.; visualization, X.Y.; supervision, X.Y.; funding acquisition, G.G., D.W., X.C. and X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (31872638, 32072382, 32100132, 32102143, 32001867), the China Postdoctoral Science Foundation (2021M702026), and the Shandong Province Natural Sciences Foundation of China (ZR2021QC008, ZR2019PC011, ZR2020QC129).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to acknowledge the funding from National Natural Science Foundation of China, China Postdoctoral Science Foundation, and Shandong Province Natural Sciences Foundation of China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Miller, W.A.; Koev, G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 2000, 273, 1–8. [Google Scholar] [CrossRef]
- Simon, A.E.; Miller, W.A. 3′ Cap-independent translation enhancers of plant viruses. Annu. Rev. Microbiol. 2013, 67, 21–42. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Korniy, N.; Klimova, M.; Karki, P.; Peng, B.Z.; Senyushkina, T.; Belardinelli, R.; Maracci, C.; Wohlgemuth, I.; Samatova, E.; et al. Translational recoding: Canonical translation mechanisms reinterpreted. Nucleic Acids Res. 2020, 48, 1056–1067. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 2017, 42, 589–611. [Google Scholar] [CrossRef] [PubMed]
- Shirokikh, N.E.; Preiss, T. Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. Wiley Interdiscip. Rev. RNA 2018, 9, e1473. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Yu, C. Data analysis on change between the ICTV 2017 release and the ICTV 9th report. Acta Phytopathol. Sin. 2019, 49, 145–150. (In Chinese) [Google Scholar] [CrossRef]
- Hong, J.; Xie, L.; Zhang, Z.K.; Zhou, X.P. Plant viruses in the new 15-rank taxonomic system of ICTV. Acta Phytopathol. Sin. 2021, 51, 143–162. (In Chinese) [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; Mielke-Ehret, N.; Muehlbach, H.P.; Martelli, G.P. ICTV virus taxonomy profile: Fimoviridae. J. Gen. Virol. 2018, 99, 1478–1479. [Google Scholar] [CrossRef]
- Chen, Z.; Anane, R.F.; Wang, Z.; Gao, L.; Chen, L.; Li, S.; Wen, G.; Zhao, M. Complete genome sequence analysis of a novel coguvirus isolated from Paris polyphylla var. yunnanensis. Arch. Virol. 2021, 166, 2045–2050. [Google Scholar] [CrossRef]
- Diaz-Lara, A.; Navarro, B.; Di Serio, F.; Stevens, K.; Hwang, M.S.; Kohl, J.; Vu, S.T.; Falk, B.W.; Golino, D.; Al Rwahnih, M. Two novel negative-sense RNA viruses infecting grapevine are members of a newly proposed genus within the family Phenuiviridae. Viruses 2019, 11, 685. [Google Scholar] [CrossRef]
- Falk, B.W.; James, H.T. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 1998, 36, 139–163. [Google Scholar] [CrossRef]
- German, T.L.; Ullman, D.E.; Moyer, J.W. Tospoviruses: Diagnosis, molecular biology, phylogeny, and vector relationships. Annu. Rev. Phytopathol. 1992, 30, 315–348. [Google Scholar] [CrossRef]
- Zhan, B.; Cao, M.; Wang, K.; Wang, X.; Zhou, X. Detection and characterization of cucumis melo cryptic virus, cucumis melo amalgavirus 1, and melon necrotic spot virus in Cucumis melo. Viruses 2019, 11, 81. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, M.; Chen, Q.; Zhu, M.; Zhou, E. A novel mycovirus closely related to viruses in the genus Alphapartitivirus confers hypovirulence in the phytopathogenic fungus Rhizoctonia solani. Virology 2014, 456, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Lin, Y.H.; Kondo, H.; Kanematsu, S.; Suzuki, N. A novel betapartitivirus RnPV6 from Rosellinia necatrix tolerates host RNA silencing but is interfered by its defective RNAs. Virus Res. 2016, 219, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Nibert, M.L.; Ghabrial, S.A.; Maiss, E.; Lesker, T.; Vainio, E.J.; Jiang, D.; Suzuki, N. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014, 188, 128–141. [Google Scholar] [CrossRef]
- Saito, M.; Kiguchi, T.; Kusume, T.; Tamada, T. Complete nucleotide sequence of the Japanese isolate S of beet necrotic yellow vein virus RNA and comparison with European isolates. Arch. Virol. 1996, 141, 2163–2175. [Google Scholar] [CrossRef] [PubMed]
- Neeleman, L.; Linthorst, H.J.; Bol, J.F. Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J. Gen. Virol. 2004, 85, 231–240. [Google Scholar] [CrossRef]
- Finetti-Sialer, M.; Gallitelli, D. Complete nucleotide sequence of Pelargonium zonate spot virus and its relationship with the family Bromoviridae. J. Gen. Virol. 2003, 84, 3143–3151. [Google Scholar] [CrossRef]
- Gallie, D.R.; Kobayashi, M. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene 1994, 142, 159–165. [Google Scholar] [CrossRef]
- Rizzo, T.M.; Palukaitis, P. Nucleotide sequence and evolutionary relationships of cucumber mosaic virus (CMV) strains: CMV RNA 2. J. Gen. Virol. 1988, 69, 1777–1787. [Google Scholar] [CrossRef]
- Scott, S.W.; Zimmerman, M.T.; Ge, X. The sequence of RNA 1 and RNA 2 of tobacco streak virus: Additional evidence for the inclusion of alfalfa mosaic virus in the genus Ilarvirus. Arch. Virol. 1998, 143, 1187–1198. [Google Scholar] [CrossRef]
- Grieco, F.; Dell’Orco, M.; Martelli, G.P. The nucleotide sequence of RNA1 and RNA2 of olive latent virus 2 and its relationships in the family Bromoviridae. J. Gen. Virol. 1996, 77, 2637–2644. [Google Scholar] [CrossRef] [PubMed]
- Ling, K.S.; Zhu, H.Y.; Drong, R.F.; Slightom, J.L.; McFerson, J.R.; Gonsalves, D. Nucleotide sequence of the 3’-terminal two-thirds of the grapevine leafroll-associated virus-3 genome reveals a typical monopartite closterovirus. J. Gen. Virol. 1998, 79, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Agranovsky, A.A.; Koonin, E.V.; Boyko, V.P.; Maiss, E.; Frötschl, R.; Lunina, N.A.; Atabekov, J.G. Beet yellows closterovirus: Complete genome structure and identification of a leader papain-like thiol protease. Virology 1994, 198, 311–324. [Google Scholar] [CrossRef]
- Klaassen, V.A.; Boeshore, M.L.; Koonin, E.V.; Tian, T.; Falk, B.W. Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly-transmitted, bipartite closterovirus. Virology 1995, 208, 99–110. [Google Scholar] [CrossRef]
- Yu, H.; Qi, S.; Chang, Z.; Rong, Q.; Akinyemi, I.A.; Wu, Q. Complete genome sequence of a novel velarivirus infecting areca palm in China. Arch. Virol. 2015, 160, 2367–2370. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.A.; Khalifa, M.E.; Okada, R.; Fukuhara, T.; Sabanadzovic, S. ICTV virus taxonomy profile: Endornaviridae. J. Gen. Virol. 2019, 100, 1204–1205. [Google Scholar] [CrossRef]
- Quito-Avila, D.F.; Brannen, P.M.; Cline, W.O.; Harmon, P.F.; Martin, R.R. Genetic characterization of Blueberry necrotic ring blotch virus, a novel RNA virus with unique genetic features. J. Gen. Virol. 2013, 94, 1426–1434. [Google Scholar] [CrossRef] [PubMed]
- Locali-Fabris, E.C.; Freitas-Astúa, J.; Souza, A.D.; Takita, M.A.; Astúa-Monge, G.; Antonioli-Luizon, R.; Rodrigues, V.; Targon, M.L.P.N.; Machado, M.A. Complete nucleotide sequence, genomic organization and phylogenetic analysis of Citrus leprosis virus cytoplasmic type. J. Gen. Virol. 2006, 87, 2721–2729. [Google Scholar] [CrossRef]
- Melzer, M.J.; Sether, D.M.; Borth, W.B.; Hu, J.S. Characterization of a virus infecting Citrus volkameriana with citrus leprosis-like symptoms. Phytopathology 2012, 102, 122–127. [Google Scholar] [CrossRef]
- Ziegler, A.; Natsuaki, T.; Mayo, M.A.; Jolly, C.A.; Murant, A.F. The nucleotide sequence of RNA-1 of raspberry bushy dwarf virus. J. Gen. Virol. 1992, 73, 3213–3218. [Google Scholar] [CrossRef]
- Valverde, R.A.; Sabanadzovic, S. A novel plant virus with unique properties infecting Japanese holly fern. J. Gen. Virol. 2009, 90, 2542–2549. [Google Scholar] [CrossRef] [PubMed]
- Shirako, Y.; Wilson, T.M. Complete nucleotide sequence and organization of the bipartite RNA genome of soil-borne wheat mosaic virus. Virology 1993, 195, 16–32. [Google Scholar] [CrossRef]
- Atsumi, G.; Tomita, R.; Yamashita, T.; Sekine, K. A novel virus transmitted through pollination causes ring-spot disease on gentian (Gentiana triflora) ovaries. J. Gen. Virol. 2015, 96, 431–439. [Google Scholar] [CrossRef]
- Gustafson, G.; Armour, S.L. The complete nucleotide sequence of RNA beta from the type strain of barley stripe mosaic virus. Nucleic Acids Res. 1986, 14, 3895–3909. [Google Scholar] [CrossRef]
- Herzog, E.; Guilley, H.; Manohar, S.K.; Dollet, M.; Richards, K.; Fritsch, C.; Jonard, G. Complete nucleotide sequence of peanut clump virus RNA 1 and relationships with other fungus-transmitted rod-shaped viruses. J. Gen. Virol. 1994, 75, 3147–3155. [Google Scholar] [CrossRef]
- Savenkov, E.I.; Sandgren, M.; Valkonen, J.P.T. Complete sequence of RNA 1 and the presence of tRNA-like structures in all RNAs of Potato mop-top virus, genus Pomovirus. J. Gen. Virol. 1999, 80, 2779–2784. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goelet, P.; Lomonossoff, G.P.; Butler, P.J.; Akam, M.E.; Gait, M.J.; Karn, J. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. USA 1982, 79, 5818–5822. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, S.A. Molecular biology of the tobraviruses. J. Gen. Virol. 1999, 80, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Cao, K.; Zhang, Z.; Wang, L.; Li, S. Identification and characterization of a novel rhabdovirus infecting peach in China. Virus Res. 2020, 280, 197905. [Google Scholar] [CrossRef]
- Stenger, D.C.; Burbank, L.P.; Wang, R.; Stewart, A.A.; Mathias, C.; Goodin, M.M. Lost and found: Rediscovery and genomic characterization of sowthistle yellow vein virus after a 30+ year hiatus. Virus Res. 2020, 284, 197987. [Google Scholar] [CrossRef]
- Tsai, C.W.; Redinbaugh, M.G.; Willie, K.J.; Reed, S.; Goodin, M.; Hogenhout, S.A. Complete genome sequence and in planta subcellular localization of maize fine streak virus proteins. J. Virol. 2005, 79, 5304–5314. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dietzgen, R.G.; Callaghan, B.; Wetzel, T.; Dale, J.L. Completion of the genome sequence of Lettuce necrotic yellows virus, type species of the genus Cytorhabdovirus. Virus Res. 2006, 118, 16–22. [Google Scholar] [CrossRef]
- Kondo, H.; Maruyama, K.; Chiba, S.; Andika, I.B.; Suzuki, N. Transcriptional mapping of the messenger and leader RNAs of orchid fleck virus, a bisegmented negative-strand RNA virus. Virology 2014, 452, 166–174. [Google Scholar] [CrossRef]
- Sasaya, T.; Ishikawa, K.; Koganezawa, H. The nucleotide sequence of RNA1 of Lettuce big-vein virus, genus Varicosavirus, reveals its relation to nonsegmented negative-strand RNA viruses. Virology 2002, 297, 289–297. [Google Scholar] [CrossRef]
- Rastgou, M.; Habibi, M.K.; Izadpanah, K.; Masenga, V.; Milne, R.G.; Wolf, Y.I.; Koonin, E.V.; Turina, M. Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 2009, 90, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Ran, M.; Li, Z.; Hu, M.; Zheng, L.; Liu, W.; Jin, P.; Miao, W.; Zhou, P.; Shen, W.; et al. Analysis of the complete genomic sequence of a novel virus, areca palm necrotic spindle-spot virus, reveals the existence of a new genus in the family Potyviridae. Arch. Virol. 2018, 163, 3471–3475. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Kwak, H.R.; Kim, M.K.; Kim, J.S.; Choi, H.S. The complete genome sequence of a novel virus, bellflower veinal mottle virus, suggests the existence of a new genus within the family Potyviridae. Arch. Virol. 2017, 162, 2457–2461. [Google Scholar] [CrossRef] [PubMed]
- Susaimuthu, J.; Tzanetakis, I.E.; Gergerich, R.C.; Martin, R.R. A member of a new genus in the Potyviridae infects Rubus. Virus Res. 2008, 131, 145–151. [Google Scholar] [CrossRef]
- Geng, G.W.; Yu, C.M.; Wang, D.Y.; Gu, K.; Shi, K.R.; Li, X.D.; Tian, Y.P.; Yuan, X.F. Evolutional analysis of two new isolates of Wheat yellow mosaic virus from Shandong Province, China. Acta Phytopathol. Sin. 2017, 47, 348–356. (In Chinese) [Google Scholar] [CrossRef]
- Rose, H.; Döring, I.; Vetten, H.J.; Menzel, W.; Richert-Pöggeler, K.R.; Maiss, E. Complete genome sequence and construction of an infectious full-length cDNA clone of celery latent virus—An unusual member of a putative new genus within the Potyviridae. J. Gen. Virol. 2019, 100, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Colinet, D.; Kummert, J.; Lepoivre, P. The nucleotide sequence and genome organization of the whitefly transmitted sweetpotato mild mottle virus: A close relationship with members of the family Potyviridae. Virus Res. 1998, 53, 187–196. [Google Scholar] [CrossRef]
- Badge, J.; Robinson, D.J.; Brunt, A.A.; Foster, G.D. 3’-Terminal sequences of the RNA genomes of narcissus latent and Maclura mosaic viruses suggest that they represent a new genus of the Potyviridae. J. Gen. Virol. 1997, 78, 253–257. [Google Scholar] [CrossRef]
- Wylie, S.J.; Tan, A.J.; Li, H.; Dixon, K.W.; Jones, M.G. Caladenia virus A, an unusual new member of the family Potyviridae from terrestrial orchids in Western Australia. Arch. Virol. 2012, 157, 2447–2452. [Google Scholar] [CrossRef]
- Robaglia, C.; Durand-Tardif, M.; Tronchet, M.; Boudazin, G.; Astier-Manifacier, S.; Casse-Delbart, F. Nucleotide sequence of potato virus Y (N Strain) genomic RNA. J. Gen. Virol. 1989, 70, 935–947. [Google Scholar] [CrossRef]
- Mollov, D.; Lockhart, B.; Zlesak, D. Complete nucleotide sequence of rose yellow mosaic virus, a novel member of the family Potyviridae. Arch. Virol. 2013, 158, 1917–1923. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, P.; Verbeek, M.; Bos, I. Mite-borne virus isolates from cultivated Allium species, and their classification into two new rymoviruses in the family Potyviridae. Neth. J. Plant Pathol. 1991, 97, 381–399. [Google Scholar] [CrossRef]
- Choi, I.R.; Horken, K.M.; Stenger, D.C.; French, R. Mapping of the P1 proteinase cleavage site in the polyprotein of Wheat streak mosaic virus (genus Tritimovirus). J. Gen. Virol. 2002, 83, 443–450. [Google Scholar] [CrossRef]
- King, A.M.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011; Volume 9, pp. 881–899. [Google Scholar]
- Andrew, M.Q.K.; Michael, J.A.; Eric, B.C.; Elliot, J.L. Order-Picornavirales. In Virus Taxonomy; Elsevier: Amsterdam, The Netherlands, 2012; pp. 835–839. [Google Scholar] [CrossRef]
- Andrew, M.Q.K.; Michael, J.A.; Eric, B.C.; Elliot, J.L. Family-Secoviridae. In Virus Taxonomy; Elsevier: Amsterdam, The Netherlands, 2012; pp. 881–899. [Google Scholar] [CrossRef]
- Fuchs, M.; Schmitt-Keichinger, C.; Sanfaçon, H. Chapter Two—A Renaissance in Nepovirus Research Provides New Insights into their Molecular Interface with Hosts and Vectors. Adv. Virus Res. 2017, 97, 61–105. [Google Scholar] [CrossRef]
- Christopher, J.B.; Colin, R.H.; Frederick, A.M. Chapter 24—Reoviruses. In Fenner and White’s Medical Virology, 5th ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 345–353. [Google Scholar] [CrossRef]
- Hull, R. Chapter 6-Genome composition, organization, and expression. In Plant Virology, 5th ed.; Hull, R., Ed.; Academic Press: Boston, MA, USA, 2014; pp. 247–339. [Google Scholar]
- Lecoq, H.; Desbiez, C. Chapter 3-Viruses of Cucurbit Crops in the Mediterranean Region: An Ever-Changing Picture. Adv. Virus Res. 2012, 84, 67–126. [Google Scholar] [CrossRef]
- Meier, M.; Olspert, A.; Sarmiento, C.; Truve, E. Sobemovirus, Encyclopedia of Virology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2008; pp. 644–652. [Google Scholar] [CrossRef]
- Zavriev, S.K. Allexivirus, Encyclopedia of Virology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2008; pp. 96–98. [Google Scholar] [CrossRef]
- Ryu, K.H.; Hong, J.S. Potexvirus, Encyclopedia of Virology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2008; pp. 310–313. [Google Scholar] [CrossRef]
- Wylie, S.; Li, H.; Jones, M.G. Donkey orchid symptomless virus: A viral ‘platypus’ from Australian terrestrial orchids. PLoS ONE 2013, 8, e79587. [Google Scholar] [CrossRef]
- Huisman, M.J.; Linthorst, H.J.; Bol, J.F.; Cornelissen, J.C. The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus-stranded RNA viruses. J. Gen. Virol. 1988, 69, 1789–1798. [Google Scholar] [CrossRef]
- Kondo, H.; Yoshida, N.; Fujita, M.; Maruyama, K.; Hyodo, K.; Hisano, H.; Tamada, T.; Andika, I.B.; Suzuki, N. Identification of a Novel Quinvirus in the Family Betaflexiviridae That Infects Winter Wheat. Front. Microbiol. 2021, 12, 715545. [Google Scholar] [CrossRef]
- Yoshikawa, N. Capillovirus, Foveavirus, Trichovirus, Vitivirus, Encyclopedia of Virology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2008; pp. 419–427. [Google Scholar] [CrossRef]
- Adams, I.P.; Skelton, A.; Macarthur, R.; Hodges, T.; Hinds, H.; Flint, L.; Nath, P.D.; Boonham, N.; Fox, A. Carrot yellow leaf virus is associated with carrot internal necrosis. PLoS ONE 2014, 9, e109125. [Google Scholar] [CrossRef]
- Veerakone, S.; Liefting, L.W.; Khan, S.; Pal, C.; Tang, J.; Ward, L.I. Partial biological and molecular characterization of a novel citrivirus from Nandina domestica. Arch. Virol. 2021, 166, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Wylie, S.; Jones, M. Hardenbergia virus A, a novel member of the family Betaflexiviridae from a wild legume in Southwest Australia. Arch. Virol. 2011, 156, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Eerakone, S.; Liefting, L.W.; Tang, J.; Ward, L.I. The complete nucleotide sequence and genome organisation of a novel member of the family Betaflexiviridae from Actinidia chinensis. Arch. Virol. 2018, 163, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Rubino, L.; Russo, M.; De Stradis, A.; Martelli, G.P. Tepovirus, a novel genus in the family Betaflexiviridae. Arch. Virol. 2012, 157, 1629–1633. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Zhang, P.; Liu, W.; Ren, Y.; Cao, M.; Wang, X. The complete nucleotide sequence and genome organization of a novel betaflexivirus infecting Citrullus lanatus. Arch. Virol. 2017, 162, 3239–3242. [Google Scholar] [CrossRef] [PubMed]
- Bejerman, N.; Giolitti, F.; Trucco, V.; de Breuil, S.; Dietzgen, R.G.; Lenardon, S. Complete genome sequence of a new enamovirus from Argentina infecting alfalfa plants showing dwarfism symptoms. Arch. Virol. 2016, 161, 2029–2032. [Google Scholar] [CrossRef]
- Martin, R.R.; Keese, P.K.; Young, M.J.; Waterhouse, P.M.; Gerlach, W.L. Evolution and molecular biology of luteoviruses. Annu. Rev. Phytopathol. 1990, 28, 341–363. [Google Scholar] [CrossRef]
- Shang, Q.X.; Xiang, H.Y.; Han, C.G.; Li, D.W.; Yu, J.L. Distribution and molecular diversity of three cucurbit-infecting poleroviruses in China. Virus Res. 2009, 145, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.P.; Sabanadzovic, S.; Abou Ghanem-Sabanadzovic, N.; Saldarelli, P. Maculavirus, a new genus of plant viruses. Arch. Virol. 2002, 147, 1847–1853. [Google Scholar] [CrossRef]
- Edwards, M.C.; Zhang, Z.; Weiland, J.J. Oat blue dwarf marafivirus resembles the tymoviruses in sequence, genome organization, and expression strategy. Virology 1997, 232, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Briand, J.P.; Keith, G.; Guilley, H. Nucleotide sequence at the 5’ extremity of turnip yellow mosaic virus genome RNA. Proc. Natl. Acad. Sci. USA 1978, 75, 3168–3172. [Google Scholar] [CrossRef] [PubMed]
- Morch, M.D.; Boyer, J.C.; Haenni, A.L. Overlapping open reading frames revealed by complete nucleotide sequencing of turnip yellow mosaic virus genomic RNA. Nucleic Acids Res. 1988, 16, 6157–6173. [Google Scholar] [CrossRef] [PubMed]
- Andrew, M.Q.K.; Michael, J.; Adams, E.B.; Carstens, E.J.L. Genus-Umbravirus. In Virus Taxonomy; Elsevier: Amsterdam, The Netherlands, 2012; pp. 1191–1195. [Google Scholar] [CrossRef]
- Monger, W.; Jeffries, C. A new virus, classifiable in the family Tombusviridae, found infecting Solanum tuberosum in the UK. Arch. Virol. 2018, 163, 1585–1594. [Google Scholar] [CrossRef]
- Guilley, H.; Carrington, J.C.; Balàzs, E.; Jonard, G.; Richards, K.; Morris, T.J. Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res. 1985, 13, 6663–6677. [Google Scholar] [CrossRef][Green Version]
- Šafářová, D.; Vavroušková, K.; Candresse, T.; Navrátil, M. Molecular characterization of a novel Aureusvirus infecting elderberry (Sambucus nigra L.). PLoS ONE 2018, 13, e0200506. [Google Scholar] [CrossRef]
- Ishii, A.; Thomas, Y.; Moonga, L.; Nakamura, I.; Ohnuma, A.; Hang’ombe, B.; Takada, A.; Mweene, A.; Sawa, H. Novel arenavirus, Zambia. Emerg. Infect. Dis. 2011, 17, 1921. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Heaton, L.A.; Zuidema, D.; Hillman, B.I.; Morris, T.J. The genome structure of turnip crinkle virus. Virology 1989, 170, 219–226. [Google Scholar] [CrossRef]
- Ciuffreda, P.; Rubino, L.; Russo, M. Molecular cloning and complete nucleotide sequence of galinsoga mosaic virus genomic RNA. Arch. Virol. 1998, 143, 173–180. [Google Scholar] [CrossRef]
- Nagamani, S.; Ankita, T.; Dikshit, H.K.; Bikash, M.; Jain, R.K. Seed transmission of a distinct soybean yellow mottle mosaic virus strain identified from India in natural and experimental hosts. Virus Res. 2020, 280, 197903. [Google Scholar] [CrossRef]
- Scheets, K.; Jordan, R.; White, K.A.; Hernández, C. Pelarspovirus, a proposed new genus in the family Tombusviridae. Arch. Virol. 2015, 160, 2385–2393. [Google Scholar] [CrossRef]
- Stenger, D.C.; French, R. Complete nucleotide sequence of a maize chlorotic mottle virus isolate from Nebraska. Arch. Virol. 2008, 153, 995–997. [Google Scholar] [CrossRef]
- Mart, K. Plant Satellite Viruses (Albetovirus, Aumaivirus, Papanivirus, Virtovirus), Encyclopedia of Virology, 4th ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 581–585. [Google Scholar] [CrossRef]
- Blanco-Pérez, M.; Pérez-Cañamás, M.; Ruiz, L.; Hernández, C. Efficient translation of Pelargonium line pattern virus RNAs relies on a TED-like 3’-translational enhancer that communicates with the corresponding 5’-region through a long-distance RNA-RNA interaction. PLoS ONE 2016, 11, e0152593. [Google Scholar] [CrossRef][Green Version]
- Fabian, M.R.; White, K.A. 5′-3′ RNA-RNA interaction facilitates cap-and poly (A) tail-independent translation of tomato bushy stunt virus mRNA: A potential common mechanism for Tombusviridae. J. Biol. Chem. 2004, 279, 28862–28872. [Google Scholar] [CrossRef] [PubMed]
- Louie, R.; Redinbaugh, M.G.; Gordon, D.T.; Abt, J.J.; Anderson, R.J. Maize necrotic streak virus, a New Maize Virus with Similarity to Species of the Family Tombusviridae. Plant Dis. 2000, 84, 1133–1139. [Google Scholar] [CrossRef]
- Mizumoto, H.; Tatsuta, M.; Kaido, M.; Mise, K.; Okuno, T. Cap-independent translational enhancement by the 3′ untranslated region of Red clover necrotic mosaic virus RNA1. J. Virol. 2003, 77, 12113–12121. [Google Scholar] [CrossRef][Green Version]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Jang, S.K.; Kräusslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef]
- Danthinne, X.; Seurinck, J.; Meulewaeter, F.; Montagu, M.V.; Cornelissenl, M. The 3’ untranslated region of satellite Tobacco necrosis virus RNA stimulates translation in vitro. Mol. Cell. Biol. 1993, 13, 3340–3349. [Google Scholar] [CrossRef]
- Shatsky, I.N.; Terenin, I.M.; Smirnova, V.V.; Andreev, D.E. Cap-Independent Translation: What’s in a Name? Trends Biochem. Sci. 2018, 43, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Dennis, M.D.; Shenberger, J.S.; Stanley, B.A.; Kimball, S.R.; Jefferson, L.S. Hyperglycemia mediates a shift from cap-dependent to cap-independent translation via a 4E-BP1-dependent mechanism. Diabetes 2013, 62, 2204–2214. [Google Scholar] [CrossRef]
- Lacerda, R.; Menezes, J.; Romao, L. More than just scanning: The importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell. Mol. Life Sci. 2017, 74, 1659–1680. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Hinson, A.; Miller, R.A.; Garcia, G.G. Cap-independent translation: A shared mechanism for lifespan extension by rapamycin, acarbose, and 17α-estradiol. Aging Cell 2021, 20, e13345. [Google Scholar] [CrossRef] [PubMed]
- Kieft, J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008, 33, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Tian, S.; Fujii, K.; Kladwang, W.; Barna, M. Rnaregulons in hox 5′UTRs confer ribosome specificity to gene regulation. Nature 2014, 517, 33–38. [Google Scholar] [CrossRef]
- Zhang, J.; Roberts, R.; Rakotondrafara, A.M. The role of the 5′ untranslated regions of Potyviridae in translation. Virus Res. 2015, 206, 74–81. [Google Scholar] [CrossRef]
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016, 351, 240. [Google Scholar] [CrossRef] [PubMed]
- Geng, G.W.; Yu, C.M.; Li, X.D.; Yuan, X.F. A unique internal ribosome entry site (IRES) representing a dynamic equilibrium state of RNA tertiary structure in the 5’UTR of Wheat yellow mosaic virus RNA1. Nucleic Acids Res. 2020, 48, 390–404. [Google Scholar] [CrossRef]
- Hellen, C.U.; de Breyne, S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: Evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007, 81, 5850–5863. [Google Scholar] [CrossRef]
- Martinez-Salas, E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008, 16, 230–237. [Google Scholar] [CrossRef]
- Belsham, G.J. Divergent picornavirus IRES elements. Virus Res. 2009, 139, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Lozano, G.; Martínez-Salas, E. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015, 12, 113–120. [Google Scholar] [CrossRef]
- Jang, S.K.; Wimmer, E. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990, 4, 1560–1572. [Google Scholar] [CrossRef] [PubMed]
- Luz, N.; Beck, E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1991, 65, 6486–6494. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Boddeker, N.; Andino, R. Translation and replication of human rhinovirus type 14 and mengovirus in Xenopus oocytes. J. Virol. 2000, 74, 11983–11987. [Google Scholar] [CrossRef]
- Sean, P.; Nguyen, J.H.; Semler, B.L. Altered interactions between stem-loop IV within the 5′ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: Effects on IRES-mediated translation and viral infectivity. Virology 2009, 389, 45–58. [Google Scholar] [CrossRef]
- Pilipenko, E.V.; Pestova, T.V.; Kolupaeva, V.G.; Khitrina, E.V.; Poperechnaya, A.N.; Agol, V.I.; Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000, 14, 2028–2045. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Li, M.L.; Shih, S.R. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009, 37, 47–59. [Google Scholar] [CrossRef]
- Pineiro, D.; Fernandez, N.; Ramajo, J.; Martinez-Salas, E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 2013, 41, 1017–1028. [Google Scholar] [CrossRef]
- Filbin, M.E.; Kieft, J.S. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 2009, 19, 267–276. [Google Scholar] [CrossRef]
- Melchers, W.J.; Hoenderop, J.G.; Bruins Slot, H.J.; Pleij, C.W.; Pilipenko, E.V.; Agol, V.I.; Galama, J.M. Kissing of the two predominant hairpin loops in the Coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 1997, 71, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Todd, S.; Towner, J.S.; Semler, B.L. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology 1997, 229, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Duque, H.; Palmenberg, A.C. Phenotypic characterization of three phylogenetically conserved stem-loop motifs in the mengovirus 3′untranslated region. J. Virol. 2001, 75, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- Saiz, M.; Gomez, S.; Martinez-Salas, E.; Sobrino, F. Deletion or substitution of the aphthovirus 3′ NCR abrogates infectivity and virus replication. J. Gen. Virol. 2001, 82, 93–101. [Google Scholar] [CrossRef]
- Dobrikova, E.; Florez, P.; Bradrick, S.; Gromeier, M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′nontranslated region. Proc. Natl. Acad. Sci. USA 2003, 100, 15125–15130. [Google Scholar] [CrossRef]
- Serrano, P.; Pulido, M.R.; Saiz, M.; Martinez-Salas, E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 2006, 87, 3013–3022. [Google Scholar] [CrossRef]
- Romero-Lopez, C.; Barroso-Deljesus, A.; Garcia-Sacristan, A.; Briones, C.; Berzal-Herranz, A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′ X-tail region. Nucleic Acids Res. 2014, 42, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Freed, D.D. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 1990, 64, 1590–1597. [Google Scholar] [CrossRef] [PubMed]
- Levis, C.; Astier-Manifacier, S. The 5′ untranslated regionof PVY RNA, even located in an internal position, enables initiationof translation. Virus Genes 1993, 7, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Basso, J.; Dallaire, P.; Charest, P.J.; Devantier, Y.; Laliberte, J.F. Evidence for an internal ribosome entry site within the 5′non-translated region of Turnip mosaic potyvirus RNA. J. Gen. Virol. 1994, 75, 3157–3165. [Google Scholar] [CrossRef]
- Roberts, R.; Zhang, J.; Mayberry, L.K.; Tatineni, S.; Browning, K.S.; Rakotondrafara, A.M. A unique 5′ translation element discovered in Triticum mosaic virus. J. Virol. 2015, 89, 12427–12440. [Google Scholar] [CrossRef]
- Eskelin, K.; Hafrén, A.; Rantalainen, K.I.; Mäkinen, K. Potyviral VPg enhances viral RNA Translation and inhibits reporter mRNA translation in planta. J. Virol. 2011, 85, 9210–9221. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Miyoshi, H.; Gallie, D.R.; Goss, D.J. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: Interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 2008, 283, 1340–1349. [Google Scholar] [CrossRef]
- Miyoshi, H.; Okade, H.; Muto, S.; Suehiro, N.; Nakashima, H.; Tomoo, K.; Natsuaki, T. Turnip mosaic virus VPg interacts with Arabidopsis thaliana eIF(iso)4E and inhibits in vitro translation. Biochimie 2008, 90, 1427–1434. [Google Scholar] [CrossRef]
- Saha, S.; Mäkinen, K. Insights into the functions of eIF4E-binding motif of VPg in potato virus A infection. Viruses 2020, 12, 197. [Google Scholar] [CrossRef]
- Wang, A.; Krishnaswamy, S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 2012, 13, 795–803. [Google Scholar] [CrossRef]
- Goodfellow, I.; Chaudhry, Y.; Gioldasi, I.; Gerondopoulos, A.; Natoni, A.; Labrie, L.; Laliberté, L.; Roberts, L. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 2005, 6, 968–972. [Google Scholar] [CrossRef]
- Langereis, M.A.; Feng, Q.; Nelissen, F.H.T.; Virgen-Slane, R.; van der Heden vanNoort, G.J.; Maciejewski, S.; Filippov, D.V.; SemLer, B.L.; van Delft, F.L.; van Kuppeveld, F.J. Modification of picornavirus genomic RNA using ‘click’ chemistry shows that unlinking of the VPg peptide is dispensable for translation and replication of the incoming viral RNA. Nucleic Acids Res. 2013, 42, 2473–2482. [Google Scholar] [CrossRef]
- Virgen-Slane, R.; Rozovics, J.M.; Fitzgerald, K.D.; Ngo, T.; Chou, W.; van der Heden van Noort, G.J.; Filippov, D.V.; Gershon, P.D.; Semler, B.L. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc. Natl. Acad. Sci. USA 2012, 109, 14634–14639. [Google Scholar] [CrossRef] [PubMed]
- Koziel, M.G.; Hari, V.; Siegel, A. In vitro translation of Tobacco etch virus RNA. Virology 1980, 106, 177–179. [Google Scholar] [CrossRef]
- Allison, R.; Johnston, R.E.; Dougherty, W.G. The nucleotide sequence of the coding region of Tobacco etch virus genomic RNA: Evidence for the synthesis of a single polyprotein. Virology 1986, 154, 9–20. [Google Scholar] [CrossRef]
- Niepel, M.; Gallie, D.R. Identification and characterization of the functional elements within the Tobacco etch virus 5′leader required forcap-independent translation. J. Virol. 1999, 73, 9080–9088. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R.; Tanguay, R.L.; Leathers, V. The Tobacco etch viral 5′leader and poly (A) tail are functionally synergistic regulators of translation. Gene 1995, 165, 233–238. [Google Scholar] [CrossRef]
- Zeenko, V.; Gallie, D.R. Cap-independent translation of Tobacco etch virus is conferred by an RNA pseudoknot in the 5′leader. J. Biol. Chem. 2005, 280, 26813–26824. [Google Scholar] [CrossRef]
- Gallie, D.R. Cap-independent translation conferred by the 5′ leader of Tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 2001, 75, 12141–12152. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Yumak, H.; Domashevskiy, A.; Khan, M.A.; Gallie, D.R.; Goss, D.J. Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J. Biol. Chem. 2006, 281, 35826–35834. [Google Scholar] [CrossRef]
- Yang, L.J.; Hidaka, M.; Sonoda, J.; Masaki, H.; Uozumi, T. Mutational analysis of the potato virus Y 5′ untranslated region for alteration in translationalenhancement in tobacco protoplasts. Biosci. Biotechnol. Biochem. 1997, 61, 2131–2133. [Google Scholar] [CrossRef]
- Seifers, D.L.; Martin, T.J.; Harvey, T.L.; Fellers, G.P.; Stack, J.P.; Ryba-White, M.; Haber, S.; Krokhin, O.; Spicer, V.; Lovat, N.; et al. Triticum mosaic virus: A new virus isolated from wheat in Kansas. Plant Dis. 2008, 92, 808–817. [Google Scholar] [CrossRef]
- Tatineni, S.; Ziems, A.D.; Wegulo, S.N.; French, R. Triticum mosaic virus: A distinct member of the family Potyviridae with an unusually long leader sequence. Phytopathology 2009, 99, 943–950. [Google Scholar] [CrossRef]
- Roberts, R.; Mayberry, L.K.; Browning, K.S.; Rakotondrafara, A.M. The Triticum mosaic virus 5′leader binds to both eIF4G and eIFiso4G for translation. PLoS ONE 2017, 12, e0169602. [Google Scholar] [CrossRef]
- Hafren, A.; Eskelin, K.; Makinen, K. Ribosomal protein P0 promotes Potato virus A infection and functions in viral translation together with VPg and eIF(iso)4E. J. Virol. 2013, 87, 4302–4312. [Google Scholar] [CrossRef]
- May, J.; Johnson, P.; Saleem, H.; Simon, A.E. A sequence-independent, unstructured internal ribosome entry site is responsible for internal expression of the coat protein of Turnip crinkle virus. J. Virol. 2017, 91, e02421-16. [Google Scholar] [CrossRef] [PubMed]
- Dorokhov, Y.L.; Skulachev, M.V.; Ivanov, P.A.; Zvereva, S.D.; Tjulkina, L.G.; Merits, A.; Gleba, Y.Y.; Hohn, T.; Atabekov, J.G. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. USA 2002, 99, 5301–5306. [Google Scholar] [CrossRef]
- Koh, D.C.Y.; Wong, S.M.; Liu, D.X. Synergism of the 3′-untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. J. Biol. Chem. 2003, 278, 20565–20573. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Miragall, O.; Hernández, C. An internal ribosome entry site directs translation of the 3′-gene from Pelargonium flower break virus genomic RNA: Implications for infectivity. PLoS ONE 2011, 6, e22617. [Google Scholar] [CrossRef]
- Karetnikov, A.; Lehto, K. The RNA2 5′ leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J. Gen. Virol. 2007, 88, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Jaag, H.M.; Kawchuk, L.; Rohde, W.; Fischer, R.; Emans, N.; Prüfer, D. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. USA 2003, 100, 8939–8944. [Google Scholar] [CrossRef]
- Komar, A.A.; Hatzoglou, M. Internal ribosome entry sites in cellular mRNAs: Mystery of their existence. J. Biol. Chem. 2005, 280, 23425–23428. [Google Scholar] [CrossRef] [PubMed]
- Komar, A.A.; Hatzoglou, M. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 2011, 10, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Holcik, M. Strong Eukaryotic IRESs Have Weak Secondary Structure. PLoS ONE 2009, 4, e4136. [Google Scholar] [CrossRef] [PubMed]
- Leppek, K.; Das, R.; Barna, M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- King, H.A.; Cobbold, L.C.; Willis, A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010, 38, 1581–1586. [Google Scholar] [CrossRef]
- Faye, M.D.; Holcik, M. The role of IRES trans-acting factors in carcinogenesis. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2014, 1849, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Boerneke, M.A.; Dibrov, S.M.; Gu, J.; Wyles, D.L.; Hermann, T. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc. Natl. Acad. Sci. USA 2014, 111, 15952–15957. [Google Scholar] [CrossRef]
- Dibrov, S.M.; Ding, K.; Brunn, N.D.; Parker, M.A.; Bergdahl, B.M.; Wyles, D.L.; Hermann, T. Structure of a Hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc. Natl. Acad. Sci. USA 2012, 109, 5223–5228. [Google Scholar] [CrossRef]
- Gasparian, A.V.; Neznanov, N.; Jha, S.; Galkin, O.; Moran, J.J.; Gudkov, A.V.; Komar, A.A. Inhibition of encephalomyocarditis virus and poliovirus replication by quinacrine: Implications for the design and discovery of novel antiviral drugs. J. Virol. 2010, 84, 9390–9397. [Google Scholar] [CrossRef]
- Liu, S.; Nelson, C.A.; Xiao, L.; Lu, L.; Seth, P.P.; Davis, D.R.; Hagedorn, C.H. Measuring antiviral activity of benzimidazole molecules that alter IRES RNA structure with an infectious Hepatitis C virus chimera expressing Renilla luciferase. Antiv. Res. 2011, 89, 54–63. [Google Scholar] [CrossRef]
- Lozano, G.; Trapote, A.; Ramajo, J.; Elduque, X.; Grandas, A.; Robles, J.; Martínez-Salas, E. Local RNA flexibility perturbation of the IRES element induced by a novel ligand inhibits viral RNA translation. RNA Biol. 2015, 12, 555–568. [Google Scholar] [CrossRef]
- Seth, P.P.; Miyaji, A.; Jefferson, E.A.; Sannes-Lowery, K.A.; Osgood, S.A.; Propp, S.S.; Ranken, R.; Massire, C.; Sampath, R.; Ecker, D.J. SAR by MS: Discovery of a new class of RNA-binding small molecules for the Hepatitis C virus: Internal ribosome entry site IIA subdomain. J. Med. Chem. 2005, 48, 7099–7102. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Wang, Z.; Yu, C.; Geng, G.; Su, C.; Yuan, X. Effect of untranslated regions of S3 and S10 from rice black-streaked dwarf virus on translation in the absence or presence of 5’cap. Acta Phytopathol. Sin. 2019, 49, 355–361. (In Chinese) [Google Scholar] [CrossRef]
- Truniger, V.; Miras, M.; Aranda, M.A. Structural and functional diversity of plant virus 3’-cap-independent translation enhancers (3’-CITEs). Front. Plant Sci. 2017, 8, 2047. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.B.; Sanfaçon, H. Mapping of sequences in the 5′region and 3′UTR of tomato ringspot virus RNA2 that facilitate cap-independent translation of reporter transcripts in vitro. PLoS ONE 2021, 16, e0249928. [Google Scholar] [CrossRef]
- Keiper, B.D. Cap-independent mRNA translation in germ cells. Int. J. Mol. Sci. 2019, 20, 173. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Man, C.; Xiang, S.; Yao, L.; Wang, X.; Fan, Y. Circular RNAs cap-independent translation protein and its roles in carcinomas. Mol. Cancer 2021, 20, 119. [Google Scholar] [CrossRef]
- Timmer, R.T.; Benkowski, L.A.; Schodin, D.; Lax, S.R.; Metz, A.M.; Ravel, J.M.; Browning, K.S. The 5′and 3′untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′cap structure. J. Biol. Chem. 1993, 268, 9504–9510. [Google Scholar] [CrossRef]
- Meulewaeter, F.; Van, M.M.; Cornelissen, M. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA 1998, 4, 1347–1356. [Google Scholar] [CrossRef][Green Version]
- Wang, S.; Browning, K.S.; Miller, W.A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997, 16, 4107–4116. [Google Scholar] [CrossRef]
- Wang, Z.; Kraft, J.J.; Hui, A.Y.; Miller, W.A. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 2010, 402, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Kraft, J.; Treder, K.; Peterson, M.S.; Miller, W.A. Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with e IF4G. Nucleic Acids Res. 2013, 41, 3398–3413. [Google Scholar] [CrossRef] [PubMed]
- Batten, J.S.; Desvoyes, B.; Yamamura, Y.; Scholthof, K.B.G. A translational enhancer element on the 3’-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006, 580, 2591–2597. [Google Scholar] [CrossRef]
- Wang, Z.; Treder, K.; Miller, W.A. Structure of a viral cap-independent translation element that functions via high affinity binding to the e IF4E subunit of eIF4F. J. Biol. Chem. 2009, 284, 14189–14202. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.; Shi, K.; Yuan, X.; Simon, A.E. Long-distance kissing loop interactions between a 3’ proximal Y-shaped structure and apical loops of 5’ hairpins enhance translation of Saguaro cactus virus. Virology 2011, 417, 113–125. [Google Scholar] [CrossRef]
- Chattopadhyay, M.; Kuhlmann, M.M.; Kumar, K.; Simon, A.E. Position of the kissing-loop interaction associated with PTE-type 3’CITEs can affect enhancement of cap-independent translation. Virology 2014, 458, 43–52. [Google Scholar] [CrossRef]
- McCormack, J.C.; Yuan, X.; Yingling, Y.G.; Kasprzak, W.; Zamor, R.E.; Shapiro, B.A.; Simon, A.E. Structural domains within the 3’ untranslated region of Turnip crinkle virus. J. Virol. 2008, 82, 8706–8720. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Wang, J.; Yu, P.; Eyler, D.; Xu, H.; Starich, M.R.; Tiede, D.M.; Simon, A.E.; Kasprzak, W.; Schwieters, C.D.; et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′UTR of Turnip crinkle virus. Proc. Natl. Acad. Sci. USA 2010, 107, 1385–1390. [Google Scholar] [CrossRef]
- Gao, F.; Kasprzak, W.; Stupina, V.A.; Shapiro, B.A.; Simon, A.E. A ribosome-binding, 3′ translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction. J. Virol. 2012, 86, 9828–9842. [Google Scholar] [CrossRef]
- Gao, F.; Gulay, S.P.; Kasprzak, W.; Dinman, J.D.; Shapiro, B.A.; Simon, A.E. The kissing-loop T-shaped structure translational enhancer of Pea enation mosaic virus can bind simultaneously to ribosomes and a 5’ proximal hairpin. J. Virol. 2013, 87, 11987–12002. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Nieto, C.; Gonzalez-Ibeas, D.; Aranda, M. Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): Cap-independent translation of a viral RNA controlled in cis by an (a) virulence determinant. Plant J. 2008, 56, 716–727. [Google Scholar] [CrossRef]
- Nicholson, B.L.; Wu, B.; Chevtchenko, I.; White, K.A. Tombusvirus recruitment of host translational machinery via the 3′UTR. RNA 2010, 16, 1402–1419. [Google Scholar] [CrossRef][Green Version]
- Miras, M.; Truniger, V.; Silva, C.; Verdaguer, N.; Aranda, M.A.; Querol-Audí, J. Structure of eIF4E in complex with an eIF4G peptide supports a universal bipartite binding mode for protein translation. Plant Physiol. 2017, 174, 1476–1491. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; White, K.A. Analysis of a 3′-translation enhancer in a Tombusvirus: A dynamic model for RNA-RNA interactions of m RNA termini. RNA 2006, 12, 1304–1314. [Google Scholar] [CrossRef]
- Miras, M.; Sempere, R.N.; Kraft, J.J.; Miller, W.A.; Aranda, M.A.; Truniger, V. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014, 202, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Meulewaeter, F.; Danthinne, X.; Va, M.; Cornelissen, M. 5′-and 3′-sequences of satellite tobacco necrosis virus RNA promoting translation in tobacco. Plant J. 1998, 14, 169–176. [Google Scholar] [CrossRef]
- Lipzig, R.; Gultyaev, A.P.; Pleij, C.W.; Van Montagu, M.; Cornelissen, M.; Meulewaeter, F. The 5’and 3’extremities of the satellite tobacco necrosis virus translational enhancer domain contribute differentially to stimulation of translation. RNA 2002, 8, 229–236. [Google Scholar] [CrossRef]
- Lipzig, R.; Montagu, M.V.; Cornelissen, M.; Meulewaeter, F. Functionality of the STNV translational enhancer domain correlates with affinity for two wheat germ factors. Nucleic Acids Res. 2001, 29, 1080–1086. [Google Scholar] [CrossRef]
- Gazo, B.M.; Murphy, P.; Gatchel, J.R.; Browning, K.S.A. novel interaction of cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 2004, 279, 13584–13592. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Edwards, A.; Miller, W.A. Structure and function of a cap-independent translation element that functions in either the 3’ or the 5’ untranslated region. RNA 2000, 6, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, Q.; Miller, W.A.; Goss, D.J. Eukaryotic translation initiation factor 4G (eIF4G) coordinates interactions with eIF4A, eIF4B, and eIF4E in binding and translation of the barley yellow dwarf virus 3’cap-independent translation element (BTE). J. Biol. Chem. 2017, 292, 5921–5931. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, U.; Powell, P.; Goss, D.J. Eukaryotic initiation factor (eIF) 3 mediates barley yellow dwarf viral mRNA 3’-5’UTR interactions and 40S ribosomal subunit binding to facilitate cap-independent translation. Nucleic Acids Res. 2019, 47, 6225–6235. [Google Scholar] [CrossRef] [PubMed]
- Treder, K.; Kneller, E.L.; Allen, E.M.; Wang, Z.; Browning, K.S.; Miller, W.A. The 3’ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 2008, 14, 134–147. [Google Scholar] [CrossRef]
- Kraft, J.J.; Hoy, J.A.; Miller, W.A. Crystallization and preliminary X-ray diffraction analysis of the barley yellow dwarf virus cap-independent translation element. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yu, C.; Liu, S.; Wang, G.; Shi, K.; Li, X.; Yuan, X. Structural evolution of a BYDV-like translation element (BTE) attenuating expression of p35 in three new isolates of Tobacco bushy top virus with mild virulence. Sci. Rep. 2017, 7, 4213. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M.; Du, Z.; Simon, A.E. Opium poppy mosaic virus has an Xrn-Resistant, translated subgenomic RNA and a BTE 3’CITE. J. Virol. 2021, 95, e02109-20. [Google Scholar] [CrossRef]
- Carino, E.; Scheets, K.; Miller, W.A. The RNA of maize chlorotic mottle virus-the essential virus in maize lethal necrosis disease-is translated via a panicum mosaic virus-like cap-independent translation element. J. Virol. 2020, 94, e01005-20. [Google Scholar] [CrossRef]
- Yuan, X.; Shi, K.; Meskauskas, A.; Simon, A.E. The 3’ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA 2009, 15, 1849–1864. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Shi, K.; Simon, A.E. A local, interactive network of 3’ RNA elements supports translation and replication of Turnip crinkle virus. J. Virol. 2012, 86, 4065–4081. [Google Scholar] [CrossRef]
- Miras, M.; Truniger, V.; Querol-Audi, J.; Aranda, M.A. Analysis of the interacting partners eIF4F and 3′CITE required for Melon necrotic spot virus cap-independent translation. Mol. Plant Pathol. 2017, 18, 635–648. [Google Scholar] [CrossRef]
- Nicholson, B.L.; Zaslaver, O.; Mayberry, L.K.; Browning, K.S.; White, K.A. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 2013, 87, 1872–1883. [Google Scholar] [CrossRef]
- Ryabova, L.A.; Pooggin, M.M.; Hohn, T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006, 119, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.G.; Wilusz, J.E. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017, 31, 1717–1731. [Google Scholar] [CrossRef]
- Fuller, F.; Bhown, A.S.; Bishop, D.H. Bunyavirus nucleoprotein, N, and a non-structural protein, NSs, are coded by overlapping reading frames in the S RNA. J. Gen. Virol. 1983, 64, 1705–1714. [Google Scholar] [CrossRef]
- Kozak, M. Pushing the limits of the scanning mechanism for initiation of translation. Gene 2002, 299, 1–34. [Google Scholar] [CrossRef]
- Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef]
- Fütterer, J.; Hohn, T. Translation in plants-rules and exceptions. Plant Mol. Biol. 1996, 32, 159–189. [Google Scholar] [CrossRef]
- Nakagawa, S.; Niimura, Y.; Gojobori, T.; Tanaka, H.; Miura, K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008, 36, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Firth, A.E.; Brierley, I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012, 93, 1385–1409. [Google Scholar] [CrossRef]
- Dinesh-Kumar, S.P.; Miller, W.A. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell 1993, 5, 679–692. [Google Scholar] [CrossRef]
- Herzog, E.; Guilley, H.; Fritsch, C. Translation of the second gene of peanut clump virus RNA 2 occurs by leaky scanning in vitro. Virology 1995, 208, 215–225. [Google Scholar] [CrossRef][Green Version]
- Zhou, H.; Jackson, A.O. Expression of the barley stripe mosaic virus RNA beta “triple gene block”. Virology 1996, 216, 367–379. [Google Scholar] [CrossRef]
- Fütterer, J.; Rothnie, H.M.; Hohn, T.; Potrykus, I. Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J. Virol. 1997, 71, 7984–7989. [Google Scholar] [CrossRef]
- Verchot, J.; Angell, S.M.; Baulcombe, D.C. In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. J. Virol. 1998, 72, 8316–8320. [Google Scholar] [CrossRef]
- Castaño, A.; Ruiz, L.; Hernández, C. Insights into the translational regulation of biologically active open reading frames of Pelargonium line pattern virus. Virology 2009, 386, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Ling, R.; Pate, A.E.; Carr, J.P.; Firth, A.E. An essential fifth coding ORF in the sobemoviruses. Virology 2013, 446, 397–408. [Google Scholar] [CrossRef]
- Curran, J.; Kolakofsky, D. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988, 7, 245–251. [Google Scholar] [CrossRef]
- Touriol, C.; Bornes, S.; Bonnal, S.; Audigier, S.; Prats, H.; Prats, A.C.; Vagner, S. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell 2003, 95, 169–178. [Google Scholar] [CrossRef]
- Kozak, M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 1990, 87, 8301–8305. [Google Scholar] [CrossRef] [PubMed]
- Clyde, K.; Harris, E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 2006, 80, 2170–2182. [Google Scholar] [CrossRef]
- Turina, M.; Desvoyes, B.; Scholthof, K.B. A gene cluster encoded by panicum mosaic virus is associated with virus movement. Virology 2000, 266, 120–128. [Google Scholar] [CrossRef]
- Scheets, K.; Blinkova, O.; Melcher, U.; Palmer, M.W.; Wiley, G.B.; Ding, T.; Roe, B.A. Detection of members of the Tombusviridae in the Tallgrass Prairie Preserve, Osage County, Oklahoma, USA. Virus Res. 2011, 160, 256–263. [Google Scholar] [CrossRef]
- Hizi, A.; Henderson, L.E.; Copeland, T.D.; Sowder, R.C.; Hixon, C.V.; Oroszlan, S. Characterisation of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc. Natl. Acad. Sci. USA 1987, 84, 7041–7045. [Google Scholar] [CrossRef] [PubMed]
- Giedroc, D.P.; Theimer, C.A.; Nixon, P.L. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal fameshiting. J. Mol. Biol. 2000, 298, 167–185. [Google Scholar] [CrossRef] [PubMed]
- Harger, J.W.; Meskauskas, A.; Dinman, J.D. An ‘integrated model’ of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002, 27, 448–454. [Google Scholar] [CrossRef]
- Stahl, G.; McCart, G.P.; Farabaugh, P.J. Ribosome structure: Revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 2002, 27, 178–183. [Google Scholar] [CrossRef]
- Plant, E.P. Ribosomal Frameshift Signals in Viral Genomes. Viral Genomes-Mol. Struct. Divers. Gene Expr. Mech. Host-Virus Interact. 2012, 57, 91–122. [Google Scholar] [CrossRef]
- Jacks, T.; Varmus, H.E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science 1985, 230, 1237–1242. [Google Scholar] [CrossRef]
- Jacks, T.; Madhani, H.D.; Masiarz, F.R.; Varmus, H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 1988, 55, 447–458. [Google Scholar] [CrossRef]
- Di, R.; Dinesh-Kumar, S.P.; Miller, W.A. Translational frameshifting by barley yellow Dwarf virus RNA (PAV serotype) in Escherichia coli and in eukaryotic cell-free extracts. Mol. Plant-Microbe Interact. MPMI 1993, 6, 444–452. [Google Scholar] [CrossRef][Green Version]
- Baranov, P.V.; Gesteland, R.F.; Atkins, J.F. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 2004, 10, 221–230. [Google Scholar] [CrossRef]
- Giedroc, D.P.; Cornish, P.V. Frameshifting RNA pseudokllots: Structure and mechanism. Virus Res. 2009, 139, 193–208. [Google Scholar] [CrossRef]
- Brierley, I.; Gilbert, R.J.C.; Pennell, S. Pseudoknot-dependent programmed -1 ribosomal frameshifting: Structures, mechanisms and models. In Recoding: Expansion of Decoding Rules Enriches Gene Expression; Atkins, J.F., Gesteland, R.F., Eds.; Springer: Heidelberg, Germany, 2010; pp. 149–174. [Google Scholar] [CrossRef]
- Plant, E.P.; Jacobs, K.L.; Harger, J.W.; Meskauskas, A.; Jacobs, J.L.; Baxter, J.L.; Petrov, A.N.; Dinman, J.D. The 9Å solution: How mRNA pseudoknots promote efficient programmed-1 ribosomal frameshifting. RNA 2003, 9, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Barry, J.K.; Miller, W.A. A -1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. USA 2002, 99, 11133–11138. [Google Scholar] [CrossRef] [PubMed]
- Paul, C.P.; Barry, J.K.; Dinesh-Kumar, S.P.; Brault, V.; Miller, W.A. A sequence required for -1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 2001, 310, 987–999. [Google Scholar] [CrossRef]
- Tajima, Y.; Iwakawa, H.; Kaido, M.; Mise, K.; Okuno, T. A long-distance RNA-RNA interaction plays an important role in programmed -1 ribosomal frameshifting in the translation of p88 replicase protein of Red clover necrotic mosaic virus. Virology 2011, 417, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Rohde, W.; Gramstat, A.; Schmitz, J.; Tacke, E.; Prüfer, D. Plant viruses as model systems for the study of non-canonical translation mechanisms in higher plants. J. Gen. Virol. 1994, 75, 2141–2149. [Google Scholar] [CrossRef]
- Chung, B.Y.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef]
- Gao, F.; Simon, A.E. Multiple Cis-acting elements modulate programmed-1 ribosomal frameshifting in Pea enation mosaic virus. Nucleic Acids Res. 2015, 44, 878–895. [Google Scholar] [CrossRef][Green Version]
- Karasev, A.V.; Boyko, V.P.; Gowda, S.; Nikolaeva, O.V.; Hilf, M.E.; Koonin, E.V.; Niblett, C.L.; Cline, K.; Gumpf, D.J.; Lee, R.F.; et al. Complete sequence of the citrus tristeza virus RNA genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef]
- Salem, N.M.; Miller, W.A.; Rowhani, A.; Golino, D.A.; Moyne, A.L.; Falk, B.W. Rose spring dwarf-associated virus has RNA structural and gene-expression features like those of Barley yellow dwarf virus. Virology 2008, 375, 354–360. [Google Scholar] [CrossRef]
- Yu, C.; Wang, G.; Wang, D.; Shi, K.; Yuan, X. Application of Renilla luciferase-fused virus vector on the study of translational frameshift and readthrough. Acta Phytopathol. Sin. 2016, 46, 846–849. (In Chinese) [Google Scholar] [CrossRef]
- Hancock, J.M.; Chaleeprom, W.; Dale, J.; Gibbs, A. Replication slippage in the evolution of potyviruses. J. Gen. Virol. 1995, 76, 3229–3232. [Google Scholar] [CrossRef] [PubMed]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef] [PubMed]
- Olspert, A.; Chung, B.Y.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef]
- Mingot, A.; Valli, A.; Rodamilans, B.; San, L.D.; Baulcombe, D.C.; Garcia, J.A.; López-Moya, J.J. The P1N-PISPO trans-frame gene of sweet potato feathery mottle potyvirus is produced during virus infection and functions as RNA silencing suppressor. J. Virol. 2016, 90, 3543–3557. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.; Wills, N.M.; Nelson, C.; Atkins, J.F.; Gesteland, R.F. Nonlinearity in genetic decoding: Homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl. Acad. Sci. USA 2000, 97, 1683–1688. [Google Scholar] [CrossRef]
- Penno, C.; Sansonetti, P.; Parsot, C. Frameshifting by transcriptional slippage is involved in production of Mxi E, the transcription activator regulated by the activity of the type III secretion apparatus in Shigella lexneri. Mol. Microbiol. 2005, 56, 204–214. [Google Scholar] [CrossRef]
- Penno, C.; Sharma, V.; Coakley, A.; Motherway, M.O.C.; van Sinderen, D.; Lubkowska, L.; Kireeva, M.L.; Kashlev, M.; Baranov, P.V.; Atkins, J.F. Productive mRNA stem loop-mediated transcriptional slippage: Crucial features in common with intrinsic terminators. Proc. Natl. Acad. Sci. USA 2015, 112, E1984–E1993. [Google Scholar] [CrossRef]
- Baranov, P.V.; Hammer, A.W.; Zhou, J.; Gesteland, R.F.; Atkins, J.F. Transcriptional slippage in bacteria: Distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 2005, 6, R25. [Google Scholar] [CrossRef]
- Lin, C.P.; Ko, C.Y.; Kuo, C.I.; Liu, M.S.; Schaleitner, R.; Chen, L.F. Transcriptional slippage and RNA editing increase the diversity of transcripts in chloroplasts: Insight from deep sequencing of Vigna radiata genome and transcriptome. PLoS ONE 2015, 10, e0129396. [Google Scholar] [CrossRef]
- McCaughan, K.K.; Brown, C.M.; Dalphin, M.E.; Berry, M.J.; Tate, W.P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA 1995, 92, 5431–5435. [Google Scholar] [CrossRef]
- Bertram, G.; Innes, S.; Minella, O.; Richardson, J.; Stansfield, I. Endless possibilities: Translation termination and stop codon recognition. Microbiology 2001, 147, 255–269. [Google Scholar] [CrossRef]
- Bruening, G.; Beachy, T.N.; Scalla, R.; Zaitlin, M. In vitro and in vivo translation of the ribonucleic acids of a cowpea strain of tobacco mosaic virus. Virology 1976, 71, 498–517. [Google Scholar] [CrossRef]
- Beier, H.; Barciszewska, M.; Krupp, G.; Mitnacht, R.; Gross, H.J. UAG readthrough during TMV RNA translation: Isolation and sequence of two tRNAs xy’ with suppressor activity from tobacco plants. EMBO J. 1984, 3, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Beier, H.; Barciszewska, M.; Sickinger, H.D. The molecular basis for the differential translation of TMV RNA in tobacco protoplasts and wheat germ extracts. EMBO J. 1984, 3, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Skuzeski, J.M.; Nichols, L.M.; Gesteland, R.F.; Atkins, J.F. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 1991, 218, 365–373. [Google Scholar] [CrossRef]
- Beier, H.; Grimm, M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001, 29, 4767–4782. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, B.; Fu, L.; Moon, J.; Bedwell, D.M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995, 251, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Tork, S.; Hatin, I.; Rousset, J.P.; Fabret, C. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 2004, 32, 415–421. [Google Scholar] [CrossRef]
- Qu, X.; Wen, J.D.; Lancaster, L.; Noller, H.F.; Bustamante, C.; Tinoco, I.J. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 2011, 475, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Napthine, S.; Yek, C.; Powell, M.L.; Brown, T.D.; Brierley, I. Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA. RNA 2012, 18, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Pelham, H.R.B. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature 1978, 272, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Malpica-López, N.; Rajeswaran, R.; Beknazariants, D.; Seguin, J.; Golyaev, V.; Farinelli, L.; Pooggin, M.M. Revisiting the Roles of Tobamovirus Replicase Complex Proteins in Viral Replication and Silencing Suppression. Mol. Plant-Microbe Interact. 2018, 31, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.M.; Dinesh-Kumar, S.P.; Miller, W.A. Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J. Virol. 1996, 70, 5884–5892. [Google Scholar] [CrossRef]
- Newburn, L.R.; Nicholson, B.L.; Yosefi, M.; Cimino, P.A.; White, K.A. Translational readthrough in Tobacco necrosis virus-D. Virology 2014, 450, 258–265. [Google Scholar] [CrossRef]
- Liu, S.; Sivakumar, S.; Wang, Z.; Bryony, C.B.; Allen, M.W. The readthrough domain of pea enation mosaic virus coat protein is not essential for virus stability in the hemolymph of the pea aphid. Arch. Virol. 2009, 154, 469–479. [Google Scholar] [CrossRef]
- Brault, V.; van den Heuvel, J.F.; Verbeek, M.; Ziegler-Graff, V.; Reutenauer, A.; Herrbach, E.; Garaud, J.C.; Guilley, H.; Richards, K.; Jonard, G. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 1995, 14, 650–659. [Google Scholar] [CrossRef]
- Gray, S.; Gildow, F.E. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 2003, 41, 539–566. [Google Scholar] [CrossRef]
- Cimino, P.A.; Nicholson, B.L.; Wu, B.; Xu, W.; White, K.A. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog. 2011, 7, e1002423. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).