Sequence Analysis of Egyptian Foot-and-Mouth Disease Virus Field and Vaccine Strains: Intertypic Recombination and Evidence for Accidental Release of Virulent Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Samples and Inactivated Vaccine
2.2. FTA Card Storage Experiment
2.3. RNA Extraction
2.3.1. Liquid Samples
2.3.2. FTA Cards
2.4. FMDV Real-Time RT-PCR
2.5. Virus Isolation
2.6. Viral Genome Sequencing
2.6.1. VP1-Coding Region
2.6.2. Full-Length Viral Genome
2.6.3. Vaccine Composition Analysis
2.6.4. Nucleotide Sequence Alignments and Phylogenetic Analysis
3. Results and Discussion
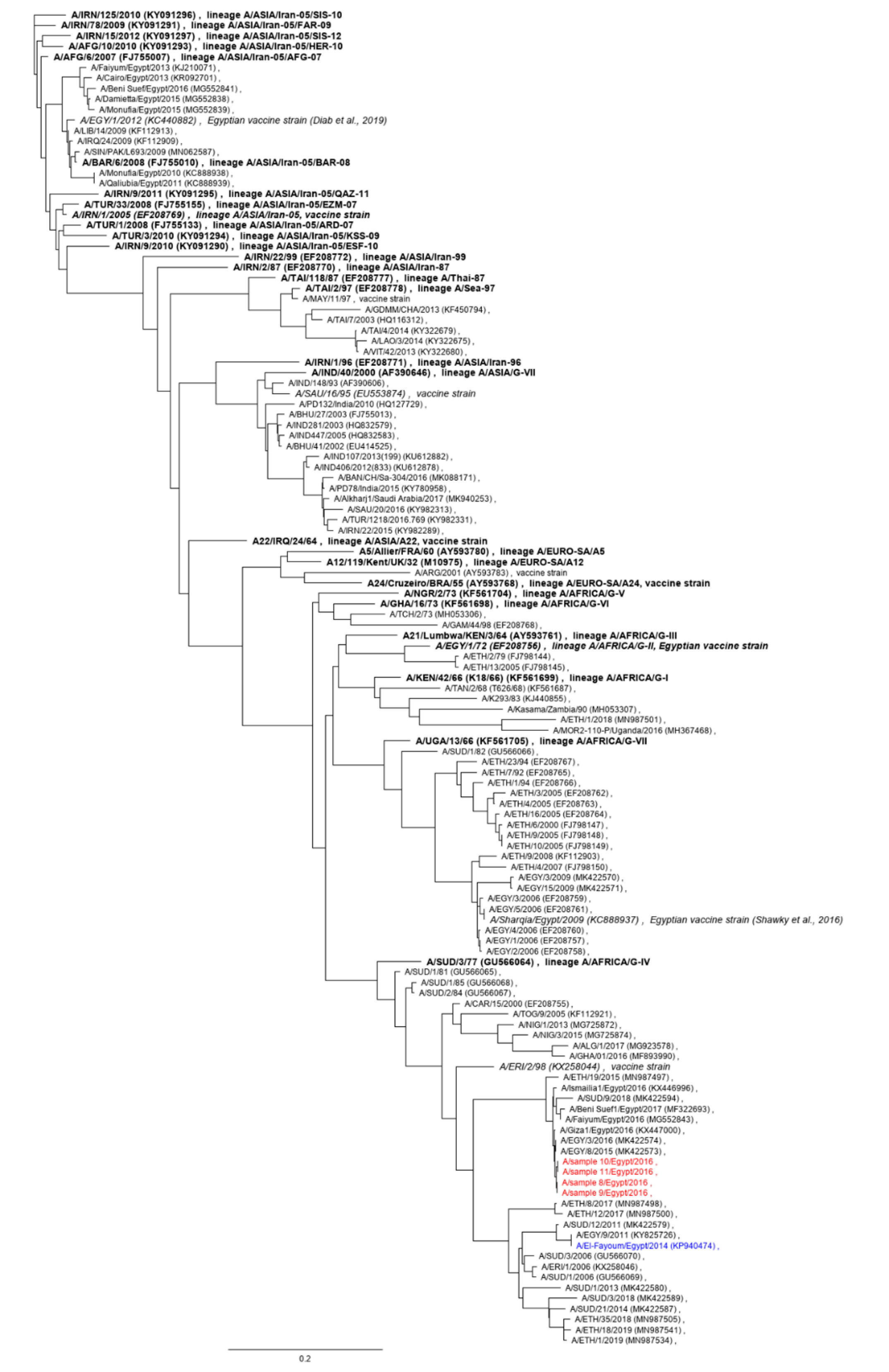
3.1. Animal Samples
3.2. Full-Length Sequence of Serotype A Isolate from 2016
3.3. FTA Card Storage Experiment
3.4. Composition Analysis of 2019 Inactivated FMDV Vaccine
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grubman, M.J.; Baxt, B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004, 17, 465–493. [Google Scholar] [CrossRef]
- Brito, B.P.; Rodriguez, L.L.; Hammond, J.M.; Pinto, J.; Perez, A.M. Review of the Global Distribution of Foot-and-Mouth Disease Virus from 2007 to 2014. Transbound. Emerg. Dis. 2017, 64, 316–332. [Google Scholar] [CrossRef]
- Carrillo, C.; Tulman, E.R.; Delhon, G.; Lu, Z.; Carreno, A.; Vagnozzi, A.; Kutish, G.F.; Rock, D.L. Comparative genomics of foot-and-mouth disease virus. J. Virol. 2005, 79, 6487–6504. [Google Scholar] [CrossRef] [PubMed]
- Arzt, J.; Juleff, N.; Zhang, Z.; Rodriguez, L.L. The pathogenesis of foot-and-mouth disease I: Viral pathways in cattle. Transbound. Emerg. Dis. 2011, 58, 291–304. [Google Scholar] [CrossRef]
- Diaz-San Segundo, F.; Medina, G.N.; Stenfeldt, C.; Arzt, J.; de Los Santos, T. Foot-and-mouth disease vaccines. Vet. Microbiol. 2017, 206, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Jamal, S.M.; Nazem Shirazi, M.H.; Ozyoruk, F.; Parlak, U.; Normann, P.; Belsham, G.J. Evidence for multiple recombination events within foot-and-mouth disease viruses circulating in West Eurasia. Transbound. Emerg. Dis. 2020, 67, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Pezzoni, G.; Bregoli, A.; Grazioli, S.; Barbieri, I.; Madani, H.; Omani, A.; Sadaoui, H.; Bouayed, N.; Wadsworth, J.; Bachanek-Bankowska, K.; et al. Foot-and-mouth disease outbreaks due to an exotic virus serotype A lineage (A/AFRICA/G-IV) in Algeria in 2017. Transbound. Emerg. Dis. 2019, 66, 7–13. [Google Scholar] [CrossRef]
- Forth, L.F.; Höper, D.; Beer, M.; Eschbaumer, M. High-Resolution Composition Analysis of an Inactivated Polyvalent Foot-and-Mouth Disease Vaccine. Pathogens 2020, 9, 63. [Google Scholar] [CrossRef]
- Callahan, J.D.; Brown, F.; Osorio, F.A.; Sur, J.H.; Kramer, E.; Long, G.W.; Lubroth, J.; Ellis, S.J.; Shoulars, K.S.; Gaffney, K.L.; et al. Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J. Am. Vet. Med. Assoc. 2002, 220, 1636–1642. [Google Scholar] [CrossRef]
- Dill, V.; Eschbaumer, M. Reliable detection, sequencing, and transfection of foot-and-mouth disease virus RNA from badly preserved vesicular epithelium. J. Vet. Diagn. Investig. 2019, 31, 778–782. [Google Scholar] [CrossRef]
- LaRocco, M.; Krug, P.W.; Kramer, E.; Ahmed, Z.; Pacheco, J.M.; Duque, H.; Baxt, B.; Rodriguez, L.L. A continuous bovine kidney cell line constitutively expressing bovine alphavbeta6 integrin has increased susceptibility to foot-and-mouth disease virus. J. Clin. Microbiol. 2013, 51, 1714–1720. [Google Scholar] [CrossRef] [PubMed]
- Dill, V.; Beer, M.; Hoffmann, B. Simple, quick and cost-efficient: A universal RT-PCR and sequencing strategy for genomic characterisation of foot-and-mouth disease viruses. J. Virol. Methods 2017. [Google Scholar] [CrossRef] [PubMed]
- Chrzastek, K.; Lee, D.H.; Smith, D.; Sharma, P.; Suarez, D.L.; Pantin-Jackwood, M.; Kapczynski, D.R. Use of Sequence-Independent, Single-Primer-Amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 2017, 509, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Scheuch, M.; Höper, D.; Beer, M. RIEMS: A software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinform. 2015, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Dill, V.; Eschbaumer, M. Cell culture propagation of foot-and-mouth disease virus: Adaptive amino acid substitutions in structural proteins and their functional implications. Virus Genes 2020, 56, 1–15. [Google Scholar] [CrossRef]
- WRLFMD. Genotyping Report WRLFMD/2017/00014. Available online: https://perma.cc/5M8F-9MUS (accessed on 21 July 2020).
- WRLFMD. FMD Vaccine Matching Strain Differentiation Report WRLFMD/2016/00024. Available online: https://perma.cc/T468-FUD4 (accessed on 21 July 2020).
- WRLFMD. FMD Vaccine Matching Strain Differentiation Report WRLFMD/2017/00014. Available online: https://perma.cc/JB26-FWVQ (accessed on 21 July 2020).
- Shafik, N.G.; Darwish, D.M.; Abousenna, M.S.; Galal, M.; Ahmed, A.R.; Attya, M.; Saad, M.A.; Abdelhakim, M. Efficacy of a Commercial Local Trivalent Foot and Mouth Disease (FMD) Vaccine against Recently Isolated O-EA3. Int. J. Vet. Sci. 2019, 8, 33–35. [Google Scholar]
- MEVAC. Ruminants Vaccines. Available online: https://perma.cc/BZ4T-HZ8W (accessed on 21 July 2020).
- WRLFMD. Quarterly Report 2020 Quarter 1 (Jan–Mar). Available online: https://perma.cc/J4BF-LPH3 (accessed on 22 July 2020).
- Ludi, A.B.; Horton, D.L.; Li, Y.; Mahapatra, M.; King, D.P.; Knowles, N.J.; Russell, C.A.; Paton, D.J.; Wood, J.L.; Smith, D.J.; et al. Antigenic variation of foot-and-mouth disease virus serotype A. J. Gen. Virol. 2014, 95, 384–392. [Google Scholar] [CrossRef]
- Brehm, K.E.; Kumar, N.; Thulke, H.H.; Haas, B. High potency vaccines induce protection against heterologous challenge with foot-and-mouth disease virus. Vaccine 2008, 26, 1681–1687. [Google Scholar] [CrossRef] [PubMed]
- El-Bagoury, G.F.; Sharawi, S.S.A.; El-Nahas, E.M.; Darwish, D.M.; Saad, M.A. Evaluation of cross-protection between FMD serotypes O and A local Egyptian isolate with vaccinal strains in the local commercial and imported vaccines by challenge test. Benha Vet. Med. J. 2015, 28, 241–246. [Google Scholar] [CrossRef]
- OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, Chapter 3.1.8. Foot and Mouth Disease. Available online: https://perma.cc/EG94-MX4N (accessed on 27 August 2020).
- VSVRI. Bivalent Inactivated Foot and Mouth Disease Oil Vaccine (Types O1&A). Available online: https://perma.cc/D4NQ-R3JN (accessed on 22 July 2020).
- Knowles, N.J.; Wadsworth, J.; Reid, S.M.; Swabey, K.G.; El-Kholy, A.A.; Abd El-Rahman, A.O.; Soliman, H.M.; Ebert, K.; Ferris, N.P.; Hutchings, G.H.; et al. Foot-and-mouth disease virus serotype A in Egypt. Emerg. Infect. Dis. 2007, 13, 1593–1596. [Google Scholar] [CrossRef] [PubMed]
- Shawky, S.M.; Thabet, N.S.; Orabi, S.H.; Nayel, M.A. Comparative Study on the Hemato-Biochemical and Immunological Effects of the Hexavalent FMD Vaccine Alone or in Combination with Trivalent FMD Vaccine in Cattle. J. Biosci. Med. 2016, 4, 16–26. [Google Scholar] [CrossRef][Green Version]
- WRLFMD. Genotyping Report WRLFMD/2019/00005. Available online: https://perma.cc/V6HM-9CP9 (accessed on 22 July 2020).
- Sobhy, N.M.; Bayoumi, Y.H.; Mor, S.K.; El-Zahar, H.I.; Goyal, S.M. Outbreaks of foot and mouth disease in Egypt: Molecular epidemiology, evolution and cardiac biomarkers prognostic significance. Int. J. Vet. Sci. Med. 2018, 6, 22–30. [Google Scholar] [CrossRef]
- Bachanek-Bankowska, K.; Di Nardo, A.; Wadsworth, J.; Mioulet, V.; Pezzoni, G.; Grazioli, S.; Brocchi, E.; Kafle, S.C.; Hettiarachchi, R.; Kumarawadu, P.L.; et al. Reconstructing the evolutionary history of pandemic foot-and-mouth disease viruses: The impact of recombination within the emerging O/ME-SA/Ind-2001 lineage. Sci. Rep. 2018, 8, 14693. [Google Scholar] [CrossRef]
- Heath, L.; van der Walt, E.; Varsani, A.; Martin, D.P. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 2006, 80, 11827–11832. [Google Scholar] [CrossRef]
- Mwiine, F.N.; Velazquez-Salinas, L.; Ahmed, Z.; Ochwo, S.; Munsey, A.; Kenney, M.; Lutwama, J.J.; Maree, F.F.; Lobel, L.; Perez, A.M.; et al. Serological and phylogenetic characterization of foot and mouth disease viruses from Uganda during cross-sectional surveillance study in cattle between 2014 and 2017. Transbound. Emerg. Dis. 2019, 66, 2011–2024. [Google Scholar] [CrossRef]
- Abdul-Hamid, N.F.; Firat-Sarac, M.; Radford, A.D.; Knowles, N.J.; King, D.P. Comparative sequence analysis of representative foot-and-mouth disease virus genomes from Southeast Asia. Virus Genes 2011, 43, 41–45. [Google Scholar] [CrossRef]
- Jamal, S.M.; Ferrari, G.; Ahmed, S.; Normann, P.; Belsham, G.J. Molecular characterization of serotype Asia-1 foot-and-mouth disease viruses in Pakistan and Afghanistan; emergence of a new genetic Group and evidence for a novel recombinant virus. Infect. Genet. Evol. 2011, 11, 2049–2062. [Google Scholar] [CrossRef]
- McCahon, D.; King, A.M.; Roe, D.S.; Slade, W.R.; Newman, J.W.; Cleary, A.M. Isolation and biochemical characterization of intertypic recombinants of foot-and-mouth disease virus. Virus Res. 1985, 3, 87–100. [Google Scholar] [CrossRef]
- Wright, C.F.; Knowles, N.J.; Di Nardo, A.; Paton, D.J.; Haydon, D.T.; King, D.P. Reconstructing the origin and transmission dynamics of the 1967-68 foot-and-mouth disease epidemic in the United Kingdom. Infect. Genet. Evol. 2013, 20, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Diab, E.; Bazid, A.I.; Fawzy, M.; El-Ashmawy, W.R.; Fayed, A.A.; El-Sayed, M.M. Foot-and-mouth disease outbreaks in Egypt during 2013-2014: Molecular characterization of serotypes A, O, and SAT2. Vet. World 2019, 12, 190–197. [Google Scholar] [CrossRef]
- Samuel, A.R.; Knowles, N.J.; Mackay, D.K. Genetic analysis of type O viruses responsible for epidemics of foot-and-mouth disease in North Africa. Epidemiol. Infect. 1999, 122, 529–538. [Google Scholar] [CrossRef] [PubMed]
- VSVRI. Polyvalent Inactivated Foot and Mouth Disease Oil Vaccine (Types O1,A&SAT2). Available online: https://perma.cc/A3NC-8V7Y (accessed on 22 July 2020).
- WRLFMD. Country Reports: Egypt, Samples Tested. Available online: https://perma.cc/KDK8-CQ9F (accessed on 22 July 2020).
- Knowles, N.J.; Samuel, A.R.; Davies, P.R.; Midgley, R.J.; Valarcher, J.F. Pandemic strain of foot-and-mouth disease virus serotype O. Emerg. Infect. Dis. 2005, 11, 1887–1893. [Google Scholar] [CrossRef]
- Abu-Elnaga, H.I.; Rizk, S.A.; Daoud, H.M.; Mohamed, A.A.; Mossad, W.; Gamil, M.A.; Soudy, A.F.; El-Shehawy, L.I. Comparative nucleotide sequencing of the VP1 capsid gene of recent isolates of foot-and-mouth disease virus serotype O from Egypt. Arch. Virol. 2020. [Google Scholar] [CrossRef]
- Beck, E.; Strohmaier, K. Subtyping of European foot-and-mouth disease virus strains by nucleotide sequence determination. J. Virol. 1987, 61, 1621–1629. [Google Scholar] [CrossRef]
- Cottam, E.M.; Wadsworth, J.; Shaw, A.E.; Rowlands, R.J.; Goatley, L.; Maan, S.; Maan, N.S.; Mertens, P.P.; Ebert, K.; Li, Y.; et al. Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathog. 2008, 4, e1000050. [Google Scholar] [CrossRef]
- Carrillo, C.; Dopazo, J.; Moya, A.; Gonzalez, M.; Martinez, M.A.; Saiz, J.C.; Sobrino, F. Comparison of vaccine strains and the virus causing the 1986 foot-and-mouth disease outbreak in Spain: Epizootiological analysis. Virus Res. 1990, 15, 45–55. [Google Scholar] [CrossRef]
| ID | Serotype | Sample Material | Origin | Year | Host | Sequence | Accession Number |
|---|---|---|---|---|---|---|---|
| Type | |||||||
| sample 8 | A | saliva | Dakahlia | 2016 | cow | 1D/VP1 | MT863264 |
| sample 9 | A | saliva | Damietta | 2016 | cow | 1D/VP1 | MT863265 |
| sample 10 | A | saliva | Damietta | 2016 | buffalo | 1D/VP1 | MT863266 |
| culture supernatant | - | - | - | whole genome | MT863268 | ||
| sample 11 | A | saliva | Dakahlia | 2016 | buffalo | 1D/VP1 | MT863267 |
| FTA card | O | inactivated vaccine | Egypt | 2019 | whole genome | MT863269 |
| Isolate | ORF | L | 1A | 1B | 1C | 1D | 2A | 2B | 2C | 3A | 3B1 | 3B2 | 3B3 | 3C | 3D |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/El-F/2014 | 87.3 | 88.1 | 89.8 | 89.3 | 90.5 | 86.6 | 98.2 | 93.9 | 83.9 | 78.2 | 76.8 | 79.2 | 83.3 | 87.2 | 88.3 |
| O/AMU_200 | 85.9 | 76.3 | 85.1 | 72.5 | 73.9 | 64.9 | 94.4 | 94.4 | 94.9 | 91.9 | 98.6 | 87.5 | 94.4 | 95.3 | 94.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd El Rahman, S.; Hoffmann, B.; Karam, R.; El-Beskawy, M.; Hamed, M.F.; Forth, L.F.; Höper, D.; Eschbaumer, M. Sequence Analysis of Egyptian Foot-and-Mouth Disease Virus Field and Vaccine Strains: Intertypic Recombination and Evidence for Accidental Release of Virulent Virus. Viruses 2020, 12, 990. https://doi.org/10.3390/v12090990
Abd El Rahman S, Hoffmann B, Karam R, El-Beskawy M, Hamed MF, Forth LF, Höper D, Eschbaumer M. Sequence Analysis of Egyptian Foot-and-Mouth Disease Virus Field and Vaccine Strains: Intertypic Recombination and Evidence for Accidental Release of Virulent Virus. Viruses. 2020; 12(9):990. https://doi.org/10.3390/v12090990
Chicago/Turabian StyleAbd El Rahman, Sahar, Bernd Hoffmann, Reham Karam, Mohamed El-Beskawy, Mohammed F. Hamed, Leonie F. Forth, Dirk Höper, and Michael Eschbaumer. 2020. "Sequence Analysis of Egyptian Foot-and-Mouth Disease Virus Field and Vaccine Strains: Intertypic Recombination and Evidence for Accidental Release of Virulent Virus" Viruses 12, no. 9: 990. https://doi.org/10.3390/v12090990
APA StyleAbd El Rahman, S., Hoffmann, B., Karam, R., El-Beskawy, M., Hamed, M. F., Forth, L. F., Höper, D., & Eschbaumer, M. (2020). Sequence Analysis of Egyptian Foot-and-Mouth Disease Virus Field and Vaccine Strains: Intertypic Recombination and Evidence for Accidental Release of Virulent Virus. Viruses, 12(9), 990. https://doi.org/10.3390/v12090990






