Innate Immune DNA Sensing of Flaviviruses
Abstract
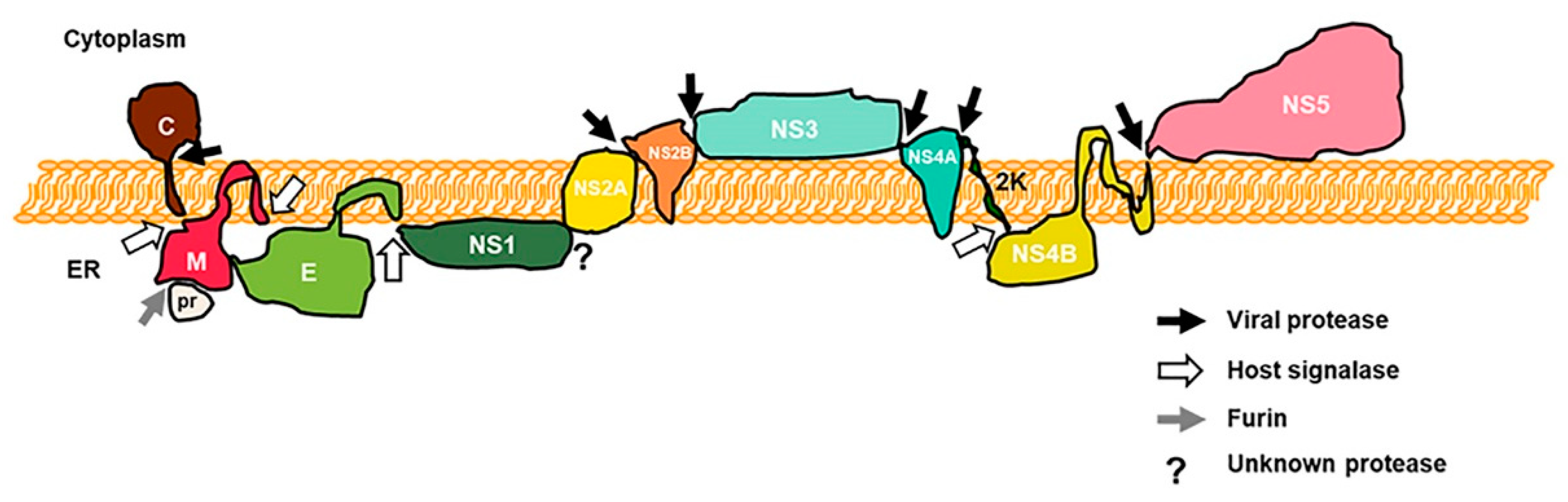
1. Introduction
2. Innate Immune System
2.1. Overview
2.2. Viral Nucleic Acid Sensing
2.3. Innate Immune DNA Sensors
2.4. cGAS/STING Pathway
3. Dengue Virus
3.1. Overview
3.2. Regulation of cGAS and STING by DENV
4. Zika Virus
4.1. Overview
4.2. Regulation of cGAS and STING by ZIKV
5. West Nile Virus
5.1. Overview
5.2. Regulation of STING by WNV
6. Other Flaviviruses
6.1. Yellow Fever Virus
6.2. Japanese Encephalitis Virus
6.3. Duck Tembusu Virus
7. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Tomori, O. Yellow fever: The recurring plague. Crit. Rev. Clin. Lab. Sci. 2004, 41, 391–427. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Gubler, D.J.; Petersen, L.R. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004, 10, S98–S109. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, M.-D.; Mazzon, M.; Jacobs, M.; Amara, A. Pathogenesis of Flavivirus Infections: Using and Abusing the Host Cell. Cell Host Microbe 2009, 5, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Tilak, R.; Ray, S.; Tilak, V.W.; Mukherji, S. Dengue, chikungunya … and the missing entity—Zika fever: A new emerging threat. Med. J. Armed. Forces India 2016, 72, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Habarugira, G.; Suen, W.W.; Hobson-Peters, J.; Hall, R.A.; Bielefeldt-Ohmann, H. West Nile Virus: An Update on Pathobiology, Epidemiology, Diagnostics, Control and “One Health” Implications. Pathogens 2020, 9, 589. [Google Scholar] [CrossRef]
- Simon, L.V.; Sandhu, D.S.; Goyal, A.; Kruse, B. Encephalitis, Japanese. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Pierson, T.C.; Diamond, M.S. Flaviviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 747–794. [Google Scholar]
- Shah, P.S.; Link, N.; Jang, G.M.; Sharp, P.P.; Zhu, T.; Swaney, D.L.; Johnson, J.R.; Von Dollen, J.; Ramage, H.R.; Satkamp, L.; et al. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 2018, 175, 1931–1945. [Google Scholar] [CrossRef] [PubMed]
- Miorin, L.; Maestre, A.M.; Fernandez-Sesma, A.; García-Sastre, A. Antagonism of type I interferon by flaviviruses. Biochem. Biophys. Res. Commun. 2017, 492, 587–596. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Chang, T.-H.; Liang, J.-J.; Chiang, R.-L.; Lee, Y.-L.; Liao, C.-L.; Lin, Y.-L. Dengue Virus Targets the Adaptor Protein MITA to Subvert Host Innate Immunity. PLoS Pathog. 2012, 8, e1002780. [Google Scholar] [CrossRef]
- Aguirre, S.; Maestre, A.M.; Pagni, S.; Patel, J.R.; Savage, T.; Gutman, D.; Maringer, K.; Bernal-Rubio, D.; Shabman, R.S.; Simon, V.; et al. DENV Inhibits Type I IFN Production in Infected Cells by Cleaving Human STING. PLoS Pathog. 2012, 8, e1002934. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Weaver, C. Janeway’s Immunobiology, 9th ed.; Garland Science: New York, NY, USA; London, UK, 2017. [Google Scholar]
- Lei, J.; Hilgenfeld, R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017, 591, 3190–3210. [Google Scholar] [CrossRef]
- Ma, Z.; Ni, G.; Damania, B. Innate Sensing of DNA Virus Genomes. Annu. Rev. Virol. 2018, 5, 341–362. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.K.; Gack, M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016, 14, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.H.; Paludan, S.R. Viral evasion of DNA-stimulated innate immune responses. Cell Mol. Immunol. 2017, 14, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xia, P.; Li, S.; Zhang, T.; Wang, T.T.; Zhu, J. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 2017, 69, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.-M.; Hoffmann, J.A. The Dorsoventral Regulatory Gene Cassette spätzle/Toll/cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Unterholzner, L.; Keating, S.E.; Baran, M.; Horan, K.A.; Jensen, S.B.; Sharma, S.; Sirois, C.M.; Jin, T.; Latz, E.; Xiao, T.S.; et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010, 11, 997–1004. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen Recognition by the Innate Immune System. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef]
- Loo, Y.-M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Y.; Liu, Z.-J.; Ouyang, S. The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell 2017, 8, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Dolasia, K.; Bisht, M.K.; Pradhan, G.; Udgata, A.; Mukhopadhyay, S. TLRs/NLRs: Shaping the landscape of host immunity. Int. Rev. Immunol. 2018, 37, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.L.; Conn, G.L. RNA regulation of the antiviral protein 2’-5’-oligoadenylate synthetase. Wiley Interdiscip. Rev. RNA 2019, 10, e1534. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, E.; Patin, E.; Laval, G.; Pajon, S.; Fornarino, S.; Crouau-Roy, B.; Quintana-Murci, L. The selective footprints of viral pressures at the human RIG-I-like receptor family. Hum. Mol. Genet. 2011, 20, 4462–4474. [Google Scholar] [CrossRef]
- Dempsey, A.; Bowie, A.G. Innate immune recognition of DNA: A recent history. Virology 2015, 479–480, 146–152. [Google Scholar] [CrossRef]
- Burdette, D.L.; Vance, R.E. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013, 14, 19–26. [Google Scholar] [CrossRef]
- Maringer, K.; Fernandez-Sesma, A. Message in a bottle: Lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev. 2014, 25, 669–679. [Google Scholar] [CrossRef]
- Patel, J.R.; García-Sastre, A. Activation and regulation of pathogen sensor RIG-I. Cytokine Growth Factor Rev. 2014, 25, 513–523. [Google Scholar] [CrossRef]
- Abe, T.; Marutani, Y.; Shoji, I. Cytosolic DNA-sensing immune response and viral infection. Microbiol. Immunol. 2019, 63, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Jin, S. The Cross-Regulation Between Autophagy and Type I Interferon Signaling in Host Defense. Adv. Exp. Med. Biol. 2019, 1209, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Jefferies, C. Role of DNA/RNA sensors and contribution to autoimmunity. Cytokine Growth Factor Rev. 2014, 25, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Li, T.; Li, X.-D.; Chen, X.; Li, Q.-Z.; Wight-Carter, M.; Chen, Z.J. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA 2015, 112, E5699–E5705. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Ren, J.; Chen, Q.; Chen, Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4612–E4620. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, H.; Wu, X.; Ma, D.; Wu, J.; Wang, L.; Jiang, Y.; Fei, Y.; Zhu, C.; Tan, R.; et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 2018, 563, 131–136. [Google Scholar] [CrossRef]
- Takaoka, A.; Shinohara, S. DNA sensors in innate immune system. Uirusu 2008, 58, 37–46. [Google Scholar] [CrossRef][Green Version]
- Unterholzner, L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology 2013, 218, 1312–1321. [Google Scholar] [CrossRef]
- Sasai, M.; Linehan, M.M.; Iwasaki, A. Bifurcation of Toll-Like Receptor 9 Signaling by Adaptor Protein 3. Science 2010, 329, 1530. [Google Scholar] [CrossRef]
- Marongiu, L.; Gornati, L.; Artuso, I.; Zanoni, I.; Granucci, F. Below the surface: The inner lives of TLR4 and TLR9. J. Leukoc. Biol. 2019, 106, 147–160. [Google Scholar] [CrossRef]
- Briggs, L.J.; Johnstone, R.W.; Elliot, R.M.; Xiao, C.Y.; Dawson, M.; Trapani, J.A.; Jans, D.A. Novel properties of the protein kinase CK2-site-regulated nuclear- localization sequence of the interferon-induced nuclear factor IFI 16. Biochem. J. 2001, 353, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Veeranki, S.; Choubey, D. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: Regulation of subcellular localization. Mol. Immunol. 2012, 49, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.R.; Bak, R.O.; Andersen, A.; Berg, R.K.; Jensen, S.B.; Jin, T.; Laustsen, A.; Hansen, K.; Østergaard, L.; Fitzgerald, K.A.; et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci. USA 2013, 110, E4571–E4580. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.R.; Paludan, S.R. IFI16: At the interphase between innate DNA sensing and genome regulation. Cytokine Growth Factor Rev. 2014, 25, 649–655. [Google Scholar] [CrossRef]
- Dell’Oste, V.; Gatti, D.; Giorgio, A.G.; Gariglio, M.; Landolfo, S.; De Andrea, M. The interferon-inducible DNA-sensor protein IFI16: A key player in the antiviral response. New Microbiol. 2015, 38, 5–20. [Google Scholar]
- Fernandes-Alnemri, T.; Yu, J.W.; Datta, P.; Wu, J.; Alnemri, E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 2009, 458, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Man, S.M.; Karki, R.; Kanneganti, T.D. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 2016, 46, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Lugrin, J.; Martinon, F. The AIM2 inflammasome: Sensor of pathogens and cellular perturbations. Immunol. Rev. 2018, 281, 99–114. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef]
- Takaoka, A.; Wang, Z.; Choi, M.K.; Yanai, H.; Negishi, H.; Ban, T.; Lu, Y.; Miyagishi, M.; Kodama, T.; Honda, K.; et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007, 448, 501–505. [Google Scholar] [CrossRef]
- Thapa, R.J.; Ingram, J.P.; Ragan, K.B.; Nogusa, S.; Boyd, D.F.; Benitez, A.A.; Sridharan, H.; Kosoff, R.; Shubina, M.; Landsteiner, V.J.; et al. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe 2016, 20, 674–681. [Google Scholar] [CrossRef]
- Kuriakose, T.; Man, S.M.; Malireddi, R.K.; Karki, R.; Kesavardhana, S.; Place, D.E.; Neale, G.; Vogel, P.; Kanneganti, T.D. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016, 1, aag2045. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mansur, D.S.; Peters, N.E.; Ren, H.; Smith, G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 2012, 1, e00047. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Kobayashi, J.; Saitoh, T.; Maruyama, K.; Ishii, K.J.; Barber, G.N.; Komatsu, K.; Akira, S.; Kawai, T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA 2013, 110, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kang, L.; Song, D.; Liu, L.; Yang, S.; Ma, L.; Guo, Z.; Ding, H.; Wang, H.; Yang, B. Ku70 Senses HTLV-1 DNA and Modulates HTLV-1 Replication. J. Immunol. 2017, 199, 2475–2482. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.H.; Macmillan, J.B.; Chen, Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 2009, 138, 576–591. [Google Scholar] [CrossRef]
- Muruve, D.A.; Pétrilli, V.; Zaiss, A.K.; White, L.R.; Clark, S.A.; Ross, P.J.; Parks, R.J.; Tschopp, J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 2008, 452, 103–107. [Google Scholar] [CrossRef]
- Ekchariyawat, P.; Hamel, R.; Bernard, E.; Wichit, S.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; Choumet, V.; Yssel, H.; Desprès, P.; et al. Inflammasome signaling pathways exert antiviral effect against Chikungunya virus in human dermal fibroblasts. Infect. Genet. Evol. 2015, 32, 401–408. [Google Scholar] [CrossRef]
- Lai, J.H.; Wang, M.Y.; Huang, C.Y.; Wu, C.H.; Hung, L.F.; Yang, C.Y.; Ke, P.Y.; Luo, S.F.; Liu, S.J.; Ho, L.J. Infection with the dengue RNA virus activates TLR9 signaling in human dendritic cells. EMBO Rep. 2018, 19, e46182. [Google Scholar] [CrossRef]
- Daniels, B.P.; Kofman, S.B.; Smith, J.R.; Norris, G.T.; Snyder, A.G.; Kolb, J.P.; Gao, X.; Locasale, J.W.; Martinez, J.; Gale, M., Jr.; et al. The Nucleotide Sensor ZBP1 and Kinase RIPK3 Induce the Enzyme IRG1 to Promote an Antiviral Metabolic State in Neurons. Immunity 2019, 50, 64–76. [Google Scholar] [CrossRef]
- Rothan, H.A.; Arora, K.; Natekar, J.P.; Strate, P.G.; Brinton, M.A.; Kumar, M. Z-DNA-Binding Protein 1 Is Critical for Controlling Virus Replication and Survival in West Nile Virus Encephalitis. Front. Microbiol. 2019, 10, 2089. [Google Scholar] [CrossRef] [PubMed]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.-Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef]
- Jin, L.; Waterman, P.M.; Jonscher, K.R.; Short, C.M.; Reisdorph, N.A.; Cambier, J.C. MPYS, a Novel Membrane Tetraspanner, Is Associated with Major Histocompatibility Complex Class II and Mediates Transduction of Apoptotic Signals. Mol. Cell. Biol. 2008, 28, 5014–5026. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Li, X.; Li, P. The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine Growth Factor Rev. 2014, 25, 641–648. [Google Scholar] [CrossRef]
- Mankan, A.K.; Schmidt, T.; Chauhan, D.; Goldeck, M.; Honing, K.; Gaidt, M.; Kubarenko, A.V.; Andreeva, L.; Hopfner, K.P.; Hornung, V. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014, 33, 2937–2946. [Google Scholar] [CrossRef]
- Cheng, Z.; Dai, T.; He, X.; Zhang, Z.; Xie, F.; Wang, S.; Zhang, L.; Zhou, F. The interactions between cGAS-STING pathway and pathogens. Signal Transduct. Target. Ther. 2020, 5, 91. [Google Scholar] [CrossRef]
- Ablasser, A.; Goldeck, M.; Cavlar, T.; Deimling, T.; Witte, G.; Röhl, I.; Hopfner, K.P.; Ludwig, J.; Hornung, V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013, 498, 380–384. [Google Scholar] [CrossRef]
- Wan, D.; Jiang, W.; Hao, J. Research Advances in How the cGAS-STING Pathway Controls the Cellular Inflammatory Response. Front. Immunol. 2020, 11, 615. [Google Scholar] [CrossRef]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef]
- Srikanth, S.; Woo, J.S.; Wu, B.; El-Sherbiny, Y.M.; Leung, J.; Chupradit, K.; Rice, L.; Seo, G.J.; Calmettes, G.; Ramakrishna, C.; et al. The Ca2+ sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 2019, 20, 152–162. [Google Scholar] [CrossRef]
- Wang, L.; Wen, M.; Cao, X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 2019, 365, eaav0758. [Google Scholar] [CrossRef] [PubMed]
- Nazmi, A.; Mukhopadhyay, R.; Dutta, K.; Basu, A. STING Mediates Neuronal Innate Immune Response Following Japanese Encephalitis Virus Infection. Sci. Rep. 2012, 2, 347. [Google Scholar] [CrossRef] [PubMed]
- Nitta, S.; Sakamoto, N.; Nakagawa, M.; Kakinuma, S.; Mishima, K.; Kusano-Kitazume, A.; Kiyohashi, K.; Murakawa, M.; Nishimura-Sakurai, Y.; Azuma, S.; et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 2013, 57, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Zevini, A.; Olagnier, D.; Hiscott, J. Crosstalk between Cytoplasmic RIG-I and STING Sensing Pathways. Trends Immunol. 2017, 38, 194–205. [Google Scholar] [CrossRef]
- Dhanwani, R.; Takahashi, M.; Sharma, S. Cytosolic sensing of immuno-stimulatory DNA, the enemy within. Curr. Opin. Immunol. 2018, 50, 82–87. [Google Scholar] [CrossRef]
- Ahn, J.; Barber, G.N. STING signaling and host defense against microbial infection. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Hopfner, K.-P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [CrossRef]
- Aguirre, S.; Luthra, P.; Sanchez-Aparicio, M.T.; Maestre, A.M.; Patel, J.; Lamothe, F.; Fredericks, A.C.; Tripathi, S.; Zhu, T.; Pintado-Silva, J.; et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2017, 2, 17037. [Google Scholar] [CrossRef]
- Dalrymple, N.A.; Cimica, V.; Mackow, E.R. Dengue Virus NS Proteins Inhibit RIG-I/MAVS Signaling by Blocking TBK1/IRF3 Phosphorylation: Dengue Virus Serotype 1 NS4A Is a Unique Interferon-Regulating Virulence Determinant. MBio 2015, 6, e00553-15. [Google Scholar] [CrossRef]
- Stabell, A.C.; Meyerson, N.R.; Gullberg, R.C.; Gilchrist, A.R.; Webb, K.J.; Old, W.M.; Perera, R.; Sawyer, S.L. Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. Elife 2018, 7, e31919. [Google Scholar] [CrossRef]
- Ding, Q.; Gaska, J.M.; Douam, F.; Wei, L.; Kim, D.; Balev, M.; Heller, B.; Ploss, A. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl. Acad. Sci. USA 2018, 115, E6310–E6318. [Google Scholar] [CrossRef]
- Su, C.I.; Kao, Y.T.; Chang, C.C.; Chang, Y.; Ho, T.S.; Sun, H.S.; Lin, Y.L.; Lai, M.M.C.; Liu, Y.H.; Yu, C.Y. DNA-induced 2′3′-cGAMP enhances haplotype-specific human STING cleavage by dengue protease. Proc. Natl. Acad. Sci. USA 2020, 117, 15947–15954. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Q.; Wu, Y.; Ma, L.; Zhang, Z.; Liu, T.; Jin, S.; She, Y.; Li, Y.-P.; Cui, J. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. EMBO J. 2018, 37, e99347. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, B.; Zeng, M.; Duan, Y.; Wu, Z.; Wu, Y.; Wang, T.; Wang, M.; Jia, R.; Zhu, D.; et al. Binding of Duck Tembusu Virus Nonstructural Protein 2A to Duck STING Disrupts Induction of Its Signal Transduction Cascade To Inhibit Beta Interferon Induction. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, W.; Wu, Y.; Wang, T.; Wu, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; et al. Binding of the Duck Tembusu Virus Protease to STING Is Mediated by NS2B and Is Crucial for STING Cleavage and for Impaired Induction of IFN-β. J. Immunol. 2019, 203, 3374–3385. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Tuiskunen Bäck, A.; Lundkvist, Å. Dengue viruses—An overview. Infect. Ecol. Epidemiol. 2013, 3, 19839. [Google Scholar] [CrossRef]
- Rothman, A.L. Cellular Immunology of Sequential Dengue Virus Infection and its Role in Disease Pathogenesis. In Dengue Virus; Rothman, A.L., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 83–98. [Google Scholar] [CrossRef]
- Malavige, G.N.; Huang, L.-C.; Salimi, M.; Gomes, L.; Jayaratne, S.D.; Ogg, G.S. Cellular and Cytokine Correlates of Severe Dengue Infection. PLoS ONE 2012, 7, e50387. [Google Scholar] [CrossRef]
- Guzman, M.G.; Alvarez, M.; Halstead, S.B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 2013, 158, 1445–1459. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Scott, T.W.; Zou, C.; Pigott, D.M.; Duda, K.A.; Bhatt, S.; Katzelnick, L.; Howes, R.E.; Battle, K.E.; et al. Global spread of dengue virus types: Mapping the 70 year history. Trends Microbiol. 2014, 22, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Marovich, M.; Grouard-Vogel, G.; Louder, M.; Eller, M.; Sun, W.; Wu, S.J.; Putvatana, R.; Murphy, G.; Tassaneetrithep, B.; Burgess, T.; et al. Human dendritic cells as targets of dengue virus infection. J. Investig. Dermatol. Symp. Proc. 2001, 6, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Madoz, J.R.; Bernal-Rubio, D.; Kaminski, D.; Boyd, K.; Fernandez-Sesma, A. Dengue virus inhibits the production of type I interferon in primary human dendritic cells. J. Virol. 2010, 84, 4845–4850. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Madoz, J.R.; Belicha-Villanueva, A.; Bernal-Rubio, D.; Ashour, J.; Ayllon, J.; Fernandez-Sesma, A. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J. Virol. 2010, 84, 9760–9774. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Jordán, J.L.; Laurent-Rolle, M.; Ashour, J.; Martínez-Sobrido, L.; Ashok, M.; Lipkin, W.I.; García-Sastre, A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005, 79, 8004–8013. [Google Scholar] [CrossRef] [PubMed]
- Ashour, J.; Laurent-Rolle, M.; Shi, P.-Y.; García-Sastre, A. NS5 of Dengue Virus Mediates STAT2 Binding and Degradation. J. Virol. 2009, 83, 5408–5418. [Google Scholar] [CrossRef]
- Muñoz-Jordán, J.L. Subversion of interferon by dengue virus. Curr. Top. Microbiol. Immunol. 2010, 338, 35–44. [Google Scholar] [CrossRef]
- Li, J.; Lim, S.P.; Beer, D.; Patel, V.; Wen, D.; Tumanut, C.; Tully, D.C.; Williams, J.A.; Jiricek, J.; Priestle, J.P.; et al. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J. Biol. Chem. 2005, 280, 28766–28774. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, L.; Sun, J.; Chen, W.; Li, S.; Wang, Q.; Yu, H.; Xia, Z.; Jin, X.; Wang, C. Endoplasmic Reticulum Protein SCAP Inhibits Dengue Virus NS2B3 Protease by Suppressing Its K27-Linked Polyubiquitylation. J. Virol. 2017, 91, e02234-16. [Google Scholar] [CrossRef]
- Aguirre, S.; Fernandez-Sesma, A. Collateral Damage during Dengue Virus Infection: Making Sense of DNA by cGAS. J. Virol. 2017, 91, e01081-16. [Google Scholar] [CrossRef]
- Pierson, T.C.; Diamond, M.S. The emergence of Zika virus and its new clinical syndromes. Nature 2018, 560, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Gubler, D.J. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastère, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P.; et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet 2016, 387, 1531–1539. [Google Scholar] [CrossRef]
- Ventura, C.V.; Maia, M.; Dias, N.; Ventura, L.O.; Belfort, R., Jr. Zika: Neurological and ocular findings in infant without microcephaly. Lancet 2016, 387, 2502. [Google Scholar] [CrossRef]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef]
- Driggers, R.W.; Ho, C.Y.; Korhonen, E.M.; Kuivanen, S.; Jääskeläinen, A.J.; Smura, T.; Rosenberg, A.; Hill, D.A.; DeBiasi, R.L.; Vezina, G.; et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N. Engl. J. Med. 2016, 374, 2142–2151. [Google Scholar] [CrossRef]
- Serman, T.M.; Gack, M.U. Evasion of Innate and Intrinsic Antiviral Pathways by the Zika Virus. Viruses 2019, 11, 970. [Google Scholar] [CrossRef]
- Shan, C.; Xie, X.; Shi, P.-Y. Zika Virus Vaccine: Progress and Challenges. Cell Host Microbe 2018, 24, 12–17. [Google Scholar] [CrossRef]
- Pattnaik, A.; Sahoo, B.R.; Pattnaik, A.K. Current Status of Zika Virus Vaccines: Successes and Challenges. Vaccines 2020, 8, 266. [Google Scholar] [CrossRef]
- Sironi, M.; Forni, D.; Clerici, M.; Cagliani, R. Nonstructural Proteins Are Preferential Positive Selection Targets in Zika Virus and Related Flaviviruses. PLoS Negl. Trop. Dis. 2016, 10, e0004978. [Google Scholar] [CrossRef]
- Liu, B.; Tang, L.; Zhang, X.; Ma, J.; Sehgal, M.; Cheng, J.; Zhang, X.; Zhou, Y.; Du, Y.; Kulp, J.; et al. A cell-based high throughput screening assay for the discovery of cGAS-STING pathway agonists. Antivir. Res. 2017, 147, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Margolis, S.R.; Wilson, S.C.; Vance, R.E. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017, 38, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Sauer, J.D.; Sotelo-Troha, K.; von Moltke, J.; Monroe, K.M.; Rae, C.S.; Brubaker, S.W.; Hyodo, M.; Hayakawa, Y.; Woodward, J.J.; Portnoy, D.A.; et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 2011, 79, 688–694. [Google Scholar] [CrossRef]
- Liu, Y.; Gordesky-Gold, B.; Leney-Greene, M.; Weinbren, N.L.; Tudor, M.; Cherry, S. Inflammation-Induced, STING-Dependent Autophagy Restricts Zika Virus Infection in the Drosophila Brain. Cell Host Microbe 2018, 24, 57–68. [Google Scholar] [CrossRef]
- Liu, Y.; Cherry, S. Zika virus infection activates sting-dependent antiviral autophagy in the Drosophila brain. Autophagy 2019, 15, 174–175. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Klionsky, D.J. Inflammatory-dependent Sting activation induces antiviral autophagy to limit zika virus in the Drosophila brain. Autophagy 2019, 15, 1–3. [Google Scholar] [CrossRef]
- Sumpter, R., Jr.; Levine, B. Autophagy and innate immunity: Triggering, targeting and tuning. Semin. Cell Dev. Biol. 2010, 21, 699–711. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef]
- Watson, R.O.; Bell, S.L.; MacDuff, D.A.; Kimmey, J.M.; Diner, E.J.; Olivas, J.; Vance, R.E.; Stallings, C.L.; Virgin, H.W.; Cox, J.S. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe 2015, 17, 811–819. [Google Scholar] [CrossRef]
- Moretti, J.; Roy, S.; Bozec, D.; Martinez, J.; Chapman, J.R.; Ueberheide, B.; Lamming, D.W.; Chen, Z.J.; Horng, T.; Yeretssian, G.; et al. STING Senses Microbial Viability to Orchestrate Stress-Mediated Autophagy of the Endoplasmic Reticulum. Cell 2017, 171, 809–823. [Google Scholar] [CrossRef]
- Diamond, M.S.; Gale, M., Jr. Cell-intrinsic innate immune control of West Nile virus infection. Trends Immunol. 2012, 33, 522–530. [Google Scholar] [CrossRef]
- Petersen, L.R.; Marfin, A.A. West Nile virus: A primer for the clinician. Ann. Intern. Med. 2002, 137, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Sejvar, J.J.; Haddad, M.B.; Tierney, B.C.; Campbell, G.L.; Marfin, A.A.; Van Gerpen, J.A.; Fleischauer, A.; Leis, A.A.; Stokic, D.S.; Petersen, L.R. Neurologic Manifestations and Outcome of West Nile Virus Infection. JAMA 2003, 290, 511–515. [Google Scholar] [CrossRef]
- Watson, J.T.; Pertel, P.E.; Jones, R.C.; Siston, A.M.; Paul, W.S.; Austin, C.C.; Gerber, S.I. Clinical characteristics and functional outcomes of West Nile Fever. Ann. Intern. Med. 2004, 141, 360–365. [Google Scholar] [CrossRef]
- Petersen, L.R.; Brault, A.C.; Nasci, R.S. West Nile virus: Review of the literature. JAMA 2013, 310, 308–315. [Google Scholar] [CrossRef]
- McGuckin Wuertz, K.; Treuting, P.M.; Hemann, E.A.; Esser-Nobis, K.; Snyder, A.G.; Graham, J.B.; Daniels, B.P.; Wilkins, C.; Snyder, J.M.; Voss, K.M.; et al. STING is required for host defense against neuropathological West Nile virus infection. PLoS Pathog. 2019, 15, e1007899. [Google Scholar] [CrossRef]
- You, F.; Wang, P.; Yang, L.; Yang, G.; Zhao, Y.O.; Qian, F.; Walker, W.; Sutton, R.; Montgomery, R.; Lin, R.; et al. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat. Immunol. 2013, 14, 1237–1246. [Google Scholar] [CrossRef]
- Chen, L.H.; Wilson, M.E. Yellow fever control: Current epidemiology and vaccination strategies. Trop. Dis. Travel. Med. Vaccines 2020, 6, 1. [Google Scholar] [CrossRef]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011, 472, 481–485, corrigendum in 2015, 525, 691–695. [Google Scholar] [CrossRef]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505, 691–695, corrigendum in 2015, 525, 691–695. [Google Scholar] [CrossRef]
- Filgueira, L.; Lannes, N. Review of Emerging Japanese Encephalitis Virus: New Aspects and Concepts about Entry into the Brain and Inter-Cellular Spreading. Pathogens 2019, 8, 111. [Google Scholar] [CrossRef]
- Kading, R.C.; Abworo, E.O.; Hamer, G.L. Rift Valley Fever Virus, Japanese Encephalitis Virus, and African Swine Fever Virus: Three Transboundary, Vector-Borne, Veterinary Biothreats With Diverse Surveillance, and Response Capacity Needs. Front. Vet. Sci. 2019, 6, 458. [Google Scholar] [CrossRef]
- Chen, S.; He, Y.; Zhang, R.; Liu, P.; Yang, C.; Wu, Z.; Zhang, J.; Wang, M.; Jia, R.; Zhu, D.; et al. Establishment of a reverse genetics system for duck Tembusu virus to study virulence and screen antiviral genes. Antivir. Res. 2018, 157, 120–127. [Google Scholar] [CrossRef]
- Wu, X.; Wu, F.H.; Wang, X.; Wang, L.; Siedow, J.N.; Zhang, W.; Pei, Z.M. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 2014, 42, 8243–8257. [Google Scholar] [CrossRef]
- Eaglesham, J.B.; Kranzusch, P.J. Conserved strategies for pathogen evasion of cGAS–STING immunity. Curr. Opin. Immunol. 2020, 66, 27–34. [Google Scholar] [CrossRef]
- Ding, Q.; Cao, X.; Lu, J.; Huang, B.; Liu, Y.J.; Kato, N.; Shu, H.B.; Zhong, J. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 2013, 59, 52–58. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Zheng, Y.; Yang, Y.; Xing, Y.; Chen, Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 2014, 5, 369–381. [Google Scholar] [CrossRef]
- Holm, C.K.; Rahbek, S.H.; Gad, H.H.; Bak, R.O.; Jakobsen, M.R.; Jiang, Z.; Hansen, A.L.; Jensen, S.K.; Sun, C.; Thomsen, M.K.; et al. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 2016, 7, 10680. [Google Scholar] [CrossRef]
- Zhao, Y.; Karijolich, J. Know Thyself: RIG-I-Like Receptor Sensing of DNA Virus Infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Chatel-Chaix, L.; Cortese, M.; Romero-Brey, I.; Bender, S.; Neufeldt, C.J.; Fischl, W.; Scaturro, P.; Schieber, N.; Schwab, Y.; Fischer, B.; et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe 2016, 20, 342–356. [Google Scholar] [CrossRef]
- Chang, D.M.; Shaio, M.F. Production of interleukin-1 (IL-1) and IL-1 inhibitor by human monocytes exposed to dengue virus. J. Infect. Dis. 1994, 170, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Aarreberg, L.D.; Esser-Nobis, K.; Driscoll, C.; Shuvarikov, A.; Roby, J.A.; Gale, M., Jr. Interleukin-1β Induces mtDNA Release to Activate Innate Immune Signaling via cGAS-STING. Mol. Cell. 2019, 74, 801–815. [Google Scholar] [CrossRef]
- Nellåker, C.; Yao, Y.; Jones-Brando, L.; Mallet, F.; Yolken, R.H.; Karlsson, H. Transactivation of elements in the human endogenous retrovirus W family by viral infection. Retrovirology 2006, 3, 44. [Google Scholar] [CrossRef]
- Volkman, H.E.; Stetson, D.B. The enemy within: Endogenous retroelements and autoimmune disease. Nat. Immunol. 2014, 15, 415. [Google Scholar] [CrossRef]
- Honda, T.; Tomonaga, K. Endogenous non-retroviral RNA virus elements evidence a novel type of antiviral immunity. Mob. Genet. Elem. 2016, 6, e1165785. [Google Scholar] [CrossRef]
- Manghera, M.; Ferguson-Parry, J.; Lin, R.; Douville, R.N. NF-κB and IRF1 Induce Endogenous Retrovirus K Expression via Interferon-Stimulated Response Elements in Its 5′ Long Terminal Repeat. J. Virol. 2016, 90, 9338. [Google Scholar] [CrossRef]
- Mu, X.; Ahmad, S.; Hur, S. Chapter Two—Endogenous Retroelements and the Host Innate Immune Sensors. In Advances in Immunology; Alt, F.W., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 132, pp. 47–69. [Google Scholar]
- Grandi, N.; Tramontano, E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Chen, S.; Hu, X.; Cui, I.H.; Wu, S.; Dou, C.; Liu, Y.; Sun, Z.; Xue, S.; Geng, T.; Liu, Z.; et al. An endogenous retroviral element exerts an antiviral innate immune function via the derived lncRNA lnc-ALVE1-AS1. Antivir. Res. 2019, 170, 104571. [Google Scholar] [CrossRef]
- Schmidt, N.; Domingues, P.; Golebiowski, F.; Patzina, C.; Tatham, M.H.; Hay, R.T.; Hale, B.G. An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17399–17408. [Google Scholar] [CrossRef]
- Shimizu, A.; Nakatani, Y.; Nakamura, T.; Jinno-Oue, A.; Ishikawa, O.; Boeke, J.D.; Takeuchi, Y.; Hoshino, H. Characterisation of cytoplasmic DNA complementary to non-retroviral RNA viruses in human cells. Sci. Rep. 2014, 4, 5074. [Google Scholar] [CrossRef] [PubMed]
- Dunker, W.; Zhao, Y.; Song, Y.; Karijolich, J. Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes. Viruses 2017, 9, 386. [Google Scholar] [CrossRef]
- Raquin, V.; Merkling, S.H.; Gausson, V.; Moltini-Conclois, I.; Frangeul, L.; Varet, H.; Dillies, M.A.; Saleh, M.C.; Lambrechts, L. Individual co-variation between viral RNA load and gene expression reveals novel host factors during early dengue virus infection of the Aedes aegypti midgut. PLoS Negl. Trop. Dis. 2017, 11, e0006152. [Google Scholar] [CrossRef]
- Suzuki, Y.; Frangeul, L.; Dickson, L.B.; Blanc, H.; Verdier, Y.; Vinh, J.; Lambrechts, L.; Saleh, M.C. Uncovering the Repertoire of Endogenous Flaviviral Elements in Aedes Mosquito Genomes. J. Virol. 2017, 91, e00571-17. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Goic, B.; Tomé-Poderti, L.; Frangeul, L.; Boussier, J.; Gausson, V.; Blanc, H.; Vallet, T.; Loyd, H.; Levi, L.I.; et al. Dicer-2-Dependent Generation of Viral DNA from Defective Genomes of RNA Viruses Modulates Antiviral Immunity in Insects. Cell Host Microbe 2018, 23, 353–365. [Google Scholar] [CrossRef]
- Suzuki, Y.; Baidaliuk, A.; Miesen, P.; Frangeul, L.; Crist, A.B.; Merkling, S.H.; Fontaine, A.; Lequime, S.; Moltini-Conclois, I.; Blanc, H.; et al. Non-retroviral Endogenous Viral Element Limits Cognate Virus Replication in Aedes aegypti Ovaries. Curr. Biol. 2020. [Google Scholar] [CrossRef]
| Genus | Species | Protein | Mechanism of cGAS/STING Antagonism | References |
|---|---|---|---|---|
| Flavivirus | DENV | NS2B | Targets cGAS for degradation through an autophagy–lysosome-dependent mechanism. | [82] |
| NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [10,11,83,84,85,86] | ||
| ZIKV | NS1 | Indirectly degrades cGAS via the enhanced stabilization of caspase-1. | [87] | |
| NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [85] | ||
| WNV | NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [85] | |
| YFV | NS4B | Unknown | [19] | |
| JEV | NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [85] | |
| DTMUV * | NS2A | Competes with duck TBK1 to bind with duck STING, thus disrupting its dimerization and inhibiting downstream IFN production. | [88] | |
| NS2B3 | Proteolytically degrades duck STING through an NS2B dependent manner with the binding of NS2B with duck STING being required for cleaving | [89] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, T.; Fernandez-Sesma, A. Innate Immune DNA Sensing of Flaviviruses. Viruses 2020, 12, 979. https://doi.org/10.3390/v12090979
Zhu T, Fernandez-Sesma A. Innate Immune DNA Sensing of Flaviviruses. Viruses. 2020; 12(9):979. https://doi.org/10.3390/v12090979
Chicago/Turabian StyleZhu, Tongtong, and Ana Fernandez-Sesma. 2020. "Innate Immune DNA Sensing of Flaviviruses" Viruses 12, no. 9: 979. https://doi.org/10.3390/v12090979
APA StyleZhu, T., & Fernandez-Sesma, A. (2020). Innate Immune DNA Sensing of Flaviviruses. Viruses, 12(9), 979. https://doi.org/10.3390/v12090979





