TRiC/CCT Complex, a Binding Partner of NS1 Protein, Supports the Replication of Zika Virus in Both Mammalians and Mosquitoes
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Viruses, Cell Lines, Mosquitoes, and Antibodies
2.3. Pull-Down Assay and Mass Spectrometry
2.4. Immunoprecipitation and Immunoblotting
2.5. Immunofluorescence Assay and Confocal Microscopy
2.6. Generation of Stable Cell Lines Suppressing CCT2
2.7. Treatment of HSF1A and LDH Cytotoxicity Assay
2.8. Gene Silencing in Mosquitoes
2.9. Quantitative Real-Time PCR
2.10. Statistical Analysis
2.11. Data Availability
3. Results
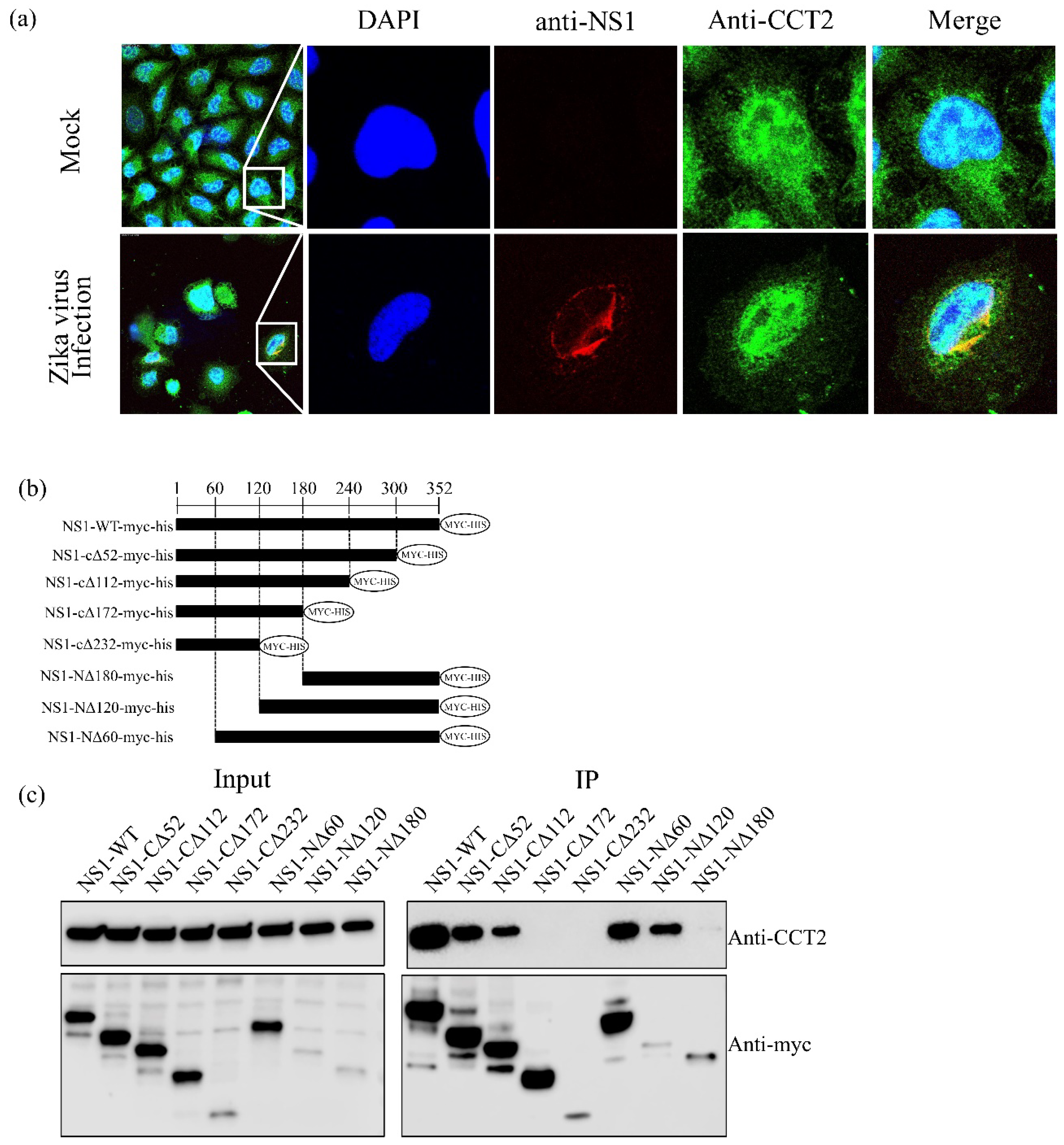
3.1. TRiC/CCT Complex Interacts with the Zika Virus NS1 Protein
3.2. CCT2 Interacts with the Central Region of Zika Virus NS1
3.3. Interaction of ZIKV NS1 with CCT2 Depends on ATP Concentration
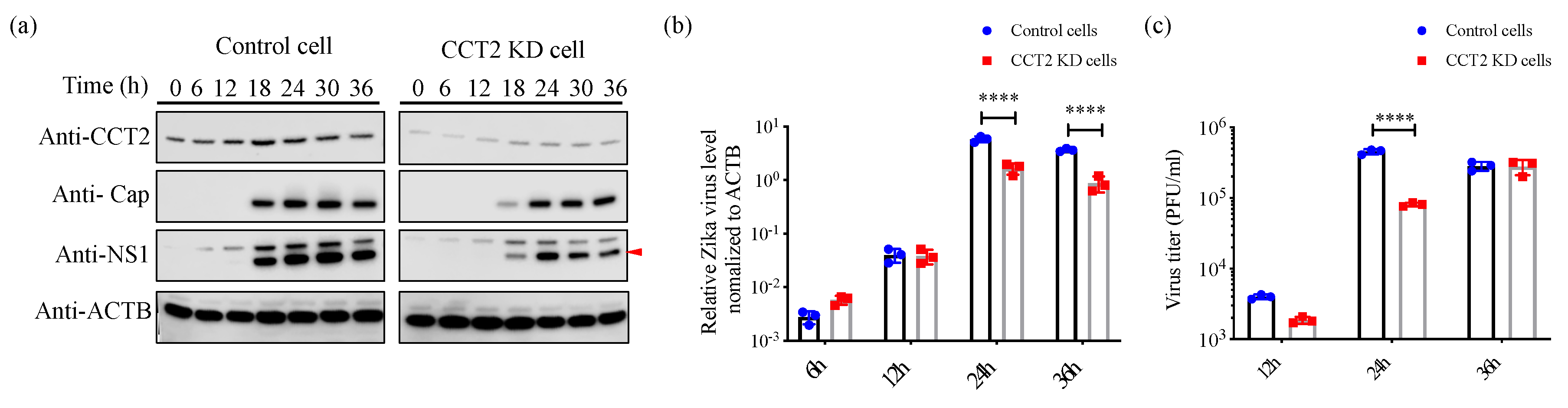
3.4. Knockdown of CCT2 Reduces Zika Virus Replication in Mammalian Cells
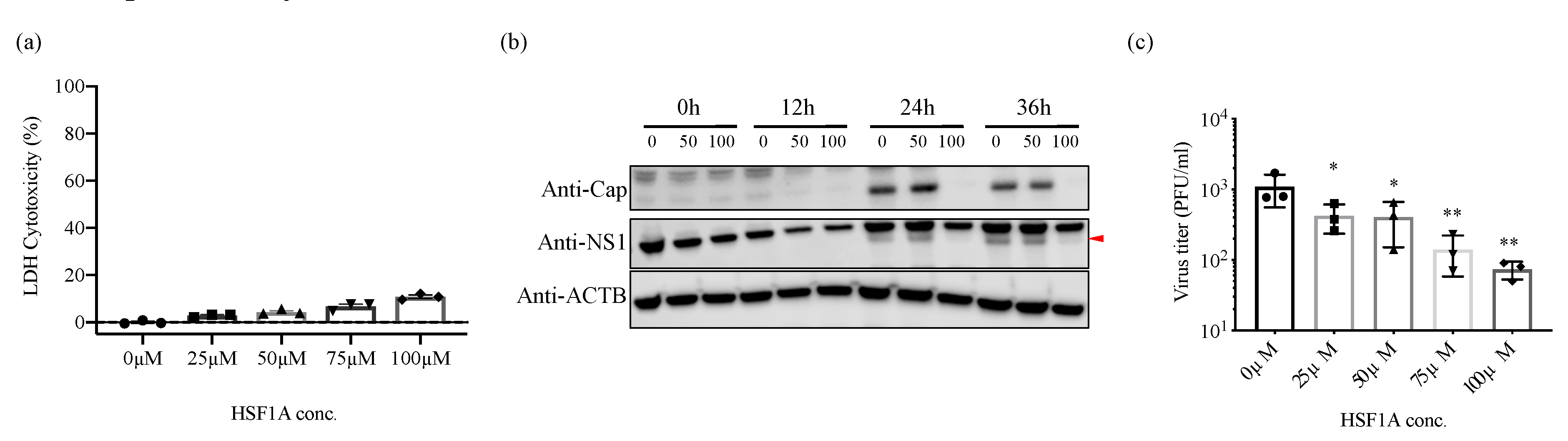
3.5. Inhibition of TRiC/CCT Complex Function Using HSF1A Reduces Zika Virus Propagation
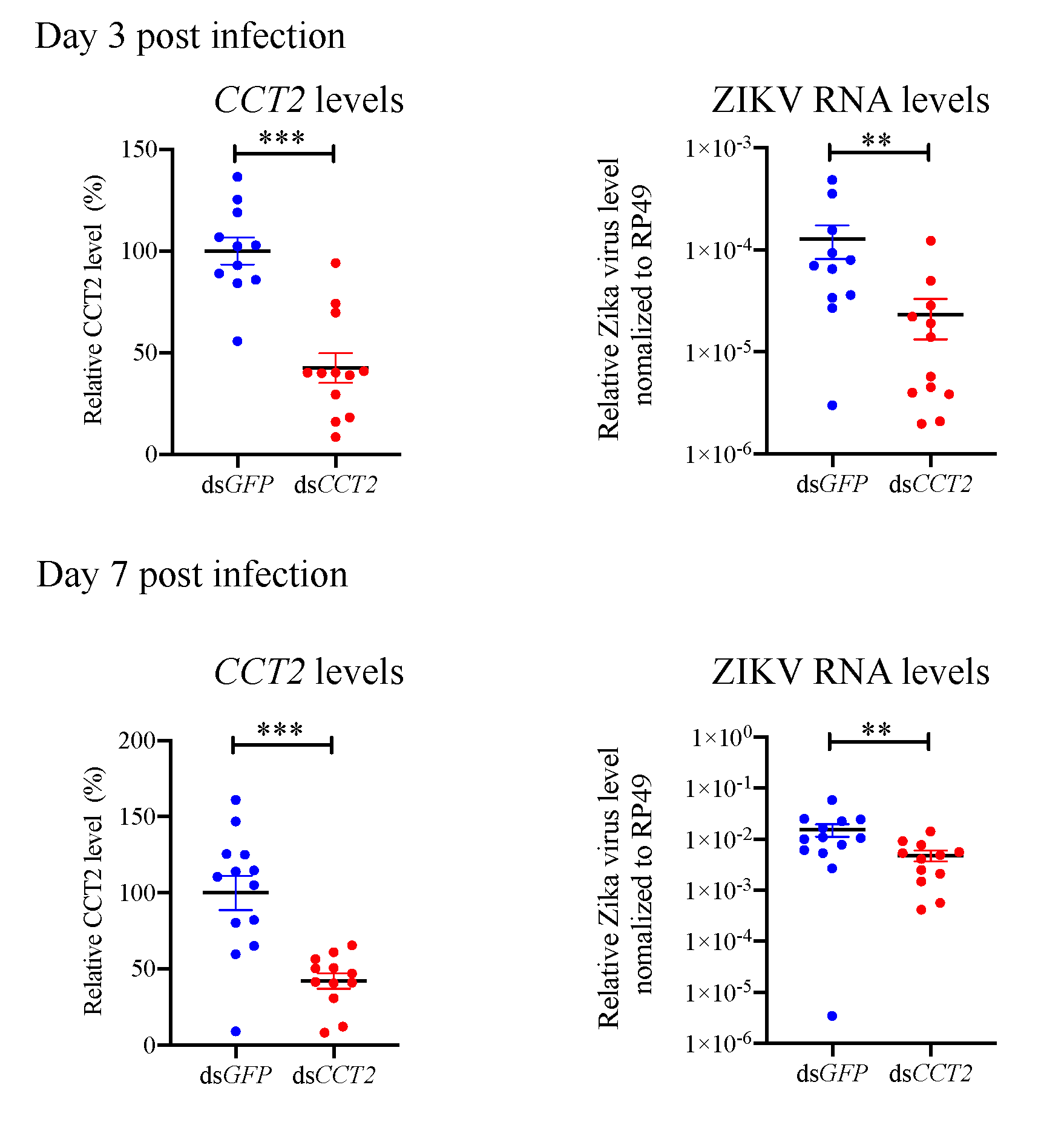
3.6. Suppression of CCT2 Reduces Zika Virus Burden in Mosquitoes
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Musso, D.; Gubler, D.J. Zika virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.R. Mosquito-borne human viral diseases: Why aedes aegypti? Am. J. Trop. Med. Hyg. 2018, 98, 1563–1565. [Google Scholar] [CrossRef] [PubMed]
- Salinas, S.; Foulongne, V.; Loustalot, F.; Fournier-Wirth, C.; Moles, J.P.; Briant, L.; Nagot, N.; Van de Perre, P.; Simonin, Y. Zika virus, an emerging threat. Med. Sci. 2016, 32, 378–386. [Google Scholar]
- Bogoch, I.I.; Brady, O.J.; Kraemer, M.U.G.; German, M.; Creatore, M.I.; Kulkarni, M.A.; Brownstein, J.S.; Mekaru, S.R.; Hay, S.I.; Groot, E.; et al. Anticipating the international spread of zika virus from brazil. Lancet 2016, 387, 335–336. [Google Scholar] [CrossRef]
- Dong, S.; Kang, S.; Dimopoulos, G. Identification of anti-flaviviral drugs with mosquitocidal and anti-zika virus activity in aedes aegypti. PLoS Negl. Trop. Dis. 2019, 13, e0007681. [Google Scholar] [CrossRef]
- Bollati, M.; Alvarez, K.; Assenberg, R.; Baronti, C.; Canard, B.; Cook, S.; Coutard, B.; Decroly, E.; de Lamballerie, X.; Gould, E.A.; et al. Structure and functionality in flavivirus ns-proteins: Perspectives for drug design. Antiviral Res. 2010, 87, 125–148. [Google Scholar] [CrossRef]
- Briant, L.; Despres, P.; Choumet, V.; Misse, D. Role of skin immune cells on the host susceptibility to mosquito-borne viruses. Virology 2014, 464–465, 26–32. [Google Scholar] [CrossRef]
- Amorim, J.H.; Alves, R.P.; Boscardin, S.B.; Ferreira, L.C. The dengue virus non-structural 1 protein: Risks and benefits. Virus Res. 2014, 181, 53–60. [Google Scholar] [CrossRef]
- Li, A.; Yu, J.; Lu, M.; Ma, Y.; Attia, Z.; Shan, C.; Xue, M.; Liang, X.; Craig, K.; Makadiya, N.; et al. A zika virus vaccine expressing premembrane-envelope-ns1 polyprotein. Nat. Commun. 2018, 9, 3067. [Google Scholar] [CrossRef]
- Bailey, M.J.; Duehr, J.; Dulin, H.; Broecker, F.; Brown, J.A.; Arumemi, F.O.; Bermudez Gonzalez, M.C.; Leyva-Grado, V.H.; Evans, M.J.; Simon, V.; et al. Human antibodies targeting zika virus ns1 provide protection against disease in a mouse model. Nat. Commun. 2018, 9, 4560. [Google Scholar] [CrossRef]
- Gestaut, D.; Limatola, A.; Joachimiak, L.; Frydman, J. The atp-powered gymnastics of tric/cct: An asymmetric protein folding machine with a symmetric origin story. Curr. Opin. Struct. Biol. 2019, 55, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Hafirassou, M.L.; Meertens, L.; Umana-Diaz, C.; Labeau, A.; Dejarnac, O.; Bonnet-Madin, L.; Kummerer, B.M.; Delaugerre, C.; Roingeard, P.; Vidalain, P.O.; et al. A global interactome map of the dengue virus ns1 identifies virus restriction and dependency host factors. Cell Rep. 2017, 21, 3900–3913. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Aizaki, H.; Hara, H.; Matsuda, M.; Ando, T.; Shimoji, T.; Murakami, K.; Masaki, T.; Shoji, I.; Homma, S.; et al. Chaperonin tric/cct participates in replication of hepatitis c virus genome via interaction with the viral ns5b protein. Virology 2011, 410, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, J.J.; Fernandez de Castro, I.; Ashbrook, A.W.; Gestaut, D.R.; Zamora, P.F.; Bauer, J.A.; Forrest, J.C.; Frydman, J.; Risco, C.; Dermody, T.S. The tric chaperonin controls reovirus replication through outer-capsid folding. Nat. Microbiol. 2018, 3, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Hayer-Hartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Spiess, C.; Meyer, A.S.; Reissmann, S.; Frydman, J. Mechanism of the eukaryotic chaperonin: Protein folding in the chamber of secrets. Trends Cell Biol. 2004, 14, 598–604. [Google Scholar] [CrossRef]
- Yam, A.Y.; Xia, Y.; Lin, H.T.; Burlingame, A.; Gerstein, M.; Frydman, J. Defining the tric/cct interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 2008, 15, 1255–1262. [Google Scholar] [CrossRef]
- Frydman, J. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu. Rev. Biochem. 2001, 70, 603–647. [Google Scholar] [CrossRef]
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar] [CrossRef]
- Neef, D.W.; Jaeger, A.M.; Gomez-Pastor, R.; Willmund, F.; Frydman, J.; Thiele, D.J. A direct regulatory interaction between chaperonin tric and stress-responsive transcription factor hsf1. Cell Rep. 2014, 9, 955–966. [Google Scholar] [CrossRef]
- Kabir, M.A.; Uddin, W.; Narayanan, A.; Reddy, P.K.; Jairajpuri, M.A.; Sherman, F.; Ahmad, Z. Functional subunits of eukaryotic chaperonin cct/tric in protein folding. J. Amino Acids 2011, 2011, 843206. [Google Scholar] [CrossRef] [PubMed]
- Leitner, A.; Joachimiak, L.A.; Bracher, A.; Monkemeyer, L.; Walzthoeni, T.; Chen, B.; Pechmann, S.; Holmes, S.; Cong, Y.; Ma, B.; et al. The molecular architecture of the eukaryotic chaperonin tric/cct. Structure 2012, 20, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Melkani, G.C.; Bhide, S.; Han, A.; Vyas, J.; Livelo, C.; Bodmer, R.; Bernstein, S.I. Tric/cct chaperonins are essential for maintaining myofibril organization, cardiac physiological rhythm, and lifespan. FEBS Lett. 2017, 591, 3447–3458. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.; Calvez, E.; Chouin-Carneiro, T.; Diallo, D.; Failloux, A.B. An overview of mosquito vectors of zika virus. Microbes Infect. 2018, 20, 646–660. [Google Scholar] [CrossRef]
- Tham, H.W.; Balasubramaniam, V.; Ooi, M.K.; Chew, M.F. Viral determinants and vector competence of zika virus transmission. Front. Microbiol. 2018, 9, 1040. [Google Scholar] [CrossRef]
- Kauffman, E.B.; Kramer, L.D. Zika virus mosquito vectors: Competence, biology, and vector control. J. Infect. Dis. 2017, 216, S976–S990. [Google Scholar] [CrossRef]
- Gutierrez-Bugallo, G.; Piedra, L.A.; Rodriguez, M.; Bisset, J.A.; Lourenco-de-Oliveira, R.; Weaver, S.C.; Vasilakis, N.; Vega-Rua, A. Vector-borne transmission and evolution of zika virus. Nat. Ecol. Evol. 2019, 3, 561–569. [Google Scholar] [CrossRef]
- Scaturro, P.; Stukalov, A.; Haas, D.A.; Cortese, M.; Draganova, K.; Plaszczyca, A.; Bartenschlager, R.; Gotz, M.; Pichlmair, A. An orthogonal proteomic survey uncovers novel zika virus host factors. Nature 2018, 561, 253–257. [Google Scholar] [CrossRef]
- Shah, P.S.; Link, N.; Jang, G.M.; Sharp, P.P.; Zhu, T.; Swaney, D.L.; Johnson, J.R.; Von Dollen, J.; Ramage, H.R.; Satkamp, L.; et al. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and zika virus pathogenesis. Cell 2018, 175, 1931–1945. [Google Scholar] [CrossRef]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus ns1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef]
- Dekker, C.; Stirling, P.C.; McCormack, E.A.; Filmore, H.; Paul, A.; Brost, R.L.; Costanzo, M.; Boone, C.; Leroux, M.R.; Willison, K.R. The interaction network of the chaperonin cct. EMBO J. 2008, 27, 1827–1839. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Baker, M.L.; Jakana, J.; Woolford, D.; Miller, E.J.; Reissmann, S.; Kumar, R.N.; Redding-Johanson, A.M.; Batth, T.S.; Mukhopadhyay, A.; et al. 4.0-a resolution cryo-em structure of the mammalian chaperonin tric/cct reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. USA 2010, 107, 4967–4972. [Google Scholar] [CrossRef] [PubMed]
- Lingappa, J.R.; Martin, R.L.; Wong, M.L.; Ganem, D.; Welch, W.J.; Lingappa, V.R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis b virus capsid, a multimeric particle. J. Cell Biol. 1994, 125, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, X.; Zan, J.; Wu, Y.; Ye, C.; Ruan, X.; Zhou, J. Cellular chaperonin cctγ contributes to rabies virus replication during infection. J. Virol. 2013, 87, 7608–7621. [Google Scholar] [CrossRef]
- Hong, S.; Choi, G.; Park, S.; Chung, A.S.; Hunter, E.; Rhee, S.S. Type d retrovirus gag polyprotein interacts with the cytosolic chaperonin tric. J. Virol. 2001, 75, 2526–2534. [Google Scholar] [CrossRef]
- Savidis, G.; McDougall, W.M.; Meraner, P.; Perreira, J.M.; Portmann, J.M.; Trincucci, G.; John, S.P.; Aker, A.M.; Renzette, N.; Robbins, D.R.; et al. Identification of zika virus and dengue virus dependency factors using functional genomics. Cell Rep. 2016, 16, 232–246. [Google Scholar] [CrossRef]
- Petrova, E.; Gracias, S.; Beauclair, G.; Tangy, F.; Jouvenet, N. Uncovering flavivirus host dependency factors through a genome-wide gain-of-function screen. Viruses 2019, 11, 68. [Google Scholar] [CrossRef]
- Scaturro, P.; Cortese, M.; Chatel-Chaix, L.; Fischl, W.; Bartenschlager, R. Dengue virus non-structural protein 1 modulates infectious particle production via interaction with the structural proteins. PLoS Pathog. 2015, 11, e1005277. [Google Scholar] [CrossRef]
- Akey, D.L.; Brown, W.C.; Jose, J.; Kuhn, R.J.; Smith, J.L. Structure-guided insights on the role of ns1 in flavivirus infection. Bioessays 2015, 37, 489–494. [Google Scholar] [CrossRef]
- Omokoko, M.D.; Pambudi, S.; Phanthanawiboon, S.; Masrinoul, P.; Setthapramote, C.; Sasaki, T.; Kuhara, M.; Ramasoota, P.; Yamashita, A.; Hirai, I.; et al. A highly conserved region between amino acids 221 and 266 of dengue virus non-structural protein 1 is a major epitope region in infected patients. Am. J. Trop. Med. Hyg. 2014, 91, 146–155. [Google Scholar] [CrossRef]
- Jiang, L.; Zhou, J.M.; Yin, Y.; Fang, D.Y.; Tang, Y.X.; Jiang, L.F. Selection and identification of b-cell epitope on ns1 protein of dengue virus type 2. Virus Res. 2010, 150, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.M.; Nybakken, G.E.; Thompson, B.S.; Engle, M.J.; Marri, A.; Fremont, D.H.; Diamond, M.S. Antibodies against west nile virus nonstructural protein ns1 prevent lethal infection through fc gamma receptor-dependent and -independent mechanisms. J. Virol. 2006, 80, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
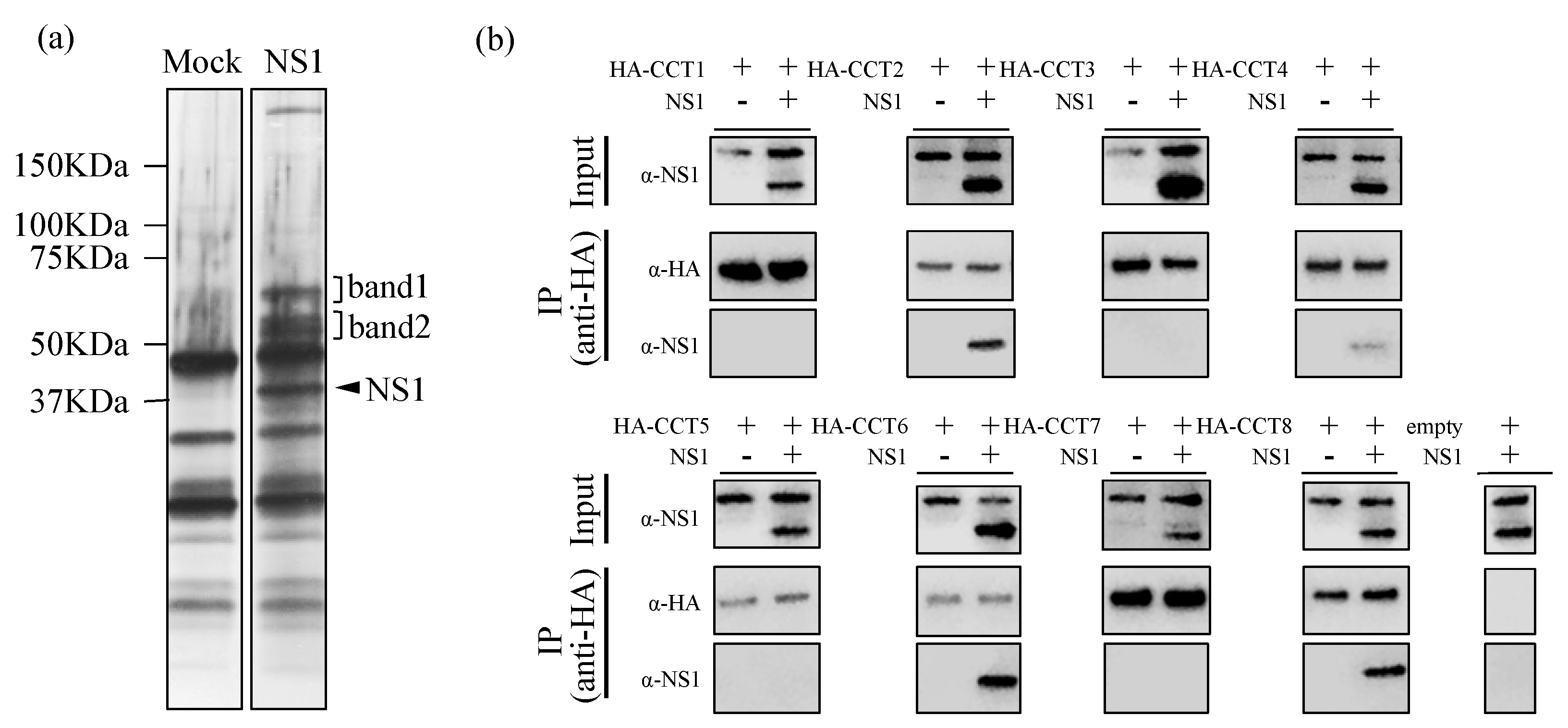

| Protein ID | Protein Name | Mass (Da) | Sequence Coverage (%) | Mascot Score |
|---|---|---|---|---|
| Band 1 | ||||
| GRP78_HUMAN | 78 kDa glucose-regulated protein Organism Name (OS) = Homo sapiens Gene Name (GN) = HSPA5 Protein Existence (PE) = 1 Sequence Version (SV) = 2 | 72,288 | 6.4 | 225 |
| ZG16B_HUMAN | Zymogen granule protein 16 homolog B OS = Homo sapiens GN = ZG16B PE = 1 SV = 3 | 22,725 | 26 | 181 |
| RORB_HUMAN | Nuclear receptor ROR-beta OS = Homo sapiens GN = RORB PE = 1 SV = 3 | 53,186 | 6 | 32 |
| WFKN2_HUMAN | WAP, Kazal, immunoglobulin, Kunitz and NTR domain-containing protein 2 OS = Homo sapiens GN = WFIKKN2 PE = 1 SV = 1 | 63,898 | 5.2 | 31 |
| COJA1_HUMAN | Collagen alpha-1(XIX) chain OS = Homo sapiens GN = COL19A1 PE = 1 SV = 3 | 115,149 | 6.5 | 29 |
| ARHGC_HUMAN | Rho guanine nucleotide exchange factor 12 OS = Homo sapiens GN = ARHGEF12 PE = 1 SV = 1 | 173,125 | 3.5 | 29 |
| ZN469_HUMAN | Zinc finger protein 469 OS = Homo sapiens GN = ZNF469 PE = 2 SV = 3 | 409,949 | 1.1 | 26 |
| KNG1_HUMAN | Kininogen-1 OS = Homo sapiens GN = KNG1 PE = 1 SV = 2 | 71,912 | 5.7 | 26 |
| EEA1_HUMAN | Early endosome antigen 1 OS = Homo sapiens GN = EEA1 PE = 1 SV = 2 | 162,367 | 3.9 | 26 |
| ATR_HUMAN | Serine/threonine-protein kinase ATR OS = Homo sapiens GN = ATR PE = 1 SV = 3 | 301,172 | 1.6 | 26 |
| CIP1_HUMAN | E3 ubiquitin-protein ligase CCNB1IP1 OS = Homo sapiens GN = CCNB1IP1 PE = 1 SV = 1 | 31,524 | 2.9 | 25 |
| ARSJ_HUMAN | Arylsulfatase J OS = Homo sapiens GN = ARSJ PE = 2 SV = 1 | 67,193 | 1.5 | 23 |
| CC180_HUMAN | Coiled-coil domain-containing protein 180 OS = Homo sapiens GN = CCDC180 PE = 2 SV = 2 | 190,979 | 0.9 | 22 |
| KI2S1_HUMAN | Killer cell immunoglobulin-like receptor 2DS1 OS = Homo sapiens GN = KIR2DS1 PE = 2 SV = 1 | 33,624 | 5.6 | 20 |
| Band 2 | ||||
| TCPB_HUMAN | T-complex protein 1 subunit beta OS = Homo sapiens GN = CCT2 PE = 1 SV = 4 | 57,452 | 47.9 | 1097 |
| TCPQ_HUMAN | T-complex protein 1 subunit theta OS = Homo sapiens GN = CCT8 PE = 1 SV = 4 | 59,583 | 35.8 | 665 |
| TCPH_HUMAN | T-complex protein 1 subunit eta OS = Homo sapiens GN = CCT7 PE = 1 SV = 2 | 59,329 | 32.2 | 627 |
| TCPA_HUMAN | T-complex protein 1 subunit alpha OS = Homo sapiens GN = TCP1 PE = 1 SV = 1 | 60,306 | 36.5 | 623 |
| TCPZ_HUMAN | T-complex protein 1 subunit zeta OS = Homo sapiens GN = CCT6A PE = 1 SV = 3 | 57,988 | 36 | 530 |
| TCPG_HUMAN | T-complex protein 1 subunit gamma OS = Homo sapiens GN = CCT3 PE = 1 SV = 4 | 60,495 | 33.2 | 476 |
| TCPE_HUMAN | T-complex protein 1 subunit epsilon OS = Homo sapiens GN = CCT5 PE = 1 SV = 1 | 59,633 | 30.9 | 458 |
| TCPD_HUMAN | T-complex protein 1 subunit delta OS = Homo sapiens GN = CCT4 PE = 1 SV = 4 | 57,888 | 18 | 360 |
| VIME_HUMAN | Vimentin OS=Homo sapiens GN = VIM PE = 1 SV = 4 | 53,619 | 12.2 | 97 |
| S10AE_HUMAN | Protein S100-A14 OS = Homo sapiens GN = S100A14 PE = 1 SV = 1 | 11,655 | 14.4 | 95 |
| CH60_HUMAN | 60 kDa heat shock protein, mitochondrial OS = Homo sapiens GN = HSPD1 PE = 1 SV = 2 | 61,016 | 3.3 | 59 |
| KPYM_HUMAN | Pyruvate kinase PKM OS = Homo sapiens GN = PKM PE = 1 SV = 4 | 57,900 | 5.1 | 45 |
| PRDX2_HUMAN | Peroxiredoxin-2 OS = Homo sapiens GN = PRDX2 PE = 1 SV = 5 | 21,878 | 10.1 | 43 |
| ACTA_HUMAN | Actin, aortic smooth muscle OS = Homo sapiens GN = ACTA2 PE = 1 SV = 1 indistinguishable | 41,982 | 4.2 | 39 |
| TBA1A_HUMAN | Tubulin alpha-1A chain OS = Homo sapiens GN = TUBA1A PE = 1 SV = 1 indistinguishable | 50,104 | 4 | 39 |
| RHG29_HUMAN | Rho GTPase-activating protein 29 OS = Homo sapiens GN = ARHGAP29 PE = 1 SV = 2 | 141,974 | 3.3 | 37 |
| ZFY27_HUMAN | Protrudin OS = Homo sapiens GN = ZFYVE27 PE = 1 SV = 1 | 45,814 | 3.4 | 35 |
| RUVB1_HUMAN | RuvB-like 1 OS = Homo sapiens GN = RUVBL1 PE = 1 SV = 1 | 50,196 | 5 | 33 |
| BLMH_HUMAN | Bleomycin hydrolase OS = Homo sapiens GN = BLMH PE = 1 SV = 1 | 52,528 | 2.4 | 33 |
| TIAM2_HUMAN | T-lymphoma invasion and metastasis-inducing protein 2 OS = Homo sapiens GN = TIAM2 PE = 2 SV = 4 | 189,985 | 0.9 | 27 |
| GON4L_HUMAN | GON-4-like protein OS = Homo sapiens GN = GON4L PE = 1 SV = 1 | 248,465 | 2.3 | 26 |
| OR6S1_HUMAN | Olfactory receptor 6S1 OS = Homo sapiens GN = OR6S1 PE = 3 SV = 2 | 36,103 | 2.4 | 25 |
| PKRI1_HUMAN | PRKR-interacting protein 1 OS = Homo sapiens GN = PRKRIP1 PE = 1 SV = 1 | 20,984 | 6.5 | 25 |
| FBXL7_HUMAN | F-box/LRR-repeat protein 7 OS = Homo sapiens GN = FBXL7 PE = 2 SV = 1 | 54,540 | 3.1 | 25 |
| RINI_HUMAN | Ribonuclease inhibitor OS = Homo sapiens GN = RNH1 PE = 1 SV = 2 | 49,941 | 2.6 | 24 |
| KI26B_HUMAN | Kinesin-like protein KIF26B OS = Homo sapiens GN = KIF26B PE = 2 SV = 1 | 223,744 | 1.8 | 23 |
| HMGB3_HUMAN | High mobility group protein B3 OS = Homo sapiens GN = HMGB3 PE = 1 SV = 4 | 22,965 | 4 | 23 |
| RNF32_HUMAN | RING finger protein 32 OS = Homo sapiens GN = RNF32 PE = 1 SV = 1 | 41,490 | 1.9 | 22 |
| BARD1_HUMAN | BRCA1-associated RING domain protein 1 OS = Homo sapiens GN = BARD1 PE = 1 SV = 2 | 86,593 | 3.3 | 21 |
| TC1D4_HUMAN | Tctex1 domain-containing protein 4 OS = Homo sapiens GN = TCTEX1D4 PE = 1 SV = 1 | 23,338 | 3.2 | 21 |
| PRA10_HUMAN | PRAME family member 10 OS = Homo sapiens GN = PRAMEF10 PE = 2 SV = 4 | 55,175 | 4 | 21 |
| GTR7_HUMAN | Solute carrier family 2, facilitated glucose transporter member 7 OS = Homo sapiens GN = SLC2A7 PE = 2 SV = 2 | 55,692 | 1.6 | 20 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Uraki, R.; Hwang, J.; Fikrig, E. TRiC/CCT Complex, a Binding Partner of NS1 Protein, Supports the Replication of Zika Virus in Both Mammalians and Mosquitoes. Viruses 2020, 12, 519. https://doi.org/10.3390/v12050519
Wang Y, Uraki R, Hwang J, Fikrig E. TRiC/CCT Complex, a Binding Partner of NS1 Protein, Supports the Replication of Zika Virus in Both Mammalians and Mosquitoes. Viruses. 2020; 12(5):519. https://doi.org/10.3390/v12050519
Chicago/Turabian StyleWang, Yuchen, Ryuta Uraki, Jesse Hwang, and Erol Fikrig. 2020. "TRiC/CCT Complex, a Binding Partner of NS1 Protein, Supports the Replication of Zika Virus in Both Mammalians and Mosquitoes" Viruses 12, no. 5: 519. https://doi.org/10.3390/v12050519
APA StyleWang, Y., Uraki, R., Hwang, J., & Fikrig, E. (2020). TRiC/CCT Complex, a Binding Partner of NS1 Protein, Supports the Replication of Zika Virus in Both Mammalians and Mosquitoes. Viruses, 12(5), 519. https://doi.org/10.3390/v12050519





