Replication of a Dog-Origin H6N1 Influenza Virus in Cell Culture and Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Cells
2.3. Plaque Assay
2.4. Viral Dynamics
2.5. MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) Assay
2.6. Chloramphenicol Acetyl Transferase Enzyme-Linked Immunosorbent Assay (CAT ELISA)
2.7. Animals
2.8. Quantitative Reverse Transcription Real Time PCR (qRT-PCR)
2.9. Histopathological Analysis and Immunohistochemistry (IHC)
2.10. Histopathological and Immunohistochemistry Scoring
2.11. Statistical Analyses
3. Results
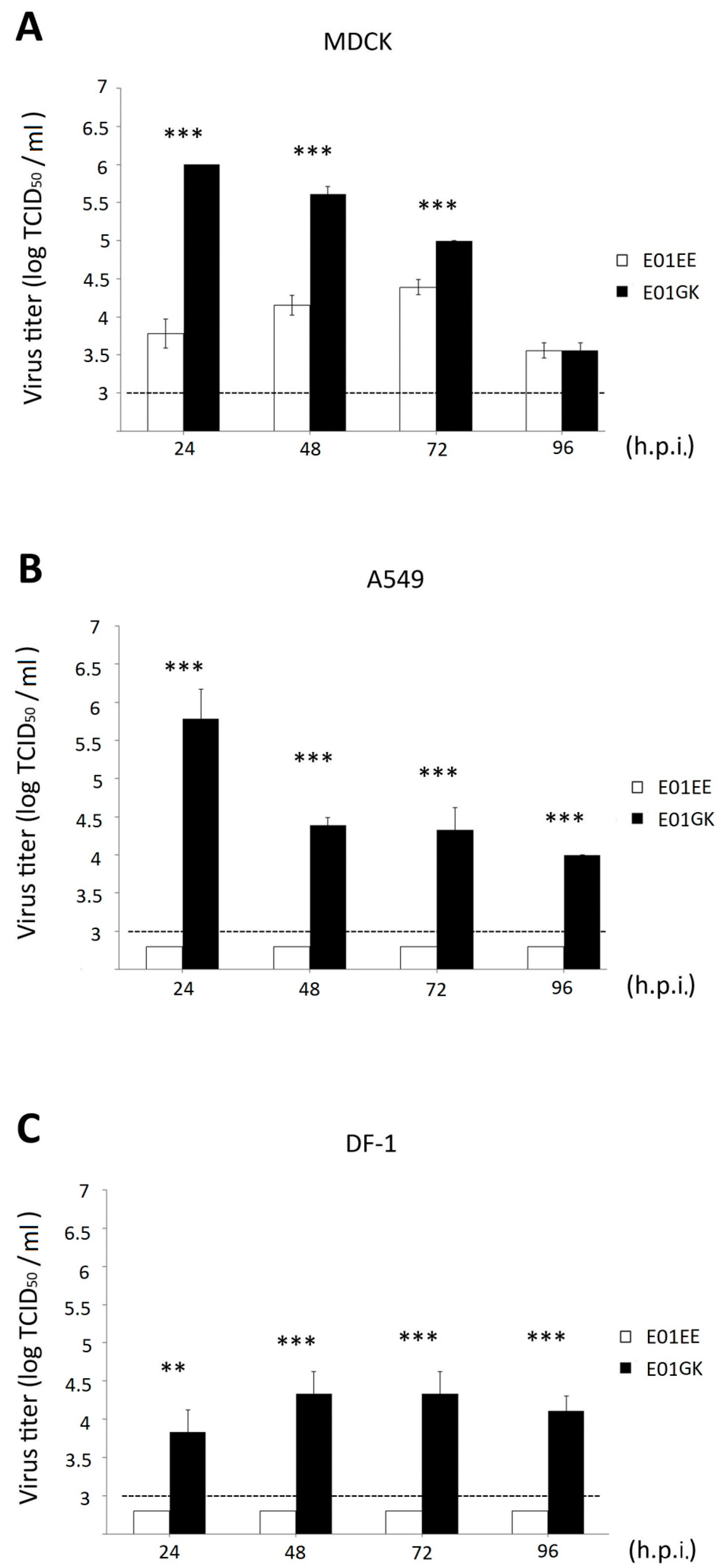
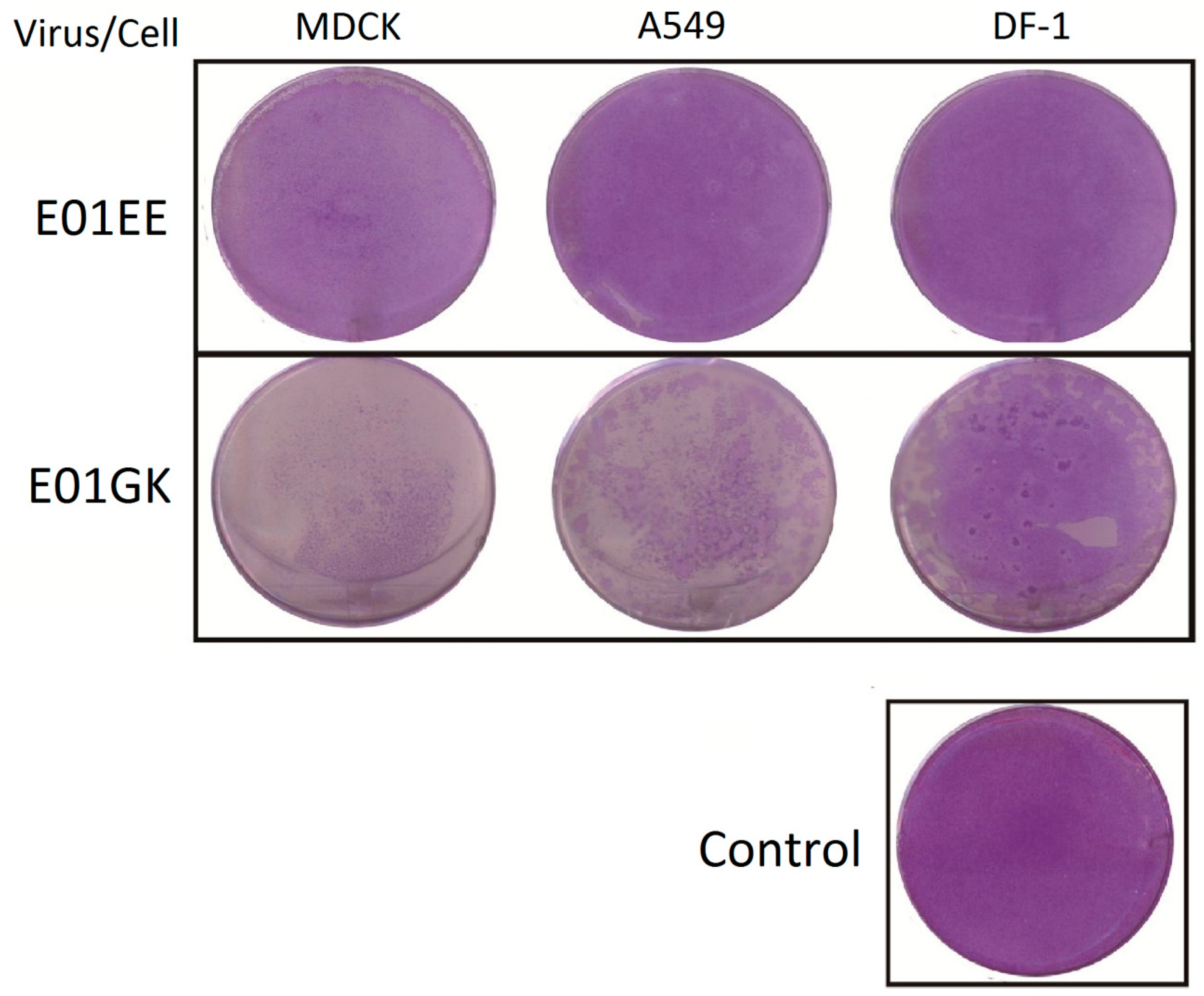
3.1. Viral Dynamics and Plaque Assay
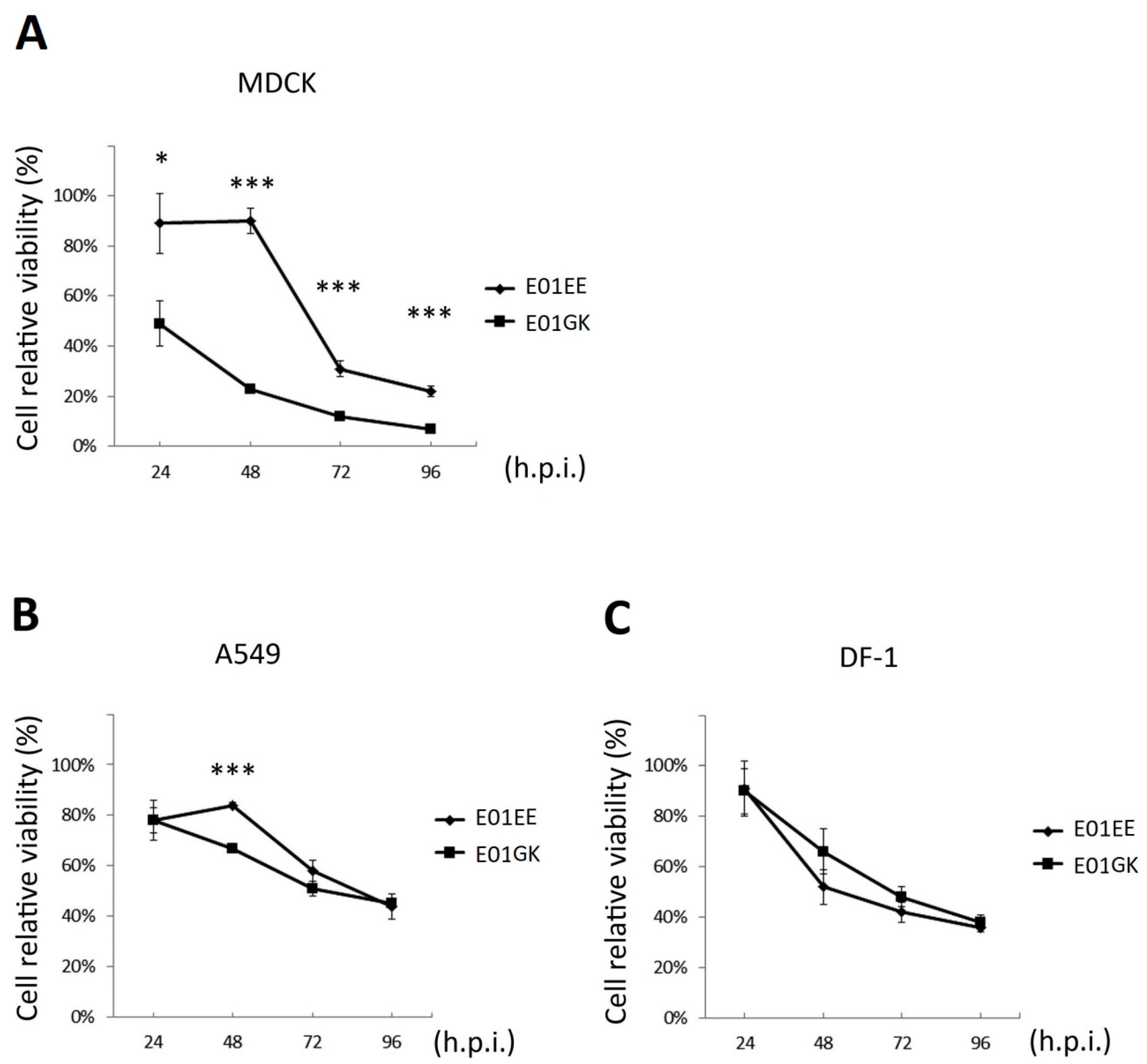
3.2. Cellular Viability Comparison
3.3. Viral Ribonucleoprotein (RNP) Activity Test
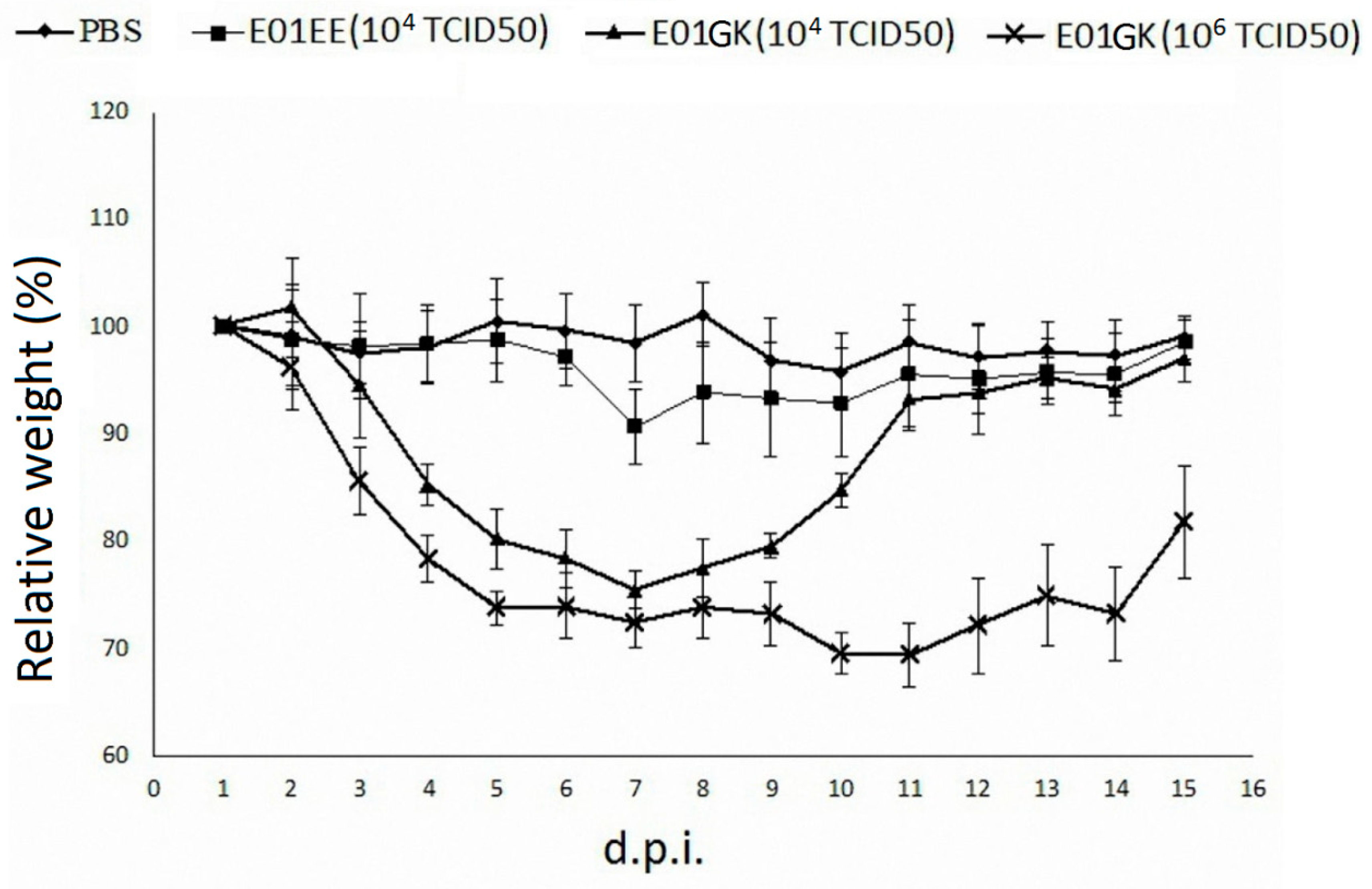
3.4. The Virus Infection of Mice
3.5. Virus Amount Quantification
3.6. Histopathological and Immunohistochemistry (IHC) Evaluation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lipatov, A.S.; Govorkova, E.A.; Webby, R.J.; Ozaki, H.; Peiris, M.; Guan, Y.; Poon, L.; Webster, R.G. Influenza: Emergence and control. J. Virol. 2004, 78, 8951–8959. [Google Scholar] [CrossRef] [PubMed]
- Crawford, P.C.; Dubovi, E.J.; Castleman, W.L.; Stephenson, I.; Gibbs, E.P.; Chen, L.; Smith, C.; Hill, R.C.; Ferro, P.; Pompey, J.; et al. Transmission of equine influenza virus to dogs. Science 2005, 310, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Kang, B.; Lee, C.; Jung, K.; Ha, G.; Kang, D.; Park, S.; Park, B.; Oh, J. Transmission of avian influenza virus (H3N2) to dogs. Emerg. Infect. Dis. 2008, 14, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Payungporn, S.; Crawford, P.C.; Kouo, T.S.; Chen, L.M.; Pompey, J.; Castleman, W.L.; Dubovi, E.J.; Katz, J.M.; Donis, R.O. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg. Infect. Dis. 2008, 14, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Lee, C.; Kang, B.; Jung, K.; Oh, T.; Kim, H.; Park, B.; Oh, J. Experimental infection of dogs with avian-origin canine influenza A virus (H3N2). Emerg. Infect. Dis. 2009, 15, 56–58. [Google Scholar] [CrossRef]
- Li, S.; Shi, Z.; Jiao, P.; Zhang, G.; Zhong, Z.; Tian, W.; Long, L.P.; Cai, Z.; Zhu, X.; Liao, M.; et al. Avian-origin H3N2 canine influenza A viruses in Southern China. Infect. Genet. Evol. 2010, 10, 1286–1288. [Google Scholar] [CrossRef]
- Zhan, G.J.; Ling, Z.S.; Zhu, Y.L.; Jiang, S.J.; Xie, Z.J. Genetic characterization of a novel influenza A virus H5N2 isolated from a dog in China. Vet. Microbiol. 2012, 155, 409–416. [Google Scholar]
- Song, Q.Q.; Zhang, F.X.; Liu, J.J.; Ling, Z.S.; Zhu, Y.L.; Jiang, S.J.; Xie, Z.J. Dog to dog transmission of a novel influenza virus (H5N2) isolated from a canine. Vet. Microbiol. 2013, 161, 331–333. [Google Scholar] [CrossRef]
- Bunpapong, N.; Nonthabenjawan, N.; Chaiwong, S.; Tangwangvivat, R.; Boonyapisitsopa, S.; Jairak, W.; Tuanudom, R.; Prakairungnamthip, D.; Suradhat, S.; Thanawongnuwech, R.; et al. Genetic characterization of canine influenza A virus (H3N2) in Thailand. Virus Genes 2014, 48, 56–63. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, S.; Ma, J.; Tan, Y.; Du, L.; Shen, Y.; Mu, Q.; Pu, J.; Lin, D.; Liu, J. Identification and characterization of avian-origin H3N2 canine influenza viruses in northern China during 2009–2010. Virology 2013, 435, 301–307. [Google Scholar] [CrossRef]
- Songserm, T.; Amonsin, A.; Jam-on, R.; Sae-Heng, N.; Pariyothorn, N.; Payungporn, S.; Theamboonlers, A.; Chutinimitkul, S.; Thanawongnuwech, R.; Poovorawan, Y. Fatal avian influenza A H5N1 in a dog. Emerg. Infect. Dis. 2006, 12, 1744–1747. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Moon, H.J.; An, D.J.; Jeoung, H.Y.; Kim, H.; Yeom, M.J.; Hong, M.; Nam, J.H.; Park, S.J.; Park, B.K.; et al. A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J. Gen. Virol. 2012, 93, 551–554. [Google Scholar] [CrossRef]
- Lee, M.S.; Chang, P.C.; Shien, J.H.; Cheng, M.C.; Chen, C.L.; Shieh, H.K. Genetic and pathogenic characterization of H6N1 avian influenza viruses isolated in Taiwan between 1972 and 2005. Avian Dis. 2006, 50, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.H.; Yang, J.R.; Wu, H.S.; Chang, M.C.; Lin, J.S.; Lin, C.Y.; Liu, Y.L.; Lo, Y.C.; Yang, C.H.; Chuang, J.H.; et al. Human infection with avian influenza A H6N1 virus: An epidemiological analysis. Lancet Respir. Med. 2013, 1, 771–778. [Google Scholar] [CrossRef]
- Lin, H.T.; Wang, C.H.; Chueh, L.L.; Su, B.L.; Wang, L.C. Influenza A(H6N1) Virus in Dogs, Taiwan. Emerg. Infect. Dis. 2015, 21, 2154–2157. [Google Scholar] [CrossRef]
- Coloma, R.; Valpuesta, J.M.; Arranz, R.; Carrascosa, J.L.; Ortin, J.; Martin-Benito, J. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009, 5, e1000491. [Google Scholar] [CrossRef]
- DesRochers, B.L.; Chen, R.E.; Gounder, A.P.; Pinto, A.K.; Bricker, T.; Linton, C.N.; Rogers, C.D.; Williams, G.D.; Webby, R.J.; Boon, A.C. Residues in the PB2 and PA genes contribute to the pathogenicity of avian H7N3 influenza A virus in DBA/2 mice. Virology 2016, 494, 89–99. [Google Scholar] [CrossRef]
- Cheng, K.; Yu, Z.; Chai, H.; Sun, W.; Xin, Y.; Zhang, Q.; Huang, J.; Zhang, K.; Li, X.; Yang, S.; et al. PB2-E627K and PA-T97I substitutions enhance polymerase activity and confer a virulent phenotype to an H6N1 avian influenza virus in mice. Virology 2014, 468, 207–213. [Google Scholar] [CrossRef]
- Yu, Z.; Cheng, K.; Sun, W.; Zhang, X.; Li, Y.; Wang, T.; Wang, H.; Zhang, Q.; Xin, Y.; Xue, L.; et al. A PB1 T296R substitution enhance polymerase activity and confer a virulent phenotype to a 2009 pandemic H1N1 influenza virus in mice. Virology 2015, 486, 180–186. [Google Scholar] [CrossRef]
- Chen, G.W.; Kuo, S.M.; Yang, S.L.; Gong, Y.N.; Hsiao, M.R.; Liu, Y.C.; Shih, S.R.; Tsao, K.C. Genomic Signatures for Avian H7N9 Viruses Adapting to Humans. PLoS ONE 2016, 11, e0148432. [Google Scholar] [CrossRef]
- Wang, C.; Lee, H.H.; Yang, Z.F.; Mok, C.K.; Zhang, Z. PB2-Q591K Mutation Determines the Pathogenicity of Avian H9N2 Influenza Viruses for Mammalian Species. PLoS ONE 2016, 11, e0162163. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Manual on Animal Influenza Diagnosis and Surveillance. Available online: https://apps.who.int/iris/handle/10665/68026 (accessed on 3 March 2016).
- Shahsavandi, S.; Ebrahimi, M.M.; Mohammadi, A.; Zarrin Lebas, N. Impact of chicken-origin cells on adaptation of a low pathogenic influenza virus. Cytotechnology 2013, 65, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, D.; Sun, W.; Liu, J.; He, L.; Hu, J.; Gu, M.; Wang, X.; Liu, X.; Hu, S.; et al. Multiplex one-step Real-time PCR by Taqman-MGB method for rapid detection of pan and H5 subtype avian influenza viruses. PLoS ONE 2017, 12, e0178634. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.C.; Huang, D.; Chen, H.W. Simultaneous subtyping and pathotyping of avian influenza viruses in chickens in Taiwan using reverse transcription loop-mediated isothermal amplification and microarray. J. Vet. Med. Sci. 2016, 78, 1223–1228. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirui, J.; Bucci, M.D.; Poole, D.S.; Mehle, A. Conserved features of the PB2 627 domain impact influenza virus polymerase function and replication. J. Virol. 2014, 88, 5977–5986. [Google Scholar] [CrossRef]
- Chin, A.W.; Li, O.T.; Mok, C.K.; Ng, M.K.; Peiris, M.; Poon, L.L. Influenza A viruses with different amino acid residues at PB2-627 display distinct replication properties in vitro and in vivo: Revealing the sequence plasticity of PB2-627 position. Virology 2014, 468, 545–555. [Google Scholar] [CrossRef]
- Lu, Y.S.; Sugimura, T.; Shieh, H.K.; Lee, Y.L.; Jong, M.H. Isolation and identification of an influenza A virus in ducks in Taiwan. J. Chin. Soc. Vet. Med. 1985, 11, 23–34. [Google Scholar]
- Lee, H.C.; Hsu, C.N.; Kuo, T.F.; Wang, C.H. Molecular epidemiology of avian influenza virus H6N1 in Taiwan from 2000 to 2003. Taiwan Vet. J. 2005, 31, 230–239. [Google Scholar]
- Beare, A.; Webster, R. Replication of avian influenza viruses in humans. Arch. Virol. 1991, 119, 37–42. [Google Scholar] [CrossRef]
- Stevens, J.; Blixt, O.; Tumpey, T.M.; Taubenberger, J.K.; Paulson, J.C.; Wilson, I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 2006, 312, 404–410. [Google Scholar] [CrossRef]
- Schrauwen, E.J.; Fouchier, R.A. Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microbes. Infect. 2014, 3, e9. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Hossain, M.J.; Hickman, D.; Perez, D.R.; Lamb, R.A. A new influenza virus virulence determinant: The NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. USA 2008, 105, 4381–4386. [Google Scholar] [CrossRef] [PubMed]
- Obenauer, J.C.; Denson, J.; Mehta, P.K.; Su, X.; Mukatira, S.; Finkelstein, D.B.; Xu, X.; Wan, J.; Ma, J.; Fan, Y.; et al. Large-scale sequence analysis of avian influenza isolates. Science 2006, 311, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Thongratsakul, S.; Suzuki, Y.; Hiramatsu, H.; Sakpuaram, T.; Sirinarumitr, T.; Poolkhet, C.; Moonjit, P.; Yodsheewan, R.; Songserm, T. Avian and human influenza A virus receptors in trachea and lung of animals. Asian Pac. J. Allergy Immunol. 2010, 28, 294–301. [Google Scholar] [PubMed]
- Viswanathan, K.; Chandrasekaran, A.; Srinivasan, A.; Raman, R.; Sasisekharan, V.; Sasisekharan, R. Glycans as receptors for influenza pathogenesis. Glycoconj. J. 2010, 27, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y. Sialobiology of influenza: Molecular mechanism of host range variation of influenza viruses. Biol. Pharm. Bull. 2005, 28, 399–408. [Google Scholar] [CrossRef]
- Ning, Z.Y.; Luo, M.Y.; Qi, W.B.; Yu, B.; Jiao, P.R.; Liao, M. Detection of expression of influenza virus receptors in tissues of BALB/c mice by histochemistry. Vet. Res. Commun. 2009, 33, 895–903. [Google Scholar] [CrossRef]
- Van Loon, A.; de Haas, N.; Zeyda, I.; Mundt, E. Alteration of amino acids in VP2 of very virulent infectious bursal disease virus results in tissue culture adaptation and attenuation in chickens. J. Gen. Virol. 2002, 83, 121–129. [Google Scholar] [CrossRef]
- Song, H.; Santi, N.; Evensen, O.; Vakharia, V.N. Molecular determinants of infectious pancreatic necrosis virus virulence and cell culture adaptation. J. Virol. 2005, 79, 10289–10299. [Google Scholar] [CrossRef]
- Miao, H.; Hollenbaugh, J.A.; Zand, M.S.; Holden-Wiltse, J.; Mosmann, T.R.; Perelson, A.S.; Wu, H.; Topham, D.J. Quantifying the early immune response and adaptive immune response kinetics in mice infected with influenza A virus. J. Virol. 2010, 84, 6687–6698. [Google Scholar] [CrossRef]
| Viruses | Amino Acid | |
|---|---|---|
| PB1 Position 739 | PB2 Position 627 | |
| E01 | E | K |
| E01EE | E | E |
| E01GK | G | K |
| PBS | 104 TCID50/mL of E01EE | 104 TCID50/mL of E01GK | 106 TCID50/mL of E01GK | |
|---|---|---|---|---|
| H&E b | 0 | 2 | 3 | 3 |
| IHC c | 0 | 1 | 2 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, S.-K.; Shih, C.-H.; Chang, H.-W.; Teng, K.-H.; Hsu, W.-E.; Lin, H.-J.; Lin, H.-Y.; Huang, C.-H.; Chen, H.-W.; Wang, L.-C. Replication of a Dog-Origin H6N1 Influenza Virus in Cell Culture and Mice. Viruses 2020, 12, 704. https://doi.org/10.3390/v12070704
Tsai S-K, Shih C-H, Chang H-W, Teng K-H, Hsu W-E, Lin H-J, Lin H-Y, Huang C-H, Chen H-W, Wang L-C. Replication of a Dog-Origin H6N1 Influenza Virus in Cell Culture and Mice. Viruses. 2020; 12(7):704. https://doi.org/10.3390/v12070704
Chicago/Turabian StyleTsai, Shou-Kuan, Cheng-Hsin Shih, Hui-Wen Chang, Kuang-Huan Teng, Wei-En Hsu, Han-Jia Lin, Han-You Lin, Ching-Huei Huang, Hui-Wen Chen, and Lih-Chiann Wang. 2020. "Replication of a Dog-Origin H6N1 Influenza Virus in Cell Culture and Mice" Viruses 12, no. 7: 704. https://doi.org/10.3390/v12070704
APA StyleTsai, S.-K., Shih, C.-H., Chang, H.-W., Teng, K.-H., Hsu, W.-E., Lin, H.-J., Lin, H.-Y., Huang, C.-H., Chen, H.-W., & Wang, L.-C. (2020). Replication of a Dog-Origin H6N1 Influenza Virus in Cell Culture and Mice. Viruses, 12(7), 704. https://doi.org/10.3390/v12070704







