Nucleocapsid Structure of Negative Strand RNA Virus
Abstract
1. Introduction
2. The Capsid Protein Fold
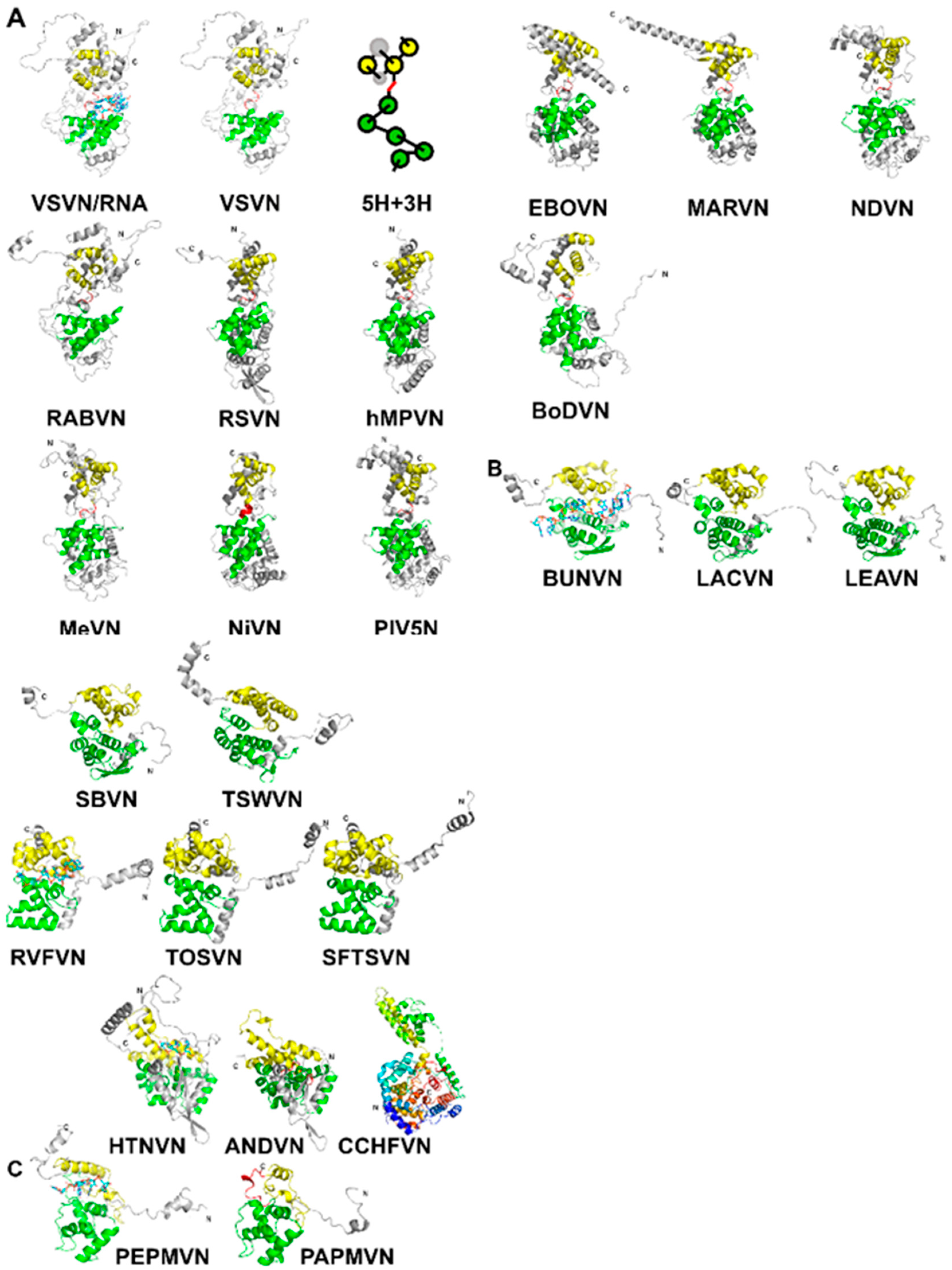
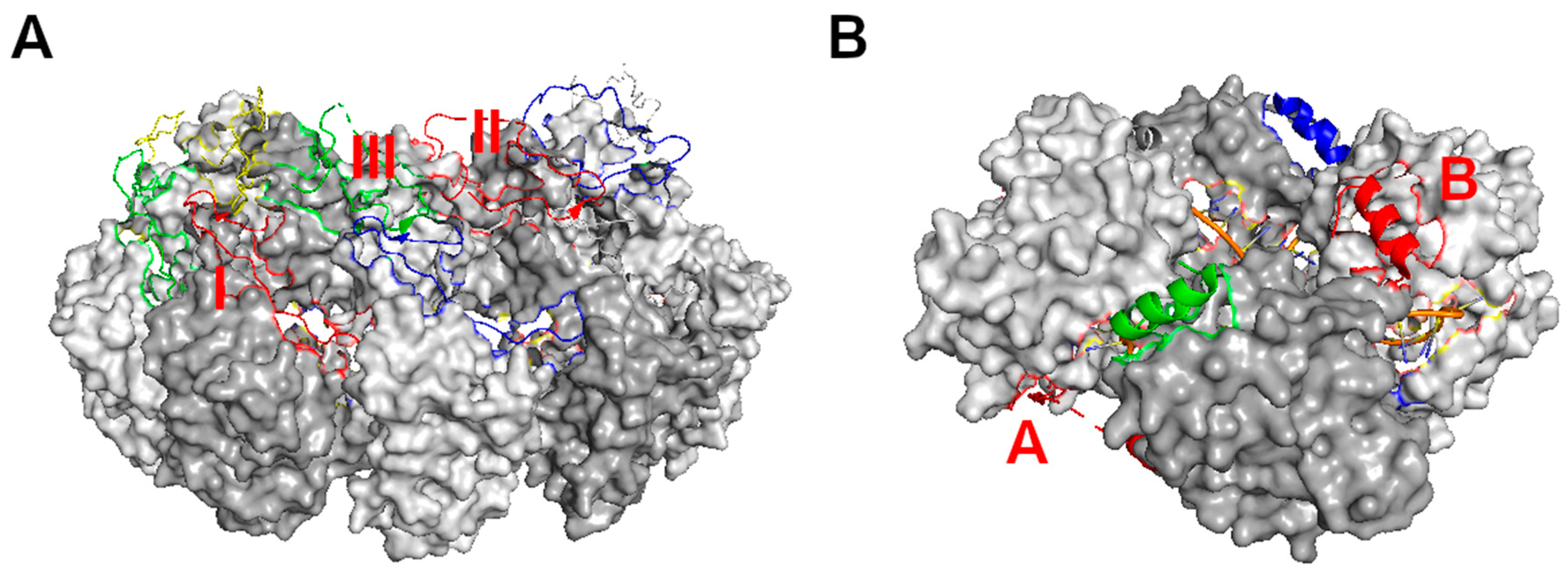
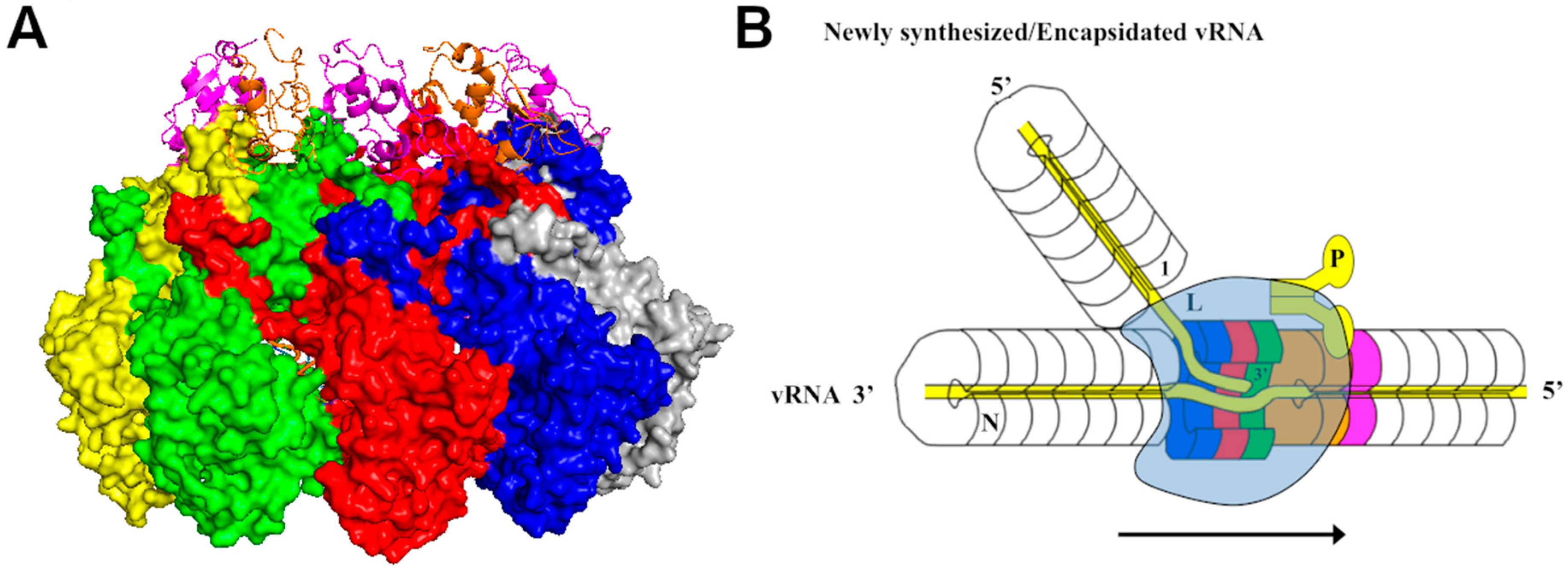
3. Assembly of the Nucleocapsid
4. Viral RNA Synthesis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Knipe, D.; Howley, P.; Fields, B.N.; Griffin, D.E. Fields’ Virilogy: Principles of Virus Structure; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2001; pp. 53–86. [Google Scholar]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, G.K.; Ayllón, M.A.; Bào, Y.; Basler, C.F.; Bavari, S.; Blasdell, K.R.; Briese, T.; Brown, P.A.; Bukreyev, A.; Balkema-Buschmann, A.; et al. Taxonomy of the order Mononegavirales: Update 2019. Arch. Virol. 2019, 164, 1967–1980. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, G.K.; Ayllón, M.A.; Bào, Y.; Basler, C.F.; Bavari, S.; Blasdell, K.R.; Briese, T.; Brown, P.A.; Bukreyev, A.; Balkema-Buschmann, A.; et al. Crystal structure of the borna disease virus nucleoprotein. Structure 2003, 11, 1219–1226. [Google Scholar] [CrossRef]
- Dong, S.; Yang, P.; Li, G.; Liu, B.; Wang, W.; Liu, X.; Xia, B.; Yang, C.; Lou, Z.; Guo, Y.; et al. Insight into the Ebola virus nucleocapsid assembly mechanism: Crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution. Protein Cell 2015, 6, 351–362. [Google Scholar] [CrossRef]
- Liu, B.; Dong, S.; Li, G.; Wang, W.; Liu, X.; Wang, Y.; Yang, C.; Rao, Z.; Guo, Y. Structural insight into nucleoprotein conformation change chaperoned by VP35 peptide in marburg virus. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Song, X.; Shan, H.; Zhu, Y.; Hu, S.; Xue, L.; Chen, Y.; Ding, W.; Niu, T.; Gu, J.; Ouyang, S.; et al. Self-capping of nucleoprotein filaments protects the Newcastle disease virus genome. eLife 2019, 8. [Google Scholar] [CrossRef]
- Yabukarski, F.; Lawrence, P.; Tarbouriech, N.; Bourhis, J.M.; Delaforge, E.; Jensen, M.R.; Ruigrok, R.W.; Blackledge, M.; Volchkov, V.; Jamin, M. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat. Struct. Mol. Biol. 2014, 21, 754–759. [Google Scholar] [CrossRef]
- Gutsche, I.; Desfosses, A.; Effantin, G.; Ling, W.L.; Haupt, M.; Ruigrok, R.W.; Sachse, C.; Schoehn, G. Structural virology: Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science 2015, 348, 704–707. [Google Scholar] [CrossRef]
- Alayyoubi, M.; Leser, G.P.; Kors, C.A.; Lamb, R.A. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc. Natl. Acad. Sci. USA 2015, 112, E1792–E1799. [Google Scholar] [CrossRef]
- Renner, M.; Bertinelli, M.; Leyrat, C.; Paesen, G.C.; Saraiva de Oliveira, L.F.; Huiskonen, J.T.; Grimes, J.M. Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein. eLife 2016, 5, e12627. [Google Scholar] [CrossRef]
- Tawar, R.G.; Duquerroy, S.; Vonrhein, C.; Varela, P.F.; Damier-Piolle, L.; Castagné, N.; MacLellan, K.; Bedouelle, H.; Bricogne, G.; Bhella, D.; et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 2009, 326, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Albertini, A.A.; Wernimont, A.K.; Muziol, T.; Ravelli, R.B.; Clapier, C.R.; Schoehn, G.; Weissenhorn, W.; Ruigrok, R.W. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 2006, 313, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Green, T.J.; Zhang, X.; Wertz, G.W.; Luo, M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 2006, 313, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Liu, T.; Li, S.; King, L.B.; Ngo, N.; Zandonatti, M.A.; Woods, V.L., Jr.; de la Torre, J.C.; Saphire, E.O. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc. Natl. Acad. Sci. USA 2011, 108, 19365–19370. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, W.; Sun, Y.; Ma, C.; Wang, X.; Wang, X.; Liu, P.; Shen, S.; Li, B.; Lin, J.; et al. Crystal structure of the core region of hantavirus nucleocapsid protein reveals the mechanism for ribonucleoprotein complex formation. J. Virol. 2016, 90, 1048–1061. [Google Scholar] [CrossRef]
- Arragain, B.; Reguera, J.; Desfosses, A.; Gutsche, I.; Schoehn, G.; Malet, H. High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms. eLife 2019, 8. [Google Scholar] [CrossRef]
- Carter, S.D.; Surtees, R.; Walter, C.T.; Ariza, A.; Bergeron, É.; Nichol, S.T.; Hiscox, J.A.; Edwards, T.A.; Barr, J.N. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J. Virol. 2012, 86, 10914–10923. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Wang, X.; Dong, H.; Ma, C.; Wang, J.; Liu, B.; Mao, Y.; Wang, Y.; Li, T.; et al. Structural and functional diversity of nairovirus-encoded nucleoproteins. J. Virol. 2015, 89, 11740–11749. [Google Scholar] [CrossRef]
- Li, B.; Wang, Q.; Pan, X.; Fernández de Castro, I.; Sun, Y.; Guo, Y.; Tao, X.; Risco, C.; Sui, S.F.; Lou, Z. Bunyamwera virus possesses a distinct nucleocapsid protein to facilitate genome encapsidation. Proc. Natl. Acad. Sci. USA 2013, 110, 9048–9053. [Google Scholar] [CrossRef]
- Reguera, J.; Malet, H.; Weber, F.; Cusack, S. Structural basis for encapsidation of genomic RNA by La Crosse Orthobunyavirus nucleoprotein. Proc. Natl. Acad. Sci. USA 2013, 110, 7246–7251. [Google Scholar] [CrossRef]
- Niu, F.; Shaw, N.; Wang, Y.E.; Jiao, L.; Ding, W.; Li, X.; Zhu, P.; Upur, H.; Ouyang, S.; Cheng, G.; et al. Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation. Proc. Natl. Acad. Sci. USA 2013, 110, 9054–9059. [Google Scholar] [CrossRef]
- Dong, H.; Li, P.; Bottcher, B.; Elliott, R.M.; Dong, C. Crystal structure of Schmallenberg orthobunyavirus nucleoprotein-RNA complex reveals a novel RNA sequestration mechanism. Rna 2013, 19, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Raymond, D.D.; Piper, M.E.; Gerrard, S.R.; Skiniotis, G.; Smith, J.L. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc. Natl. Acad. Sci. USA 2012, 109, 19008–19213. [Google Scholar] [CrossRef]
- Olal, D.; Dick, A.; Woods, V.L.; Jr Liu, T.; Li, S.; Devignot, S.; Weber, F.; Saphire, E.O.; Daumke, O. Structural insights into RNA encapsidation and helical assembly of the Toscana virus nucleoprotein. Nucleic Acids Res. 2014, 42, 6025–6037. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Ouyang, S.; Liang, M.; Niu, F.; Shaw, N.; Wu, W.; Ding, W.; Jin, C.; Peng, Y.; Zhu, Y.; et al. Structure of severe fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential. J. Virol. 2013, 87, 6829–6839. [Google Scholar] [CrossRef]
- Komoda, K.; Narita, M.; Yamashita, K.; Tanaka, I.; Yao, M. Asymmetric trimeric ring structure of the nucleocapsid protein of tospovirus. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Krug, R.M.; Tao, Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 2006, 444, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.K.; Lam, M.K.; Zhang, H.; Liu, J.; Au, S.W.; Chan, P.K.; Wang, J.; Shaw, P.C. Structural basis for RNA binding and homo-oligomer formation by influenza B virus nucleoprotein. J. Virol. 2012, 86, 6758–6767. [Google Scholar] [CrossRef] [PubMed]
- Donchet, A.; Oliva, J.; Labaronne, A.; Tengo, L.; Miloudi, M.; CA Gerard, F.; Mas, C.; Schoehn, G.; WH Ruigrok, R.; Ducatez, M.; et al. The structure of the nucleoprotein of Influenza D shows that all Orthomyxoviridae nucleoproteins have a similar NPCORE, with or without a NPTAIL for nuclear transport. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Zheng, W.; Olson, J.; Vakharia, V.; Tao, Y.J. The crystal structure and RNA-binding of an orthomyxovirus nucleoprotein. PLoS Pathog. 2013, 9, e1003624. [Google Scholar] [CrossRef]
- Yang, S.; Wang, T.; Bohon, J.; Gagné, M.È.; Bolduc, M.; Leclerc, D.; Li, H. Crystal structure of the coat protein of the flexible filamentous papaya mosaic virus. J. Mol. Biol. 2012, 422, 263–273. [Google Scholar] [CrossRef]
- Agirrezabala, X.; Méndez-López, E.; Lasso, G.; Sánchez-Pina, M.A.; Aranda, M.; Valle, M. The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses. eLife 2015, 4, e11795. [Google Scholar] [CrossRef] [PubMed]
- Green, T.J.; Cox, R.; Tsao, J.; Rowse, M.; Qiu, S.; Luo, M. Common mechanism for RNA encapsidation by negative-strand RNA viruses. J. Virol. 2014, 88, 3766–3775. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chapman, M.S.; Liljas, L. Structural folds of viral proteins. Adv. Protein Chem. 2003, 64, 125–196. [Google Scholar] [CrossRef]
- Pandit, S.B.; Skolnick, J. Fr-TM-align: A new protein structural alignment method based on fragment alignments and the TM-score. BMC Bioinform. 2008, 9, 531. [Google Scholar] [CrossRef]
- Luo, M.; Green, T.J.; Zhang, X.; Tsao, J.; Qiu, S. Structural comparisons of the nucleoprotein from three negative strand RNA virus families. J. Virol. 2007, 4, 72–78. [Google Scholar] [CrossRef][Green Version]
- Wichgers Schreur, P.J.; Kormelink, R.; Kortekaas, J. Genome packaging of the Bunyavirales. Curr. Opin. Virol. 2018, 33, 151–155. [Google Scholar] [CrossRef]
- Sherman, M.B.; Freiberg, A.N.; Holbrook, M.R.; Watowich, S.J. Single-particle cryo-electron microscopy of Rift Valley fever virus. Virology 2009, 387, 11–15. [Google Scholar] [CrossRef]
- DeLano, W.L. PyMOL Molecular Graphic System, version 1.3; Schrödinger LLC: New York, NY, USA, 2015. [Google Scholar]
- Desfosses, A.; Ribeiro, E.A., Jr.; Schoehn, G.; Blondel, D.; Guilligay, D.; Jamin, M.; Ruigrok, R.W.; Gutsche, I. Self-organization of the vesicular stomatitis virus nucleocapsid into a bullet shape. Nat. Commun. 2013, 4, 1631. [Google Scholar] [CrossRef]
- Ge, P.; Tsao, J.; Schein, S.; Green, T.J.; Luo, M.; Zhou, Z.H. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 2010, 327, 689–693. [Google Scholar] [CrossRef]
- Luo, M.; Green, T.J.; Zhang, X.; Tsao, J.; Qiu, S. Conserved characteristics of the rhabdovirus nucleoprotein. Virus Res. 2007, 129, 246–251. [Google Scholar] [CrossRef]
- Zhang, X.; Green, T.J.; Tsao, J.; Qiu, S.; Luo, M. Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J. Virol. 2008, 82, 674–682. [Google Scholar] [CrossRef][Green Version]
- Arranz, R.; Coloma, R.; Chichón, F.J.; Conesa, J.J.; Carrascosa, J.L.; Valpuesta, J.M.; Ortín, J.; Martín-Benito, J. The structure of native influenza virion ribonucleoproteins. Science 2012, 338, 1634–1637. [Google Scholar] [CrossRef]
- Moeller, A.; Kirchdoerfer, R.N.; Potter, C.S.; Carragher, B.; Wilson, I.A. Organization of the influenza virus replication machinery. Science 2012, 338, 1631–1634. [Google Scholar] [CrossRef]
- Pflug, A.; Guilligay, D.; Reich, S.; Cusack, S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 2014, 516, 355–360. [Google Scholar] [CrossRef]
- Leyrat, C.; Yabukarski, F.; Tarbouriech, N.; Ribeiro, E.A., Jr.; Jensen, M.R.; Blackledge, M.; Ruigrok, R.W.; Jamin, M. Structure of the vesicular stomatitis virus N0-P complex. PLoS Pathog. 2011, 7, e1002248. [Google Scholar] [CrossRef]
- Aggarwal, M.; Leser, G.P.; Kors, C.A.; Lamb, R.A. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein in complex with an amino-terminal peptide of the phosphoprotein. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Abelson, D.M.; Li, S.; Wood, M.R.; Saphire, E.O. Assembly of the Ebola Virus nucleoprotein from a chaperoned VP35 complex. Cell Rep. 2015, 12, 140–149. [Google Scholar] [CrossRef]
- Landeras-Bueno, S.; Oda, S.I.; Norris, M.J.; Li Salie, Z.; Guenaga, J.; Wyatt, R.T.; Saphire, E.O. Sudan Ebolavirus VP35-NP crystal structure reveals a potential target for pan-filovirus treatment. mBio 2019, 10. [Google Scholar] [CrossRef]
- Raymond, D.D.; Piper, M.E.; Gerrard, S.R.; Smith, J.L. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc. Natl. Acad. Sci. USA 2010, 107, 11769–11774. [Google Scholar] [CrossRef]
- Chenavas, S.; Estrozi, L.F.; Slama-Schwok, A.; Delmas, B.; Di Primo, C.; Baudin, F.; Li, X.; Crépin, T.; Ruigrok, R.W. Monomeric nucleoprotein of influenza A virus. PLoS Pathog. 2013, 9, e1003275. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Green, T.J.; Lu, S.; Luo, M. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 2006, 80, 2808–2814. [Google Scholar] [CrossRef] [PubMed]
- Jenni, S.; Bloyet, L.M.; Diaz-Avalos, R.; Liang, B.; Whelan, S.; Grigorieff, N.; Harrison, S.C. Structure of the vesicular stomatitis virus l protein in complex with its phosphoprotein cofactor. Cell Rep. 2020, 30, 53–60. [Google Scholar] [CrossRef]
- Ruigrok, R.W.; Crepin, T.; Kolakofsky, D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 2011, 14, 504–510. [Google Scholar] [CrossRef]
- Kolakofsky, D.; Roux, L.; Garcin, D.; Ruigrok, R.W. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: A hypothesis. J. Gen. Virol. 2005, 86, 1869–1877. [Google Scholar] [CrossRef]
- Kolakofsky, D.; Pelet, T.; Garcin, D.; Hausmann, S.; Curran, J.; Roux, L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: The rule of six revisited. J. Virol. 1998, 72, 891–899. [Google Scholar] [CrossRef]
- Iseni, F.; Baudin, F.; Garcin, D.; Marq, J.B.; Ruigrok, R.W.; Kolakofsky, D. Chemical modification of nucleotide bases and mRNA editing depend on hexamer or nucleoprotein phase in Sendai virus nucleocapsids. Rna 2002, 8, 1056–1067. [Google Scholar] [CrossRef]
- Harouaka, D.; Wertz, G.W. Mutations in the C-terminal loop of the nucleocapsid protein affect vesicular stomatitis virus RNA replication and transcription differentially. J. Virol. 2009, 83, 11429–11439. [Google Scholar] [CrossRef]
- Harouaka, D.; Wertz, G.W. Second-site mutations selected in transcriptional regulatory sequences compensate for engineered mutations in the vesicular stomatitis virus nucleocapsid protein. J. Virol. 2012, 86, 11266–11275. [Google Scholar] [CrossRef]
- Anhlan, D.; Grundmann, N.; Makalowski, W.; Ludwig, S.; Scholtissek, C. Origin of the 1918 pandemic H1N1 influenza A virus as studied by codon usage patterns and phylogenetic analysis. Rna 2011, 17, 64–73. [Google Scholar] [CrossRef]
- Cristina, J.; Moreno, P.; Moratorio, G.; Musto, H. Genome-wide analysis of codon usage bias in Ebolavirus. Virus Res. 2015, 196, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Pagan, I.; Holmes, E.C.; Simon-Loriere, E. Level of gene expression is a major determinant of protein evolution in the viral order Mononegavirales. J. Virol. 2012, 86, 5253–5263. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, C.; Tekes, G.; Mueller, S.; Paul, A.; Whelan, S.P.; Wimmer, E. Recoding of the vesicular stomatitis virus L gene by computer-aided design provides a live, attenuated vaccine candidate. mBio 2015, 6. [Google Scholar] [CrossRef]
- Rima, B.K. Nucleotide sequence conservation in paramyxoviruses; the concept of codon constellation. J. Gen. Virol. 2015, 96, 939–955. [Google Scholar] [CrossRef]
- Gumpper, R.H.; Li, W.; Luo, M. Contrains of viral RNA synthesis on codon usage of negative strand RNA virus. J. Virol. 2018. [Google Scholar] [CrossRef]
- Gumpper, R.H.; Li, W.; Castañeda, C.H.; Scuderi, M.J.; Bashkin, J.K.; Luo, M. A polyamide inhibits replication of vesicular stomatitis virus by targeting RNA in the nucleocapsid. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Reich, S.; Guilligay, D.; Pflug, A.; Malet, H.; Berger, I.; Crépin, T.; Hart, D.; Lunardi, T.; Nanao, M.; Ruigrok, R.W.; et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 2014, 516, 361–366. [Google Scholar] [CrossRef]
- Hengrung, N.; El Omari, K.; Serna Martin, I.; Vreede, F.T.; Cusack, S.; Rambo, R.P.; Vonrhein, C.; Bricogne, G.; Stuart, D.I.; Grimes, J.M.; et al. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 2015, 527, 114–117. [Google Scholar] [CrossRef]
- Fan, H.; Walker, A.P.; Carrique, L.; Keown, J.R.; Serna Martin, I.; Karia, D.; Sharps, J.; Hengrung, N.; Pardon, E.; Steyaert, J.; et al. Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Nature 2019, 573, 287–290. [Google Scholar] [CrossRef]
- Pflug, A.; Lukarska, M.; Resa-Infante, P.; Reich, S.; Cusack, S. Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res. 2017, 234, 103–117. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef]
- Reguera, J.; Gerlach, P.; Cusack, S. Towards a structural understanding of RNA synthesis by negative strand RNA viral polymerases. Curr. Opin. Struct. Biol. 2016, 36, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, P.; Malet, H.; Cusack, S.; Reguera, J. Structural Insights into bunyavirus replication and its regulation by the vRNA promoter. Cell 2015, 161, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Biedenkopf, N.; Hartlieb, B.; Hoenen, T.; Becker, S. Phosphorylation of Ebola virus VP30 influences the composition of the viral nucleocapsid complex: Impact on viral transcription and replication. J. Biol. Chem. 2013, 288, 11165–11174. [Google Scholar] [CrossRef] [PubMed]
- Kirchdoerfer, R.N.; Moyer, C.L.; Abelson, D.M.; Saphire, E.O. The Ebola Virus VP30-NP interaction is a regulator of viral RNA synthesis. PLoS Pathog. 2016, 12, e1005937. [Google Scholar] [CrossRef] [PubMed]
- Green, T.J.; Luo, M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc. Natl. Acad. Sci. USA 2009, 106, 11713–11718. [Google Scholar] [CrossRef] [PubMed]
- Kingston, R.L.; Baase, W.A.; Gay, L.S. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 2004, 78, 8630–8640. [Google Scholar] [CrossRef]
- Bourhis, J.M.; Receveur-Bréchot, V.; Oglesbee, M.; Zhang, X.; Buccellato, M.; Darbon, H.; Canard, B.; Finet, S.; Longhi, S. The intrinsically disordered C-terminal domain of the measles virus nucleoprotein interacts with the C-terminal domain of the phosphoprotein via two distinct sites and remains predominantly unfolded. Protein. Sci. 2005, 14, 1975–1992. [Google Scholar] [CrossRef]
- Kingston, R.L.; Gay, L.S.; Baase, W.S.; Matthews, B.W. Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization. J. Mol. Biol. 2008, 379, 719–731. [Google Scholar] [CrossRef]
- Cox, R.; Pickar, A.; Qiu, S.; Tsao, J.; Rodenburg, C.; Dokland, T.; Elson, A.; He, B.; Luo, M. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc. Natl. Acad. Sci. USA 2014, 111, 15208–15213. [Google Scholar] [CrossRef]
- Cox, R.; Green, T.J.; Purushotham, S.; Deivanayagam, C.; Bedwell, G.J.; Prevelige, P.E.; Luo, M. Structural and functional characterization of the mumps virus phosphoprotein. J. Virol. 2013, 87, 7558–7568. [Google Scholar] [CrossRef]
- Horwitz, J.A.; Jenni, S.; Harrison, S.C.; Whelan, S.P.J. Structure of a rabies virus polymerase complex from electron cryo-microscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Gilman, M.; Liu, C.; Fung, A.; Behera, I.; Jordan, P.; Rigaux, P.; Ysebaert, N.; Tcherniuk, S.; Sourimant, J.; Eléouët, J.F.; et al. Structure of the respiratory syncytial virus polymerase complex. Cell 2019, 179, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Pan, J.; Qian, X.; Lattmann, S.; El Sahili, A.; Yeo, T.H.; Jia, H.; Cressey, T.; Ludeke, B.; Noton, S.; et al. Structure of the human metapneumovirus polymerase phosphoprotein complex. Nature 2020, 577, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Ogino, M.; Gupta, N.; Green, T.J.; Ogino, T. A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res. 2019, 47, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Qanungo, K.R.; Shaji, D.; Mathur, M.; Banerjee, A.K. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. USA 2004, 101, 5952–5957. [Google Scholar] [CrossRef]
- Ogino, T.; Green, T.J. RNA Synthesis and capping by non-segmented negative strand RNA viral polymerases: Lessons from a prototypic virus. Front. Microbiol. 2019, 10, 2714. [Google Scholar] [CrossRef]
- Hodges, J.; Tang, X.; Landesman, M.B.; Ruedas, J.B.; Ghimire, A.; Gudheti, M.V.; Perrault, J.; Jorgensen, E.M.; Gerton, J.M.; Saffarian, S. Asymmetric packaging of polymerases within vesicular stomatitis virus. Biochem. Biophys. Res. Commun. 2013, 440, 271–276. [Google Scholar] [CrossRef]
- Liang, B.; Li, Z.; Jenni, S.; Rahmeh, A.A.; Morin, B.M.; Grant, T.; Grigorieff, N.; Harrison, S.C.; Whelan, S. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 2015, 162, 314–327. [Google Scholar] [CrossRef]
- Guryanov, S.G.; Liljeroos, L.; Kasaragod, P.; Kajander, T.; Butcher, S.J. Crystal structure of the measles virus nucleoprotein core in complex with an N-terminal region of phosphoprotein. J. Virol. 2016, 90, 2849–2857. [Google Scholar] [CrossRef]
- Severin, C.; Terrell, J.R.; Zengel, J.R.; Cox, R.; Plemper, R.K.; He, B.; Luo, M. Releasing the genomic RNA sequestered in the mumps virus nucleocapsid. J. Virol. 2016, 99, 10113–10119. [Google Scholar] [CrossRef] [PubMed]
- Green, T.J.; Rowse, M.; Tsao, J.; Kang, J.; Ge, P.; Zhou, H.; Luo, M. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J. Virol. 2011, 85, 2714–2722. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gumpper, R.H.; Uddin, Y.; Schmidt-Krey, I.; Luo, M. Mutations in the nucleocapsid protein were complemented by mutations in the L protein to restore viral RNA synthesis. J. Virol. 2018. [Google Scholar] [CrossRef]
- Abrescia, N.G.; Bamford, D.H.; Grimes, J.M.; Stuart, D.I. Structure unifies the viral universe. Annu. Rev. Biochem. 2012, 81, 795–822. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G.; Johnson, J.E. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989, 58, 533–573. [Google Scholar] [CrossRef]

| Family | Subfamily | Genus | Name | (PDB Code) |
|---|---|---|---|---|
| Subphylum:Haploviricotina;Class:Monjiviricetes;Order:Mononegavirales | ||||
| Bornaviridae | Mammalian 1 orthobornavirus | Borna disease virus (BoDV) | (1N93) [4] | |
| Filoviridae | Ebolavirus | Ebola virus (EBOV) | (4Z9P) [5] | |
| Marburgvirus | Marburg virus (MARV) | (5F5M) [6] | ||
| Paramyxoviridae | Avulaviridae | Avian orthoavulavirus 1 | Newcastle disease virus (NDV) | (6JC3) [7] |
| Orthoparamyxovirdae | Henipavirus | Nipah virus (NiV) | (4CO6) [8] | |
| Morbillivirus | Measles virus (MeV) | (4UFT) [9] | ||
| Rubulaviridae | Orrthorubulavirus | parainfluenza virus 5 (PIV5) | (4XJN) [10] | |
| Pneumoviridae | Metapneumovirus | Human metapneumovirus (hMPV) | (5FVC) [11] | |
| Orthopneumovirus | respiratory syncytial virus (RSV) | (2WJ8) [12] | ||
| Rhabdoviridae | Lyssavirus | Rabies virus (RABV) | (2GTT) [13] | |
| Vesiculovirus | Vesicular stomatitis virus (VSV) | (2GIC) [14] | ||
| Subphylum:Polyploviricotina;Class:Ellioviricetes;Order:Bunyavirales | ||||
| Arenaviridae | Mammarenavirus | Lassa Virus (LASV) | (3T5Q) [15] | |
| Hantaviridae | Mammantaviridae | Orthohantavirus | Andes virus (ANDV) | (5E04) [16] |
| Hantaan virus (HTNV) | (6I2N) [17] | |||
| Nairoviridae | Orthonairovirus | Crimean-Congo Hemorrhagic Fever Virus (CCHFV) | (4AKL) [18] | |
| Hazara virus (HAZV) | (4XZE) [19] | |||
| Kupe virus (KUPV) | (4XZC) [19] | |||
| Erve virus (ERVEV) | (4XZ8) [19] | |||
| Peribunyaviridae | Orthobunyavirus | Bunyamwera virus (BUNV) | (4IJS) [20] | |
| La Crosse virus (LACV) | (4BHH) [21] | |||
| Leanyer orthobunyavirus (LEAV) | (4J1G) [22] | |||
| Schmallenberg virus (SBV) | (4JNG) [23] | |||
| Phenuiviridae | Phlebovirus | Rift Valley Fever Virus (RVFV) | (4H5P) [24] | |
| Toscana virus (TOSV) | (4CSF) [25] | |||
| Severe fever with thrombocytopenia syndrome virus (SFTSV) | 4J4R) [26] | |||
| Tospoviridae | Orthotospovirus | Tomato spotted wilt tospovirus (TSWV) | (5IP3) [27] | |
| Subphylum:Polyploviricotina;Class:Insthoviricetes;Order:Articulavirales | ||||
| Orthomyxoviridae | Alphainfluenzavirus | Influenza A virus (IFAV) | (2IQH) [28] | |
| Betainfluenzavirus | Influenza B virus (IFBV) | (3TJ0) [29] | ||
| Deltainfluenzavirus | Influenza D virus (IFDV) | (5N2U) [30] | ||
| Isavirus | Infectious Salmon Anemia Virus (ISAV) | (4EWC) [31] | ||
| Class:Tymovirales *;Order:Alphaflexiviridae | ||||
| Potexvirus | Papaya mosaic virus (PapMV) | (4DOX) [32] | ||
| Pepino mosaic virus (PepMV) | (5FN1) [33] | |||
| RABV | RSV | hMPV | PIV5 | MeV | NiV | NDV | EBOV | MARV | BoDV | |
|---|---|---|---|---|---|---|---|---|---|---|
| VSV | 2.63/96 * | 5.07/79 | 5.17/81 | 5.34/78 | 5.30/79 | 4.73/83 | 5.20/75 | 5.18/82 | 4.20/85 | 5.09/78 |
| RABV | 5.01/76 | 4.93/78 | 5.04/73 | 5.06/76 | 4.27/81 | 5.11/73 | 4.38/78 | 5.01/82 | 5.03/75 | |
| RSV | 1.40/100 | 4.35/91 | 4.35/91 | 4.90/93 | 4.36/91 | 4.39/87 | 4.85/86 | 4.83/80 | ||
| hMPV | 4.10/94 | 4.20/94 | 4.79/92 | 4.24/94 | 4.48/89 | 4.11/90 | 4.36/78 | |||
| PIV5 | 2.47/97 | 3.59/100 | 1.82/98 | 4.87/89 | 3.77/94 | 4.51/83 | ||||
| MeV | 3.26/97 | 2.32/98 | 4.43/87 | 3.97/94 | 4.41/81 | |||||
| NiV | 3.38/99 | 4.12/92 | 3.62/91 | 5.19/80 | ||||||
| NDV | 4.79/89 | 3.92/96 | 4.76/84 | |||||||
| EBOV | 1.87/99 | 5.53/83 | ||||||||
| MARV | 5.47/84 |
| LACV | LEAV | SBV | TSWV | TOSV | SFTSV | |
|---|---|---|---|---|---|---|
| BUNV | 2.11/98 | 1.87/100 | 2.07/100 | 3.68/87 | - | - |
| LACV | 2.07/94 | 2.09/93 | 3.64/85 | - | - | |
| LEAV | 1.66/99 | 3.96/89 | - | - | ||
| SBV | 3.52/88 | - | - | |||
| RVFV | 1.88/91 | 1.83/90 | ||||
| TOSV | 2.65/94 |
| HTNV | HAZV | KUPV | ERVEV | IFBV | IFDV | ISAV | PepMV | |
|---|---|---|---|---|---|---|---|---|
| ANDV | 1.94/95 | - | - | - | - | - | - | - |
| CCHFV | 1.77/100 † | 1.90/99 | 1.56/91 | - | - | - | - | |
| HAZV | 1.56/97 | 1.32/89 | - | - | - | - | ||
| KUPV | 1.27/87 | - | - | - | - | |||
| IFAV | 2.29/95 | 3.21/94 | 3.49/84 | - | ||||
| IFBV | 3.02/93 | 3.36/78 | - | |||||
| IFDV | 3.58/76 | - | ||||||
| PapMV | 2.06/93 |
| VSV-N ≠ | CCHFV-N | HTNV-C | |
|---|---|---|---|
| BUNV-N | 4.77/82 | 3.77/82 | |
| BUNV-C | 2.88/87 | ||
| PepMV-N | 3.28/61 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, M.; Terrell, J.R.; Mcmanus, S.A. Nucleocapsid Structure of Negative Strand RNA Virus. Viruses 2020, 12, 835. https://doi.org/10.3390/v12080835
Luo M, Terrell JR, Mcmanus SA. Nucleocapsid Structure of Negative Strand RNA Virus. Viruses. 2020; 12(8):835. https://doi.org/10.3390/v12080835
Chicago/Turabian StyleLuo, Ming, James Ross Terrell, and Shelby Ashlyn Mcmanus. 2020. "Nucleocapsid Structure of Negative Strand RNA Virus" Viruses 12, no. 8: 835. https://doi.org/10.3390/v12080835
APA StyleLuo, M., Terrell, J. R., & Mcmanus, S. A. (2020). Nucleocapsid Structure of Negative Strand RNA Virus. Viruses, 12(8), 835. https://doi.org/10.3390/v12080835






