Evidence of the Mechanism by Which Polyomaviruses Exploit the Extracellular Vesicle Delivery System during Infection
Abstract
1. Introduction
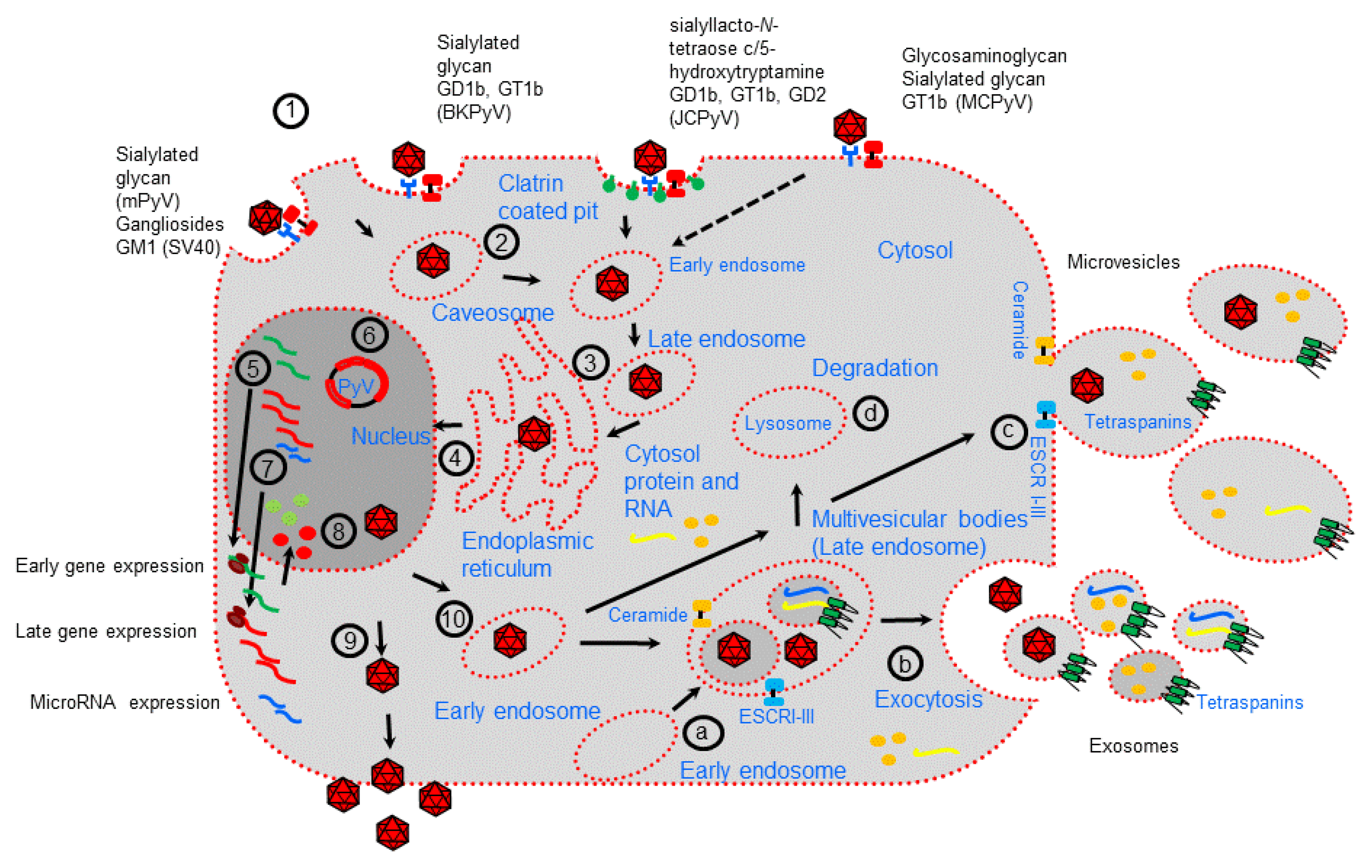
2. Polyomavirus Life Cycle and Extracellular Vesicles Biogenesis
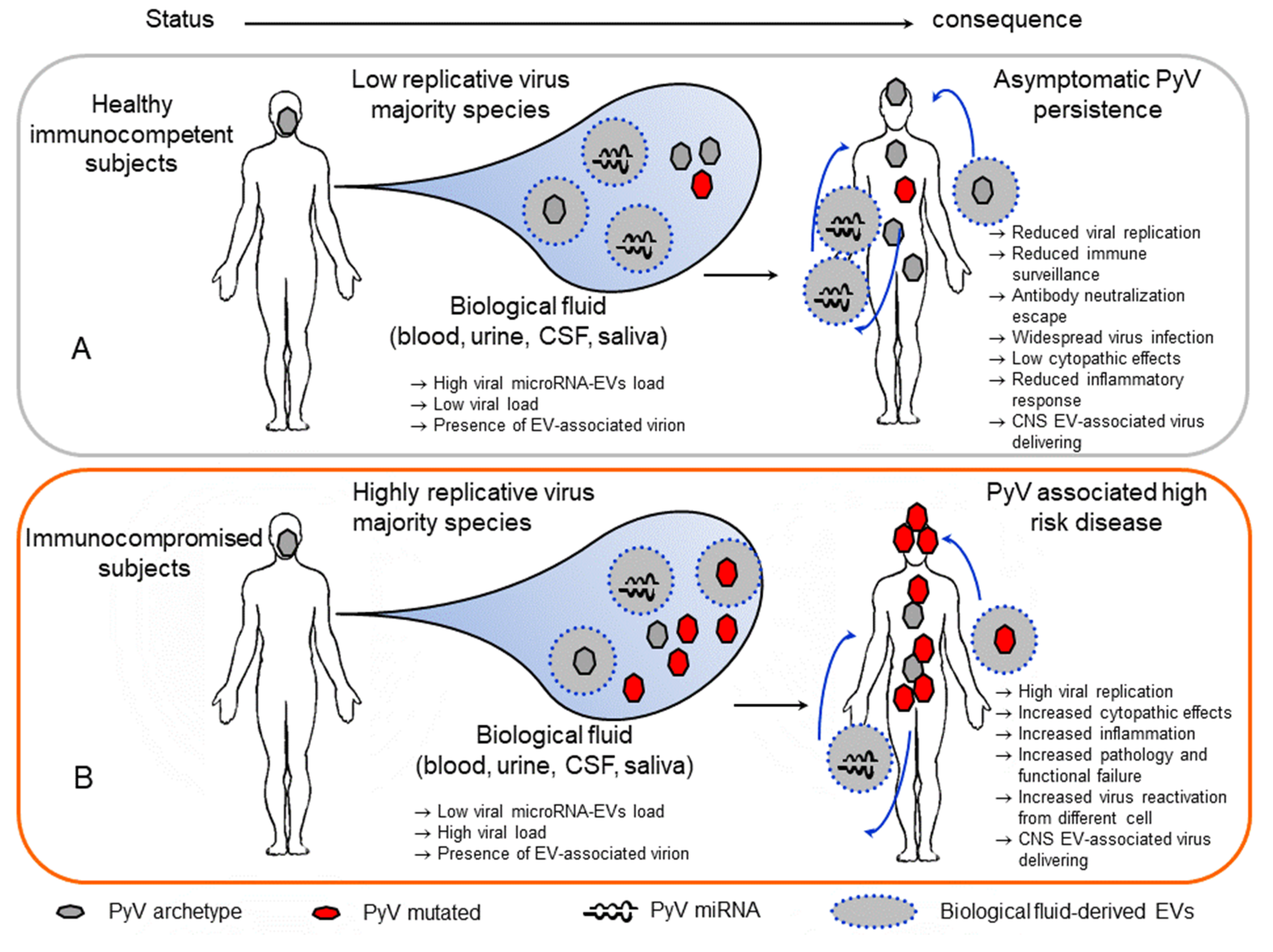
3. Polyomavirus Association with Extracellular Vesicles
4. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 297, 1257–1260. [Google Scholar] [CrossRef]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971, 297, 1253–1277. [Google Scholar] [CrossRef]
- Moens, U.; Krumbholz, A.; Ehlers, B.; Zell, R.; Johne, R.; Calvignac-Spencer, S.; Lauber, C. Biology, evolution, and medical importance of polyomaviruses: An update. Infect. Genet. Evol. 2017, 54, 18–38. [Google Scholar] [CrossRef] [PubMed]
- Calvignac-Spencer, S.; Feltkamp, M.C.; Daugherty, M.D.; Moens, U.; Ramqvist, T.; Johne, R.; Ehlers, B.A. Polyomaviridae Study Group of the International Committee on Taxonomy of Viruses, taxonomy update for the family Polyomaviridae. Arch. Virol. 2016, 161, 1739–1750. [Google Scholar] [CrossRef] [PubMed]
- Dalianis, T.; Hirsch, H.H. Human polyomaviruses in disease and cancer. Virology 2013, 437, 63–72. [Google Scholar] [CrossRef]
- Moens, U.; Van Ghelue, M.; Song, X.; Ehlers, B. Serological cross-reactivity between human polyomaviruses. Rev. Med. Virol. 2013, 23, 250–264. [Google Scholar] [CrossRef]
- Prado, J.C.M.; Monezi, T.A.; Amorim, A.T.; Lino, V.; Paladino, A.; Boccardo, E. Human polyomaviruses and cancer: An overview. Clinics 2018, 73 (Suppl. 1), e558s. [Google Scholar] [CrossRef]
- van Niel, G.; D‘Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Altan-Bonnet, N. Extracellular vesicles are the Trojan horses of viral infection. Curr. Opin. Microbiol. 2016, 32, 77–81. [Google Scholar] [CrossRef]
- Anderson, M.R.; Kashanchi, F.; Jacobson, S. Exosomes in Viral Disease. Neurotherapeutics 2016, 13, 535–546. [Google Scholar] [CrossRef]
- Feng, Z.; Muratori, C.; Santiana, M.; Ghosh, S.; Ho, B.A.; Rajasekaran, V.; Du, W.L.; Mutsafi, Y.; de Jesus-Diaz, D.A.; Sosnovtsev, S.V.; et al. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe 2018, 24, 208–220.e8. [Google Scholar] [CrossRef]
- Raab-Traub, N.; Dittmer, D.P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017, 15, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Ben-nun-Shaul, O.; Bronfeld, H.; Reshef, D.; Schueler-Furman, O.; Oppenheim, A. The SV40 capsid is stabilized by a conserved pentapeptide hinge of the major capsid protein VP1. J. Mol. Biol. 2009, 386, 1382–1391. [Google Scholar] [CrossRef]
- Hurdiss, D.L.; Morgan, E.L.; Thompson, R.F.; Prescott, E.L.; Panou, M.M.; Macdonald, A.; Ranson, N.A. New structural insights into the genome and minor capsid proteins of BK polyomavirus using Cryo-electron microscopy. Structure 2016, 24, 528–536. [Google Scholar] [CrossRef]
- Fang, C.Y.; Chen, H.Y.; Wang, M.; Chen, P.L.; Chang, C.F.; Chen, L.S.; Shen, C.H.; Ou, W.C.; Tsai, M.D.; Hsu, P.H.; et al. Global analysis of modifications of the human BK virus structural proteins by LC-MS/MS. Virology 2010, 402, 164–176. [Google Scholar] [CrossRef]
- Frearson, P.M.; Crawford, L.V. Polyoma virus basic proteins. J. Gen. Virol. 1972, 14, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Campanero-Rhodes, M.A.; Smith, A.; Chai, W.; Sonnino, S.; Mauri, L.; Childs, R.A.; Zhang, Y.; Ewers, H.; Helenius, A.; Imberty, A.; et al. N-glycolyl GM1 ganglioside as a receptor for simian virus 40. J. Virol. 2007, 81, 12846–12858. [Google Scholar] [CrossRef] [PubMed]
- Boss, I.W.; Renne, R. Viral miRNAs and immune evasion. Biochim. Biophys. Acta 2011, 1809, 708–714. [Google Scholar] [CrossRef]
- Chen, C.J.; Cox, J.E.; Azarm, K.D.; Wylie, K.N.; Woolard, K.D.; Pesavento, P.A.; Sullivan, C.S. Identification of a polyomavirus microRNA highly expressed in tumors. Virology 2015, 476, 43–53, Erratum in: Virology 2015, 482, 269. [Google Scholar] [CrossRef]
- Lagatie, O.; Tritsmans, L.; Stuyver, L.J. The miRNA world of polyomaviruses. Virol. J. 2013, 10, 268–288. [Google Scholar] [CrossRef]
- Sullivan, C.S.; Grundhoff, A.; Tevethia, S.; Treisman, R.; Pipas, J.M.; Ganem, D. Expression and function of microRNAs in viruses great and small. Cold. Spring. Harb. Symp. Quant. Biol. 2006, 71, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Bauman, Y.; Mandelboim, O. MicroRNA based immunoevasion mechanism of human polyomaviruses. RNA Biol. 2011, 8, 591–594. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, S.D.; Stehle, T.; Garcea, R. Glycan receptors of the Polyomaviridae: Structure, function, and pathogenesis. Curr. Opin. Virol. 2014, 7, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Maginnis, M.S.; Haley, S.A.; Gee, G.V.; Atwood, W.J. Role of N-linked glycosylation of the 5-HT2A receptor in JC virus infection. J. Virol. 2010, 84, 9677–9684. [Google Scholar] [CrossRef]
- Erickson, K.D.; Garcea, R.L.; Tsai, B. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J. Virol. 2009, 83, 10275–10279. [Google Scholar] [CrossRef]
- Cerqueira, C.; Schelhaas, M. Principles of polyoma- and papillomavirus uncoating. Med. Microbiol. Immunol. 2012, 201, 427–436. [Google Scholar] [CrossRef]
- Dupzyk, A.; Tsai, B. How polyomaviruses exploit the ERAD machinery to cause infection. Viruses 2016, 8, 242. [Google Scholar] [CrossRef]
- Grassel, L.; Fast, L.A.; Scheffer, K.D.; Boukhallouk, F.; Spoden, G.A.; Tenzer, S.; Boller, K.; Bago, R.; Rajesh, S.; Overduin, M.; et al. The CD63-Syntenin-1 Complex Controls PostEndocytic Trafficking of Oncogenic Human Papillomaviruses. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Kuksin, D.; Norkin, L.C. Disassociation of the SV40 genome from capsid proteins prior to nuclear entry. Virol. J. 2012, 9, 158. [Google Scholar] [CrossRef]
- DeCaprio, J.A.; Garcea, R.L. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013, 11, 264–276. [Google Scholar] [CrossRef]
- Good, P.J.; Welch, R.C.; Barkan, A.; Somasekhar, M.B.; Mertz, J.E. Both VP2 and VP3 are synthesized from each of the alternative spliced late 19S RNA species of simian virus 40. J. Virol. 1988, 62, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Safak, M.; Barrucco, R.; Darbinyan, A.; Okada, Y.; Nagashima, K.; Khalili, K. Interaction of JC virus agno protein with T antigenmodulates transcription and replication of the viral genome in glial cells. J. Virol. 2001, 75, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Jay, G.; Nomura, S.; Anderson, C.W.; Khoury, G. Identification of the SV40 agnogene product: A DNA binding protein. Nature 1981, 291, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, C.H.; Traavik, T.; Hey, A. The agnogene of the human polyomavirus BK is expressed. J. Virol. 1998, 72, 6233–6236. [Google Scholar] [CrossRef]
- Valle, M.; Chen, X.S.; Donate, L.E.; Fanning, E.; Carazo, J.M. Structural basis for the cooperative assembly of large T antigen on the origin of replication. J. Mol. Biol. 2006, 357, 1295–1305. [Google Scholar] [CrossRef]
- Saribas, A.S.; White, M.K.; Safak, M. JC virus agnoprotein enhances large T antigen binding to the origin of viral DNA replication: Evidence for its involvement in viral DNA replication. Virology 2012, 433, 12–26. [Google Scholar] [CrossRef]
- Tsang, S.H.; Wang, R.; Nakamaru-Ogiso, E.; Knight, S.A.; Buck, C.B.; You, J. The oncogenic small tumor antigen of Merkel cell polyomavirus is an iron-sulfur cluster protein that enhances viral DNA replication. J. Virol. 2015, 90, 1544–1556. [Google Scholar] [CrossRef]
- Martelli, F.; Giannecchini, S. Polyomavirus microRNAs circulating in biological fluids during viral persistence. Rev. Med. Virol. 2017, 27. [Google Scholar] [CrossRef]
- Erickson, K.D.; Bouchet-Marquis, C.; Heiser, K.; Szomolanyi-Tsuda, E.; Mishra, R.; Lamothe, B.; Hoenger, A.; Garcea, R.L. Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Allison, A.C.; Black, P.H. Lysosomal changes in lytic and nonlytic infections with the simian vacuolating virus (SV40). J. Natl. Cancer Inst. 1967, 39, 777–782. [Google Scholar]
- Evans, G.L.; Caller, L.G.; Foster, V.; Crump, C.M. Anion homeostasis is important for non-lytic release of BK polyomavirus from infected cells. Open Biol. 2015, 5, 150041. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.C.; Norton, E.; Dang, X.; Koralnik, I.J. Agnogene deletion in a novel pathogenic JC virus isolate impairs VP1 expression and virion production. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, M.; Myhre, M.R.; Dragset, M.; Tummler, C.; Moens, U. Phosphorylation of human polyomavirus BK agnoprotein at Ser-11 is mediated by PKC and has an important regulative function. Virology 2008, 379, 97–109. [Google Scholar] [CrossRef]
- Sariyer, I.K.; Saribas, A.S.; White, M.K.; Safak, M. Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virol. J. 2011, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Semba, S.; Sunden, Y.; Orba, Y.; Kobayashi, S.; Nagashima, K.; Kimura, T.; Hasegawa, H.; Sawa, H. Role of JC virus agnoprotein in virion formation. Microbiol. Immunol. 2012, 56, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Orba, Y.; Makino, Y.; Okada, Y.; Sunden, Y.; Hasegawa, H.; Hall, W.W.; Sawa, H. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc. Natl. Acad. Sci. USA 2013, 110, 18668–18673. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, M.; Walquist, M.; Gerits, N.; Dragset, M.; Spang, A.; Moens, U. BKV agnoprotein interacts with alpha-soluble N-ethylmaleimide-sensitive fusion attachment protein, and negatively influences transport of VSVG-EGFP. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Engel, S.; Heger, T.; Mancini, R.; Herzog, F.; Kartenbeck, J.; Hayer, A.; Helenius, A. Role of endosomes in simian virus 40 entry and infection. J. Virol. 2011, 85, 4198–4211. [Google Scholar] [CrossRef]
- Chen, Y.J.; Liu, X.; Tsai, B. SV40 hijacks cellular transport, membrane penetration, and disassembly machineries to promote infection. Viruses 2019, 11, 917. [Google Scholar] [CrossRef]
- Clayson, E.T.; Brando, L.V.; Compans, R.W. Release of simian virus 40 virions from epithelial cells is polarized and occurs without cell lysis. J. Virol. 1989, 63, 2278–2288. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed]
- Kubo, H. Extracellular vesicles in lung disease. Chest 2018, 153, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. Genet. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Stoorvogel, W.; Strous, G.J.; Geuze, H.J.; Oorschot, V.; Schwartz, A.L. Late endosomes derive from early endosomes by maturation. Cell 1991, 65, 417–427. [Google Scholar] [CrossRef]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell. Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell. Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef]
- Davies, B.A.; Lee, J.R.; Oestreich, A.J.; Katzmann, D.J. Membrane protein targeting to the MVB/lysosome. Chem. Rev. 2009, 109, 1575–1586. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. Hrs and endocytic sorting of ubiquitinated membrane proteins. Cell. Struct. Funct. 2002, 27, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001, 106, 145–155. [Google Scholar] [CrossRef]
- Babst, M.; Katzmann, D.J.; Snyder, W.B.; Wendland, B.; Emr, S.D. Endosome- associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002, 3, 283–289. [Google Scholar] [CrossRef]
- Tamai, K.; Tanaka, N.; Nakano, T.; Kakazu, E.; Kondo, Y.; Inoue, J.; Shiina, M.; Fukushima, K.; Hoshino, T.; Sano, K.; et al. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 2010, 399, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Goni, F.M.; Alonso, A. Effects of ceramide and other simple sphingolipids on membrane lateral structure. Biochim. Biophys. Acta 2009, 1788, 169–177. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Savina, A.; Furla´n, M.; Vidal, M.; Colombo, M.I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Piccin, A.; Murphy, W.G.; Smith, O.P. Circulating microparticles: Pathophysiology and clinical implications. Blood Rev. 2007, 21, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Antonyak, M.A.; Zhang, J.; Cerione, R.A. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene 2012, 31, 4740–4749. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Wahlgren, J.; Karlson, T.d.L.; Brisslert, M.; Vaziri Sani, F.; Telemo, E.; Sunnerhagen, P.; Valadi, H. Plasma exosomes can deliver exogenous short interfering rna to monocytes and lymphocytes. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 50-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef]
- Bolukbasi, M.F.; Mizrak, A.; Ozdener, G.B.; Madlener, S.; Ströbel, T.; Erkan, E.P.; Fan, J.B.; Breakefield, X.O.; Saydam, O. miR-1289 and “zipcode”-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids 2012, 1, e10. [Google Scholar] [CrossRef]
- Bieniasz, P.D. Late budding domains and host proteins in enveloped virus release. Virology. 2006, 344, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Hurley, J.H. Proline-rich regions and motifs in trafficking: From ESCRT interaction to viral exploitation. Traffic 2011, 12, 1282–1290. [Google Scholar] [CrossRef]
- Shen, B.; Fang, Y.; Wu, N.; Gould, S.J. Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. J. Biol. Chem. 2011, 286, 44162–44176. [Google Scholar] [CrossRef]
- Yang, J.M.; Gould, S.J. The cis-acting signals that target proteins to exosomes and microvesicles. Biochem. Soc. Trans. 2013, 41, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, M.J. Polyomavirus miRNAs: The beginning. Curr. Opin. Virol. 2014, 7, 29–32. [Google Scholar] [CrossRef]
- Sullivan, C.S.; Grundhoff, A.T.; Tevethia, S.; Pipas, J.M.; Ganem, D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 2005, 435, 682–686. [Google Scholar] [CrossRef]
- Seo, G.J.; Fink, L.H.; O’Hara, B.; Atwood, W.J.; Sullivan, C.S. Evolutionarily conserved function of a viral microRNA. J. Virol. 2008, 82, 9823–9828. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.C.; Li, Y.J.; Chen, H.C.; Wu, H.H.; Weng, C.H.; Chen, Y.C.; Lee, C.C.; Chang, M.Y.; Hsu, H.H.; Yen, T.H.; et al. Polyomavirus BK-encoded microRNA suppresses autoregulation of viral replication. Biochem. Biophys. Res. Commun. 2014, 447, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.J.; Chen, C.J.; Sullivan, C.S. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 2009, 383, 183–187. [Google Scholar] [CrossRef]
- Bauman, Y.; Nachmani, D.; Vitenshtein, A.; Tsukerman, P.; Drayman, N.; Stern-Ginossar, N.; Lankry, D.; Gruda, R.; Mandelboim, O. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 2011, 9, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Bauman, Y.; Drayman, N.; Ben-Nun-Shaul, O.; Vitenstein, A.; Yamin, R.; Ophir, Y.; Oppenheim, A.; Mandelboim, O. Downregulation of the stress-induced ligand ULBP1 following SV40 infection confers viral evasion from NK cell cytotoxicity. Oncotarget 2016, 7, 15369–15381. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.K.; Yim, H.; Andrews, E.; Benjamin, T.L. A mouse polyomavirus-encoded microRNA targets the cellular apoptosis pathway through Smad2 inhibition. Virology 2014, 470, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; Corazzari, M.; Antonioli, M.; Piacentini, M. Ambra1 at the crossroad between autophagy and cell death. Oncogene 2013, 32, 3311–3318. [Google Scholar] [CrossRef]
- Lee, S.; Paulson, K.G.; Murchison, E.P.; Afanasiev, O.K.; Alkan, C.; Leonard, J.H.; Byrd, D.R.; Hannon, G.J.; Nghiem, P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J. Clin. Virol. 2011, 52, 272–275. [Google Scholar] [CrossRef]
- Chen, C.J.; Cox, J.E.; Kincaid, R.P.; Martinez, A.; Sullivan, C.S. Divergent MicroRNA targetomes of closely related circulating strains of a polyomavirus. J. Virol. 2013, 87, 11135–11147, Erratum in: J. Virol. 2016, 90, 2157–2158. [Google Scholar] [CrossRef]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Coumans, F.A.W.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F.; et al. Methodological Guidelines to Study Extracellular Vesicles. Circ Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef]
- Giovannelli, I.; Clausi, V.; Nukuzuma, S.; Della Malva, N.; Nosi, D.; Giannecchini, S. Polyomavirus JC microRNA expression after infection in vitro. Virus Res. 2016, 213, 269–273. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Martelli, F.; Wu, Z.; Delbue, S.; Weissbach, F.H.; Giulioli, M.C.; Ferrante, P.; Hirsch, H.H.; Giannecchini, S. BK Polyomavirus MicroRNA Levels in Exosomes Are Modulated by Non-Coding Control Region Activity and Down-Regulate Viral Replication When Delivered to Non-Infected Cells Prior to Infection. Viruses 2018, 10, 466. [Google Scholar] [CrossRef]
- Rocca, A.; Martelli, F.; Delbue, S.; Ferrante, P.; Bartolozzi, D.; Azzi, A.; Giannecchini, S. The JCPYV DNA load inversely correlates with the viral microrna expression in blood and cerebrospinal fluid of patients at risk of PML. J. Clin. Virol. 2015, 70, 1–6. [Google Scholar] [CrossRef]
- Giovannelli, I.; Martelli, F.; Repice, A.; Massacesi, L.; Azzi, A.; Giannecchini, S. Detection of JCPyV microRNA in blood and urine samples of multiple sclerosis patients under natalizumab therapy. J. Neurovirol. 2015, 21, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Lee, Y.H.; Seo, J.W.; Moon, H.; Kim, J.S.; Kim, Y.G.; Jeong, K.H.; Moon, J.Y.; Lee, T.W.; Ihm, C.G.; et al. Urinary exosomal viral microRNA as a marker of BK virus nephropathy in kidney transplant recipients. PLoS ONE 2017, 12, e0190068. [Google Scholar] [CrossRef]
- Martelli, F.; Mencarini, J.; Rocca, A.; Malva, N.D.; Bartolozzi, D.; Giannecchini, S. Polyomavirus microRNA in saliva reveals persistent infectious status in the oral cavity. Virus Res. 2018, 249, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Morris-Love, J.; Gee, G.V.; O‘Hara, B.A.; Assetta, B.; Atkinson, A.L.; Dugan, A.S.; Haley, S.A.; Atwood, W.J. JC Polyomavirus uses extracellular vesicles to infect target cells. mBio 2019, 10, 00379-19. [Google Scholar] [CrossRef] [PubMed]
- Santiana, M.; Altan-Bonnet, N. Insane in the Membrane: Glial Extracellular Vesicles Transmit Polyomaviruses. mBio 2019, 10, e01024-19. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, B.A.; Morris-Love, J.; Gee, G.V.; Haley, S.A.; Atwood, W.J. JC Virus infected choroid plexus epithelial cells produce extracellular vesicles that infect glial cells independently of the virus attachment receptor. PLoS Pathog. 2020, 16. [Google Scholar] [CrossRef]
- Handala, L.; Blanchard, E.; Raynal, P.I.; Roingeard, P.; Morel, V.; Descamps, V.; Castelain, S.; Francois, C.; Duverlie, G.; Brochot, E.; et al. BK polyomavirus hijacks extracellular vesicles for en bloc transmission. J Virol. 2020, 94, JVI.01834-19. [Google Scholar] [CrossRef]
- Scribano, S.; Guerrini, M.; Arvia, R.; Guasti, D.; Nardini, P.; Romagnoli, P.; Giannecchini, S. Archetype JC polyomavirus DNA associated with extracellular vesicles circulates in human plasma samples. J. Clin. Virol. 2020, 128. [Google Scholar] [CrossRef] [PubMed]
- Stroh, L.J.; Maginnis, M.S.; Blaum, B.S.; Nelson, C.D.; Neu, U.; Gee, G.V.; O’Hara, B.A.; Motamedi, N.; DiMaio, D.; Atwood, W.J.; et al. The greater affinity of JC polyomavirus capsid for α2,6-linked lactoseries tetrasaccharide c than for other sialylated glycans is a major determinant of infectivity. J. Virol. 2015, 89, 6364–6375. [Google Scholar] [CrossRef]
- Assetta, B.; Maginnis, M.S.; Gracia, A.; Ahufinger, I.; Haley, S.A.; Gee, G.V.; Nelson, C.D.; O‘Hara, B.A.; Allen, R.; Atwood, W.J. 5-HT2 receptors facilitate JC polyomavirus entry. J. Virol. 2013, 87, 13490–13498. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Maginnis, M.S.; Palma, A.S.; Stroh, L.J.; Nelson, C.D.; Feizi, T.; Atwood, W.J.; Stehle, T. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe 2010, 8, 309–319. [Google Scholar] [CrossRef] [PubMed]
- González-López, O.; Rivera-Serrano, E.E.; Hu, F.; Hensley, L.; McKnight, K.L.; Ren, J.; Stuart, D.I.; Fry, E.E.; Lemon, S.M. Redundant late domain functions of tandem VP2 YPX3L motifs in nonlytic cellular egress of quasi-enveloped hepatitis A virus. J. Virol. 2018, 92, e01308-18. [Google Scholar] [CrossRef] [PubMed]
- Meckes, D.G., Jr.; Raab-Traub, N. Microvesicles and viral infection. J. Virol. 2011, 85, 12844–12854. [Google Scholar] [CrossRef] [PubMed]
- Bukong, T.N.; Momen-Heravi, F.; Kodys, K.; Bala, S.; Szabo, G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014, 10, e1004424. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef]
- Mao, L.; Wu, J.; Shen, L.; Yang, J.; Chen, J.; Xu, H. Enterovirus 71 transmission by exosomes establishes a productive infection in human neuroblastoma cells. Virus Genes 2016, 52, 189–194. [Google Scholar] [CrossRef]
- Takahashi, M.; Tanaka, T.; Takahashi, H.; Hoshino, Y.; Nagashima, S.; Jirintai; Mizuo, H.; Yazaki, Y.; Takagi, T.; Azuma, M.; et al. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: Characterization of HEV virions in blood circulation. J. Clin. Microbiol. 2010, 48, 1112–11125. [Google Scholar] [CrossRef]
- Assetta, B.; Atwood, W.J. The biology of JC polyomavirus. Biol Chem. 2017, 398, 839–855. [Google Scholar] [CrossRef] [PubMed]
| Features and Markers | Extracellular Vesicles | |
|---|---|---|
| Exosomes | Microvesicles | |
| Origin | endosome | Plasma membrane |
| Size | 30–200 nm | 100–1000 nm |
| Membrane markers | Tetraspanins: CD9, CD81 CD63, TSPAN6, TSPAN8, CD151, CD37, CD53, Flotilin 1 and 2 | Tetraspanins: CD9, CD63, CD81, CD82 |
| Lipids | Phosphatidylserine, cholesterol, ceramide and other sphingolipids, LBPA | Phosphatidylserine, ceramide phosphatidylethanolamine, sphingolipids |
| Cell adhesion | Integrin, lactadherin, ICAM | Integrin, PECAM1, fibronectin |
| Intracellular trafficking | Rab GTPases, annexins | Rab GTPases, annexins |
| Cell type-specific protein | MHC-I, MHC-II, APP, PMEL, TCR, FasL, CXCR4, HSPG, CD86, PrP, TFR, WNT | MHC-I, MHC-II, APP, PMEL, TCR, FasL, CXCR4, HSPG, CD86, PrP, TFR, WNT LFA1, CD14 |
| Cytoplasmic material (enzyme) | Peroxidases, pyruvate kinase, enolase, GAPDH | Tau, TDP43, GAPDH |
| Signaling molecules | Protein kinases, catenin, 14-3-3, G proteins | For example, ARF6, RAB11, ROCK |
| Biogenesis components | ALIX, TSG101, syntenin, ubiquitin, clathrin, VPS32, VPS4 | ALIX, TSG101, ERK, PLD, VPS4 |
| Chaperones | HSP70, HSP90 | HSP70, HSP90 |
| Cytoskeletal molecules | Not determined | Actin, tubulin |
| Nucleic acids | MicroRNAs and other noncoding RNAs, mRNA, DNA (and histones) (associated to the outside of the EV or part of their cargo) | MicroRNAs and other noncoding RNAs, mRNA, DNA (and histones) (associated to the outside of the EV or part of their cargo) |
| Polyomavirus | Biological Fluid | EV Extraction and Characterization Methods | EV Markers | PyV Markers | Reference |
|---|---|---|---|---|---|
| Studies on EVs and MicroRNAs | |||||
| JCPyV | COS-7 cell supernatant, KG-1 cell supernatant | Exosomes extraction kit; NTA and WB | CD63 | jcv-miR-J1-3p and -5p | [100] |
| BKPyV | COS-7 cell supernatant, RPTEC supernatant | Exosomes extraction kit; NTA and WB | CD63, CD81, annexin II | bkv-miR-B1-3p and -5p | [102] |
| JCPyV | Plasma Urine Saliva CSF | Exosomes extraction kit; NTA and WB | CD63, | jcv-miR-J1-3p and -5p | [103,104,105,106] |
| BKPyV | Plasma Urine Saliva | Exosomes extraction kit | bkv-miR-B1-3p and -5p | [105,106] | |
| MCPyV | Plasma Saliva | Exosomes extraction kit | mcv-miR-M1-5p | [106] | |
| SV40 | Plasma Saliva | Exosomes extraction kit; | sv40-miR-S1-5p | [106] | |
| Studies on EVs and whole viruses | |||||
| JCPyV | SVG-A, CPE cell supernatant | Ultracentrifugation; NTA, Immunoelectron microscopy, WB | CD9, CD81, annexin V, flotillin-1, TSG101 | Whole-virus particle | [107,108,109] |
| BKPyV | Vero supernatant RPTEC supernatant | Ultracentrifugation; Immunoelectron microscopy | CD9, CD63, CD81 | Whole-virus particle | [110] |
| JCPyV | Plasma | Exosomes extraction kit; NTA and WB, Immunoelectron microscopy | CD63, CD81, annexin II | Viral DNA and Vp1 | [111] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannecchini, S. Evidence of the Mechanism by Which Polyomaviruses Exploit the Extracellular Vesicle Delivery System during Infection. Viruses 2020, 12, 585. https://doi.org/10.3390/v12060585
Giannecchini S. Evidence of the Mechanism by Which Polyomaviruses Exploit the Extracellular Vesicle Delivery System during Infection. Viruses. 2020; 12(6):585. https://doi.org/10.3390/v12060585
Chicago/Turabian StyleGiannecchini, Simone. 2020. "Evidence of the Mechanism by Which Polyomaviruses Exploit the Extracellular Vesicle Delivery System during Infection" Viruses 12, no. 6: 585. https://doi.org/10.3390/v12060585
APA StyleGiannecchini, S. (2020). Evidence of the Mechanism by Which Polyomaviruses Exploit the Extracellular Vesicle Delivery System during Infection. Viruses, 12(6), 585. https://doi.org/10.3390/v12060585





