Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays
Abstract
1. Introduction
2. Results
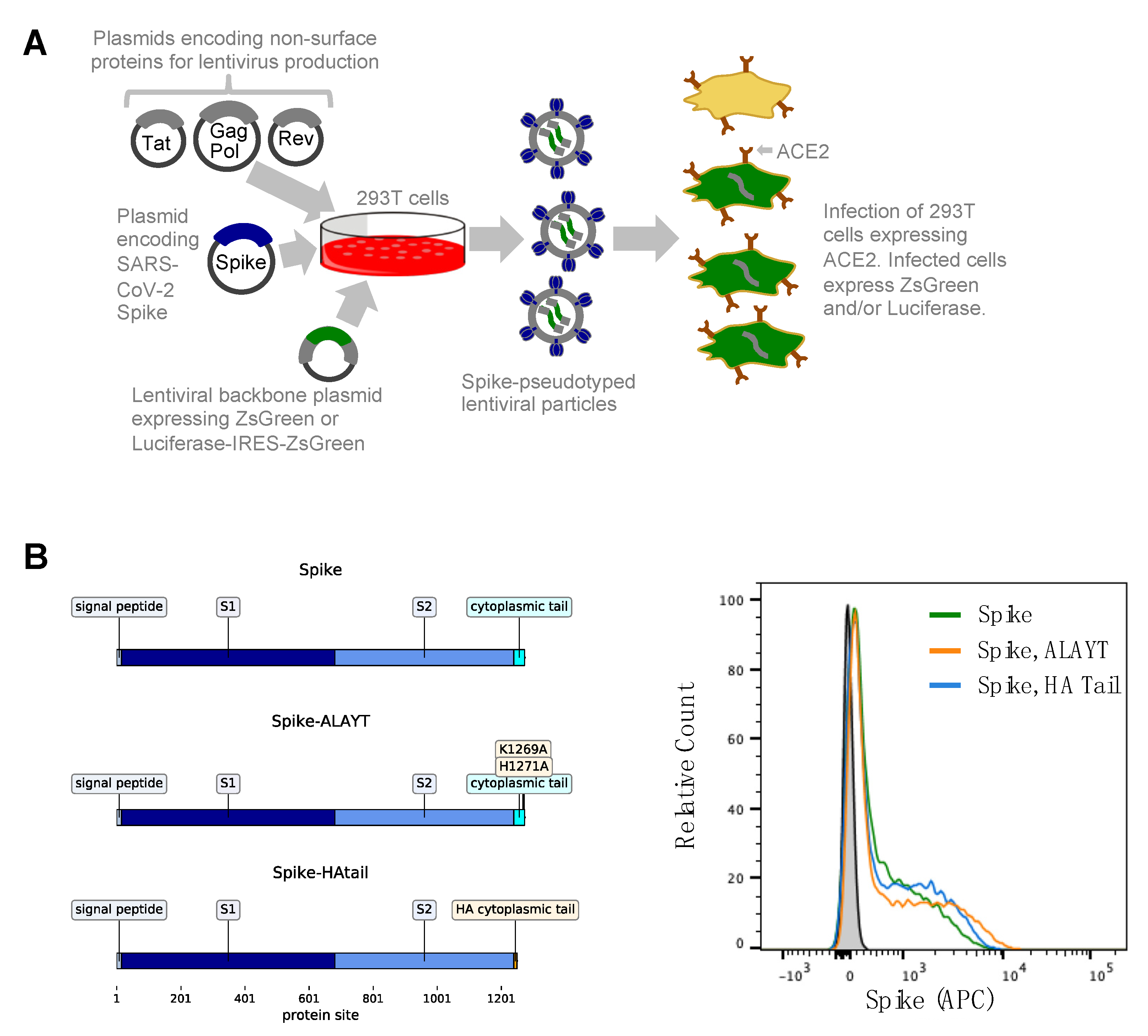
2.1. General Approach for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike
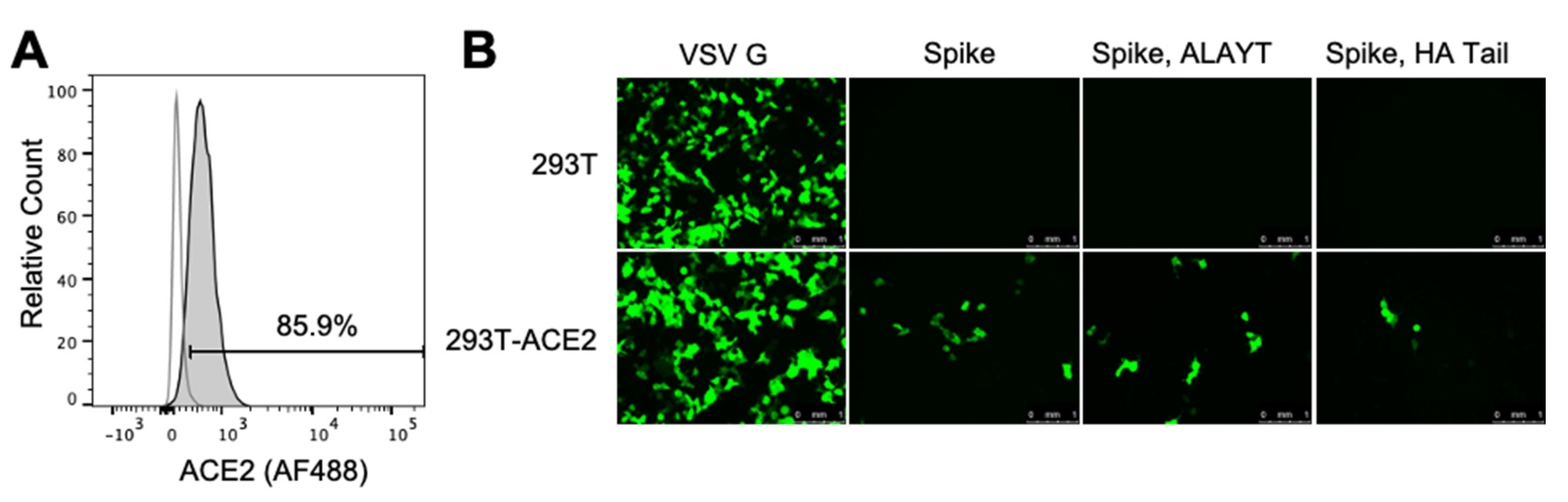
2.2. Target 293T Cells Constititutively Expressing Spike’s ACE2 Receptor
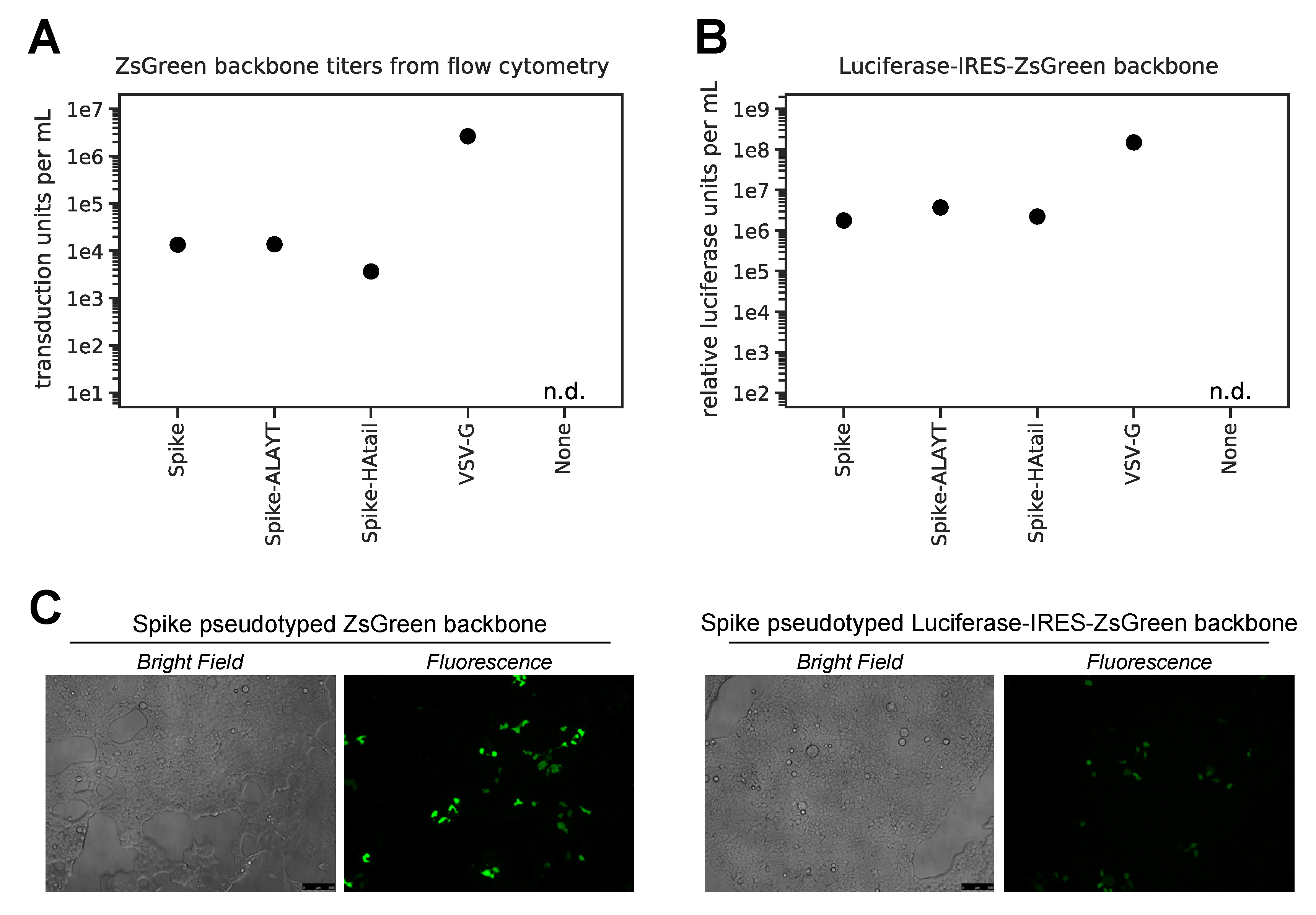
2.3. Titers of Pseudotyped Lentiviral Particles with Different Spike Cytoplasmic Tail Variants
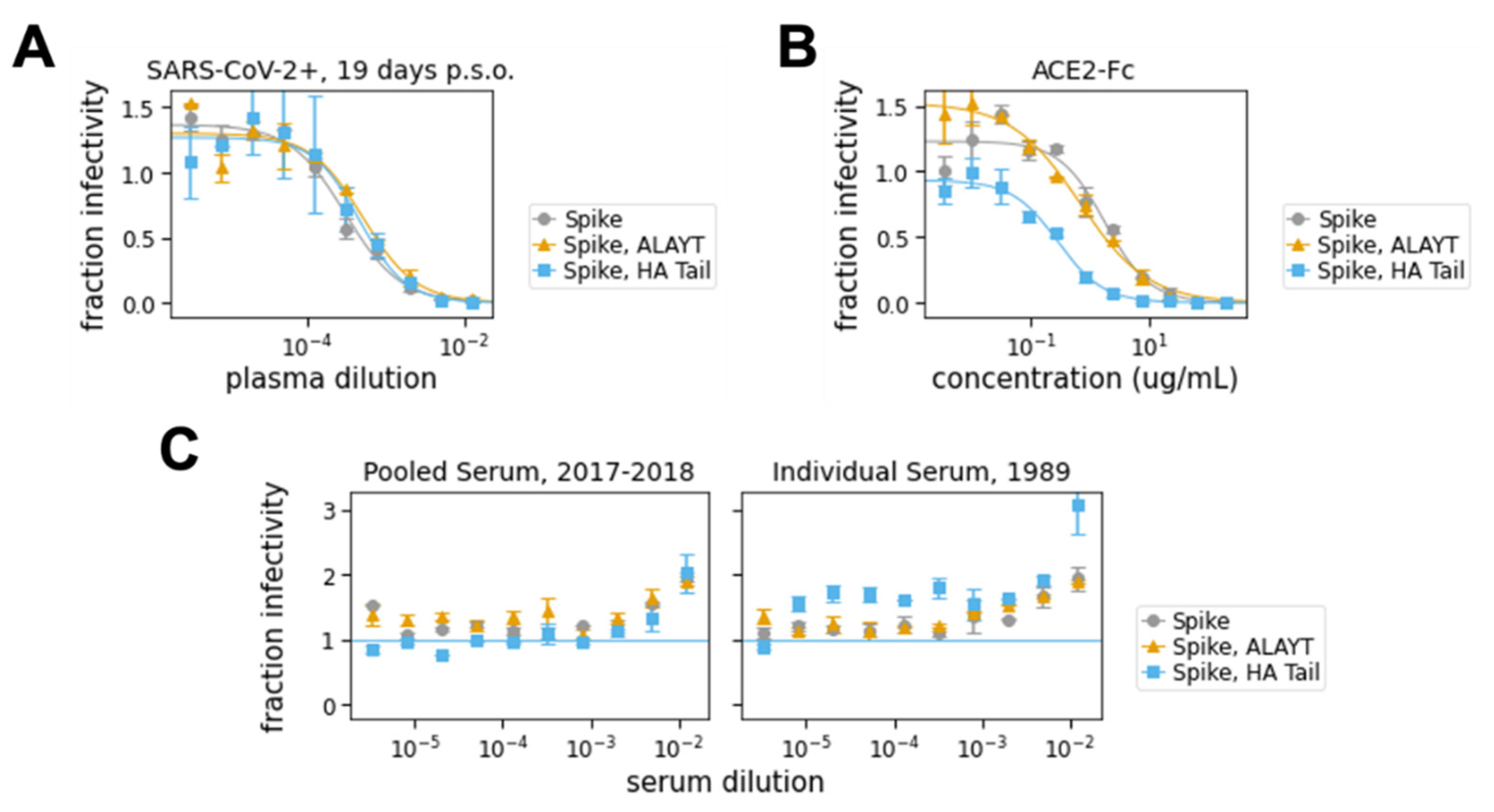
2.4. Neutralization Assays with Spike-Pseudotyped Lentiviral Particles
3. Discussion
4. Materials and Methods
4.1. Plasmids
- pHAGE2-EF1aInt-ACE2-WT (BEI catalog number NR52512): Lentiviral backbone plasmid expressing the human ACE2 gene (GenBank ID for human ACE2 is NM_021804) under an EF1a promoter with an intron to increase expression.
- HDM-IDTSpike-fixK-HA-tail (BEI catalog number NR52513): Plasmid expressing under a CMV promoter the Spike from SARS-CoV-2 strain Wuhan-Hu-1 (Genbank NC_045512) codon-optimized using IDT, with the Spike cytoplasmic tail replaced by that from the HA protein of A/WSN/1933 (H1N1) influenza, and the Kozak sequence in the plasmid fixed compared to an earlier version of this plasmid.
- HDM-IDTSpike-fixK (BEI catalog number NR-52514): Plasmid expressing under a CMV promoter the Spike from SARS-CoV-2 strain Wuhan-Hu-1 (Genbank NC_045512) codon-optimized using IDT and the Kozak sequence in the plasmid fixed compared to an earlier version of this plasmid.
- HDM-nCoV-Spike-IDTopt-ALAYT (BEI catalog number NR-52515): plasmid expressing under a CMV promoter the Spike from SARS-CoV-2 strain Wuhan-Hu-1 (Genbank NC_045512) codon-optimized using IDT, with the Spike containing two mutations in the cytoplasmic tail such that the last five amino acids are ALAYT.
- pHAGE-CMV-Luc2-IRES-ZsGreen-W (BEI catalog number NR-52516): Lentiviral backbone plasmid that uses a CMV promoter to express luciferase followed by an IRES and ZsGreen.
- HDM-Hgpm2 (BEI catalog number NR-52517): lentiviral helper plasmid expressing HIV Gag-Pol under a CMV promoter.
- HDM-tat1b (NR-52518): Lentiviral helper plasmid expressing HIV Tat under a CMV promoter.
- pRC-CMV-Rev1b (NR-52519): Lentiviral helper plasmid expressing HIV Rev under a CMV promoter.
- pHAGE2-CMV-ZsGreen-W (NR-52520): Lentiviral backbone plasmid that uses a CMV promoter to express ZsGreen.
4.2. Creation of 293T ACE2 Cells
4.3. Detailed Protocol for Generation of Pseudotyped Lentiviral Particles
- Seed 293T cells in D10 growth media (see Section 4.2 for media composition) so that they will be 50%–70% confluent the next day. For a six-well plate, this is 5 × 105 cells per well (2.5 × 105 cells per mL).
- At 16-24 h after seeding, transfect the cells with the plasmids required for lentiviral production. We transfected using BioT (Bioland Scientific, Paramount, CA, USA) following the manufacturer’s instructions and using the following plasmid mix per well of a six-well plate (plasmid amounts should be adjusted for larger plates):
- 1 μg of lentiviral backbone–we used either the ZsGreen (NR-52520) or the Luciferase-IRES-ZsGreen (NR-52516) backbone
- 0.22 μg each of plasmids HDM-Hgpm2 (NR-52517), pRC-CMV-Rev1b (NR-52519), and HDM-tat1b (NR-52518)
- 0.34 μg viral entry protein—either SARS-CoV-2 Spike (NR-52513, NR-52514, or NR-52515), VSV G (positive control), or transfection carrier DNA (E4881, Promega, Madison, WI, USA) as a negative control.
- At 18 to 24 h post-transfection, change the media to fresh, pre-warmed D10.
- At 60 h post transfection, collect virus by harvesting the supernatant from each well and filtering it through a 0.45 μm SFCA low protein-binding filter. Virus can be stored at 4 °C for immediate use or frozen at −80 °C. The titers of Spike- and VSV G-pseudotyped lentiviruses were found to be unaffected by a single freeze-thaw cycle (data not shown). Nonetheless, we recommend freezing virus in small aliquots to avoid multiple freeze-thaw cycles. All titers presented here are from virus that was frozen at −80 °C prior to use and underwent a single freeze-thaw.
4.4. Detailed Protocol for Titering Pseudotyped Lentiviral Particles
- Coat a 96-well cell-culture plate with 25 μL poly-l-lysine per well (P4707, Millipore Sigma, Burlington, MA, USA) according to the manufacturer’s protocol. Poly-L-lysine improves cell adherence and prevents cell disruption during infection.
- Seed a poly-l-lysine-coated 96-well plate with 1.25 × 104 293T-ACE2 cells per well in D10 media.
- The next day (12–24 h post-seeding), count at least two wells of cells to determine the number of cells present at infection.
- Prepare serial dilutions of the viruses to be titered in a final volume of 150 μL D10 growth media.
- For ZsGreen virus, we started with a 1:5 dilution and made three 1:5 serial dilutions.
- For Spike-pseudotyped Luciferase_IRES_ZsGreen virus, we started with undiluted virus and made three 1:3 dilutions. For VSV G-pseudotyped Luciferase_IRES_ZsGreen virus, we started with a 1:50 dilution.
- Gently remove the media from the cells and slowly add the virus dilutions.
- Add polybrene (TR-1003-G, Sigma Aldrich, St. Louis, MO, USA) to a final concentration of 5 μg/mL. We did this by adding 3 μL of polybrene diluted to 250 μg/mL to our final infection volume of 150 μL. Polybrene is a polycation that helps facilitate lentiviral infection through minimizing charge-repulsion between the virus and cells [56].
- At 48–60 h post-infection, collect cells for analysis:
- For flow cytometry:
- Look at the cells under a fluorescent microscope and select wells that appear ~1%–10% positive. Harvest cells from these wells using trypsin and transfer them to a V-bottom plate or microcentrifuge tubes. Pellet cells at 300× g for 4 min and wash twice with 3% BSA in PBS. After the final wash, resuspend in 1% BSA in PBS and analyze via flow cytometry. We used a Becton Dickinson Celesta cell analysis machine with a 530/30 filter to detect ZsGreen in the FITC channel. Resulting FCS files were analyzed using FlowJo (v10) (BD Life Sciences, Ashland, OR, USA).
- Calculate titers using the Poisson formula. If P is the percentage of cells that are ZsGreen positive, then the titer per ml is: -ln(1 − P/100) × (number of cells/well)/(volume of virus per well in mL). Note that when the percentage of cells that are ZsGreen positive is low, this formula is approximately equal to: (% ZsGreen positive/100) × (number of cells per well)/volume of virus per well in mL. Furthermore, the titers using even the Poisson equation will only be accurate if the percentage of cells that are ZsGreen positive is relatively low (ideally 1–10%).
- For luciferase:
- Thaw luciferase reagent at room temperature. We used the Bright-Glo Luciferase Assay System (E2610, Promega, Madison, WI, USA).
- Prepare cells by removing 100 μL media from each well. Accounting for evaporation, this leaves ~30 μL of media in each well.
- Add 30 μL of luciferase reagent, mix well, and transfer all 60 μL to a black-bottom plate (Costar 96-well solid black, 07-200-590, Fisher Scientific, Waltham, MA, USA).
- Incubate plate for 2 min at room temperature in the dark, then measure luminescence using a plate reader. We used a Tecan Infinite M1000 Pro plate reader with no attenuation and a luminescence integration time of 1 s.
- Plot RLUs vs. virus dilution. Select an amount of virus for further assays where there is sufficient (>1000-fold) signal above virus-only background and a linear relationship between virus added and RLU.
4.5. Detailed Protocol for Neutralization Assays
- Seed a poly-L-lysine-coated 96-well plate with 1.25 × 104 293T-ACE2 cells (BEI NR-52511) per well in 50 μL D10 (2.5 × 105 cells per mL). Plan to infect this plate 8–12 h post-seeding.
- About 1.5 h prior to infecting cells, begin preparing serum and/or ACE2 dilutions in D10:
- In a separate 96-well “setup” plate, serially dilute serum samples, leaving 60 μL diluted serum in each well. For the data in Figure 4A,C, we started at an initial serum dilution of 1:80 and did serial 2.5-fold dilutions, meaning each replicate of the assay required 5 μL of sera. For ACE2 (Figure 4B), we started with a concentration of 200 μg/mL and did serial three-fold dilutions.
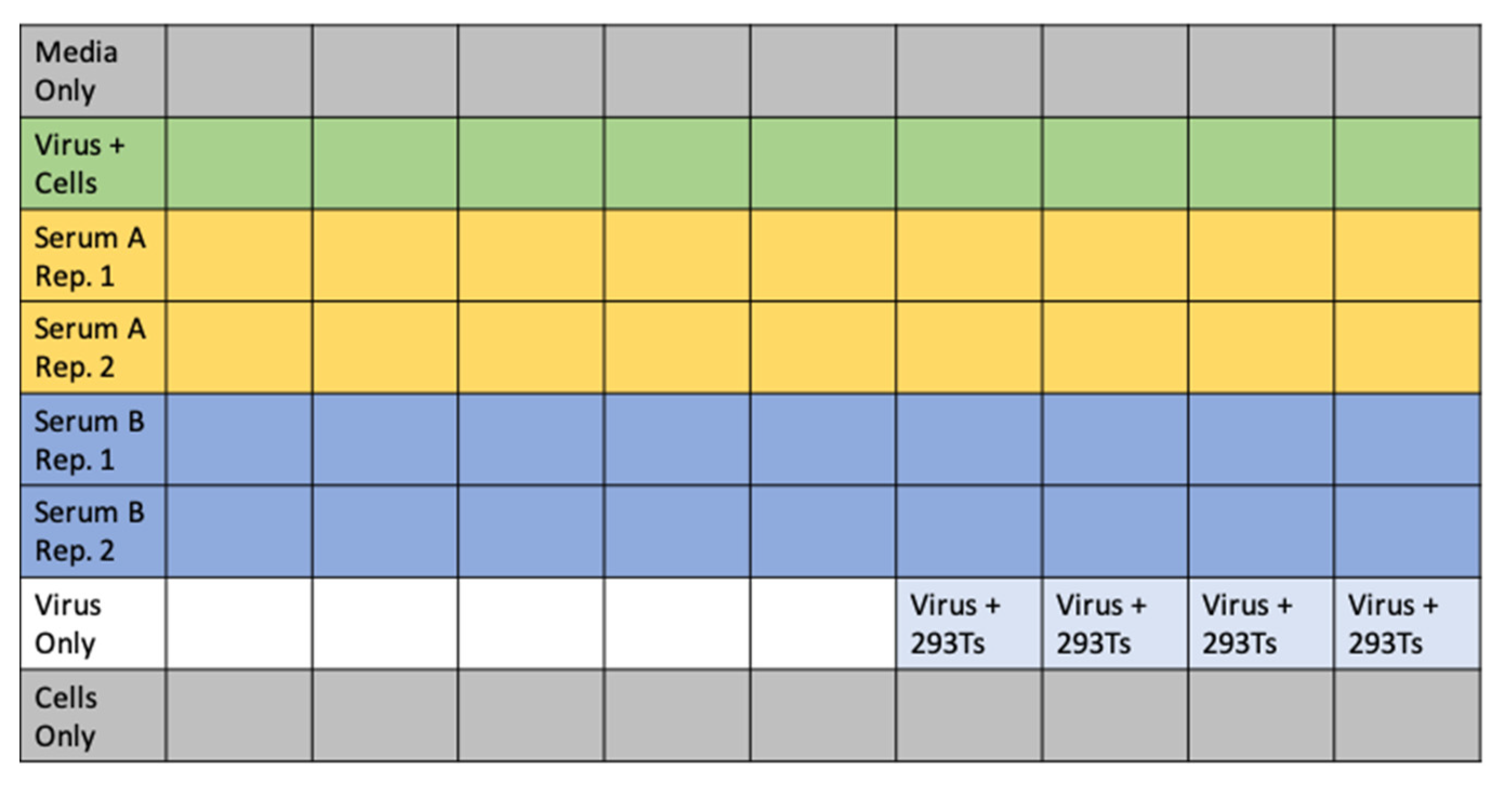
- Add 60 μL of D10 to wells corresponding to virus only and virus plus cells control wells. Add 120 μL of D10 to media only and cells only control wells. See Figure 5 for an example plate layout.
- Dilute virus to ~2–4 × 106 RLU per mL. Add 60 μL of diluted virus to all wells containing serum dilutions and the virus only and virus plus cells control wells.
- Incubate virus and serum at 37 °C for 1 h.
- Carefully add 100 μL from each well of the setup plate containing the sera and virus dilutions to the corresponding wells of the plate of 293T-ACE2 cells.
- Add polybrene (Sigma Aldrich, TR-1003-G) as described in Section 4.4 for a final concentration of 5 μg/mL in each well.
- Incubate at 37 °C for 48–60 h before reading out luminescence or fluorescence as described in Section 4.4.
- Plot the data. For our analysis, we first subtracted out the background signal (average of the “virus only” and “virus + 293Ts” wells) and then calculated the “maximum infectivity” for each plate as the average signal from the wells without serum (“virus + cells” wells). We then calculated the “fraction infectivity” for each well, as the luciferase reading from each well divided by the “maximum infectivity” for that plate. For the curves shown in Figure 4, we then fit and plotted the fraction infectivity data using the neutcurve Python package (https://jbloomlab.github.io/neutcurve/). This package fits a three-parameter Hill curve, with the top baseline being a free parameter and bottom baseline fixed to zero.
4.6. Human Plasma Sample and Soluble ACE2
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ju, B.; Zhang, Q.; Ge, X.; Wang, R.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; Tang, X.; Yu, J.; et al. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Khan, S.; Nakajima, R.; Jain, A.; de Assis, R.R.; Jasinskas, A.; Obiero, J.M.; Adenaiye, O.; Tai, S.; Hong, F.; Milton, D.K.; et al. Analysis of Serologic Cross-Reactivity Between Common Human Coronaviruses and SARS-CoV-2 Using Coronavirus Antigen Microarray. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients of Novel Coronavirus Disease 2019. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Wu, F.; Wang, A.; Liu, M.; Wang, Q.; Chen, J.; Xia, S.; Ling, Y.; Zhang, Y.; Xun, J.; Lu, L.; et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 2020. [Google Scholar] [CrossRef]
- Long, Q.; Deng, H.; Chen, J.; Hu, J.; Liu, B.; Liao, P.; Lin, Y.; Yu, L.; Mo, Z.; Xu, Y.; et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: The perspective application of serological tests in clinical practice. medRxiv 2020. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Müller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D.; et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg. Infect. Dis. 2020, 26. [Google Scholar] [CrossRef]
- Bootz, A.; Karbach, A.; Spindler, J.; Kropff, B.; Reuter, N.; Sticht, H.; Winkler, T.H.; Britt, W.J.; Mach, M. Protective capacity of neutralizing and non-neutralizing antibodies against glycoprotein B of cytomegalovirus. PLoS Pathog. 2017, 13. [Google Scholar] [CrossRef]
- Piedra, P.A.; Jewell, A.M.; Cron, S.G.; Atmar, R.L.; Paul Glezen, W. Correlates of immunity to respiratory syncytial virus (RSV) associated-hospitalization: Establishment of minimum protective threshold levels of serum neutralizing antibodies. Vaccine 2003, 21, 3479–3482. [Google Scholar] [CrossRef]
- Karlsson Hedestam, G.B.; Fouchier, R.A.M.; Phogat, S.; Burton, D.R.; Sodroski, J.; Wyatt, R.T. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 2008, 6, 143–155. [Google Scholar] [CrossRef]
- Gunn, B.M.; Yu, W.H.; Karim, M.M.; Brannan, J.M.; Herbert, A.S.; Wec, A.Z.; Halfmann, P.J.; Fusco, M.L.; Schendel, S.L.; Gangavarapu, K.; et al. A Role for Fc Function in Therapeutic Monoclonal Antibody-Mediated Protection against Ebola Virus. Cell Host Microbe 2018, 24, 221–233. [Google Scholar] [CrossRef]
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.P.M.; Leung, W.S.; So, R.T.Y.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H.; et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pinto, D.; Park, Y.-J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv 2020. [Google Scholar] [CrossRef]
- To, K.K.W.; Zhang, A.J.X.; Hung, I.F.N.; Xu, T.; Ip, W.C.T.; Wong, R.T.Y.; Ng, J.C.K.; Chan, J.F.W.; Chan, K.H.; Yuen, K.Y. High titer and avidity of nonneutralizing antibodies against influenza vaccine antigen are associated with severe influenza. Clin. Vaccine Immunol. 2012, 19, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Co, M.D.T.; Terajima, M.; Thomas, S.J.; Jarman, R.G.; Rungrojcharoenkit, K.; Fernandez, S.; Yoon, I.K.; Buddhari, D.; Cruz, J.; Ennis, F.A. Relationship of preexisting influenza hemagglutination inhibition, complement-dependent lytic, and antibody-dependent cellular cytotoxicity antibodies to the development of clinical illness in a prospective study of A(H1N1)pdm09 influenza in children. Viral Immunol. 2014, 27, 375–382. [Google Scholar] [CrossRef]
- Callow, K.A. Effect of specific humoral immunity and some non-specific factors on resistance of volunteers to respiratory coronavirus infection. J. Hyg. (Lond.) 1985, 95, 173–189. [Google Scholar] [CrossRef]
- Corti, D.; Zhao, J.; Pedotti, M.; Simonelli, L.; Agnihothram, S.; Fett, C.; Fernandez-Rodriguez, B.; Foglierini, M.; Agatic, G.; Vanzetta, F.; et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. USA 2015, 112, 10473–10478. [Google Scholar] [CrossRef]
- Menachery, V.D.; Yount, B.L.; Sims, A.C.; Debbink, K.; Agnihothram, S.S.; Gralinski, L.E.; Graham, R.L.; Scobey, T.; Plante, J.A.; Royal, S.R.; et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA 2016, 113, 3048–3053. [Google Scholar] [CrossRef]
- Rockx, B.; Corti, D.; Donaldson, E.; Sheahan, T.; Stadler, K.; Lanzavecchia, A.; Baric, R. Structural Basis for Potent Cross-Neutralizing Human Monoclonal Antibody Protection against Lethal Human and Zoonotic Severe Acute Respiratory Syndrome Coronavirus Challenge. J. Virol. 2008, 82, 3220–3235. [Google Scholar] [CrossRef]
- Subbarao, K.; McAuliffe, J.; Vogel, L.; Fahle, G.; Fischer, S.; Tatti, K.; Packard, M.; Shieh, W.-J.; Zaki, S.; Murphy, B. Prior Infection and Passive Transfer of Neutralizing Antibody Prevent Replication of Severe Acute Respiratory Syndrome Coronavirus in the Respiratory Tract of Mice. J. Virol. 2004, 78, 3572–3577. [Google Scholar] [CrossRef]
- Kapadia, S.U.; Rose, J.K.; Lamirande, E.; Vogel, L.; Subbarao, K.; Roberts, A. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology 2005, 340, 174–182. [Google Scholar] [CrossRef]
- Callow, K.A.; Parry, H.F.; Sergeant, M.; Tyrrell, D.A.J. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990, 105, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.E. The behaviour of recent isolates of human respiratory coronavirus in vitro and in volunteers: Evidence of heterogeneity among 229E-related strains. J. Med. Virol. 1984, 13, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Soo, Y.O.Y.; Cheng, Y.; Wong, R.; Hui, D.S.; Lee, C.K.; Tsang, K.K.S.; Ng, M.H.L.; Chan, P.; Cheng, G.; Sung, J.J.Y. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004, 10, 676–678. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wong, R.; Soo, Y.O.Y.; Wong, W.S.; Lee, C.K.; Ng, M.H.L.; Chan, P.; Wong, K.C.; Leung, C.B.; Cheng, G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA 2020. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Nguyen, T.; Chromikova, V.; Strohmeier, S.; Stadlbauer, D.; Javier, A.; Jiang, K.; Asthagiri-Arunkumar, G.; Polanco, J.; Bermudez-Gonzalez, M.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv 2020. [Google Scholar] [CrossRef]
- Chen, X.; Li, R.; Pan, Z.; Qian, C.; Yang, Y.; You, R.; Zhao, J.; Liu, P.; Gao, L.; Li, Z.; et al. Human monoclonal antibodies block the binding of SARS-CoV-2 Spike protein to angiotensin converting enzyme 2. medRxiv 2020. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Fukushi, S.; Mizutani, T.; Saijo, M.; Matsuyama, S.; Miyajima, N.; Taguchi, F.; Itamura, S.; Kurane, I.; Morikawa, S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005, 86, 2269–2274. [Google Scholar] [CrossRef]
- Temperton, N.J.; Chan, P.K.; Simmons, G.; Zambon, M.C.; Tedder, R.S.; Takeuchi, Y.; Weiss, R.A. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg. Infect. Dis. 2005, 11. [Google Scholar] [CrossRef] [PubMed]
- Carnell, G.; Grehan, K.; Ferrara, F.; Molesti, E.; Temperton, N. An Optimized Method for the Production Using PEI, Titration and Neutralization of SARS-CoV Spike Luciferase Pseudotypes. Bio-Protocol 2017, 7. [Google Scholar] [CrossRef]
- Yan, K.X.; Tan, W.J.; Zhang, X.M.; Wang, H.J.; Li, Y.; Ruan, L. Development and application of a safe SARS-CoV neutralization assay based on lentiviral vectors pseudotyped with SARS-CoV spike protein. Bing Du Xue Bao 2007, 23, 440–446. [Google Scholar] [PubMed]
- Grehan, K.; Ferrara, F.; Temperton, N. An optimised method for the production of MERS-CoV spike expressing viral pseudotypes. MethodsX 2015, 2, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Lester, S.; Harcourt, J.; Whitt, M.; Al-Abdely, H.M.; Midgley, C.M.; Alkhamis, A.M.; Aziz Jokhdar, H.A.; Assiri, A.M.; Tamin, A.; Thornburg, N. Middle East respiratory coronavirus (MERS-CoV) spike (S) protein vesicular stomatitis virus pseudoparticle neutralization assays offer a reliable alternative to the conventional neutralization assay in human seroepidemiological studies. Access Microbiol. 2019, 9. [Google Scholar] [CrossRef]
- Millet, J.; Whittaker, G. Murine Leukemia Virus (MLV)-based Coronavirus Spike-pseudotyped Particle Production and Infection. Bio-Protocol 2016, 6. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Quinlan, B.D.; Mou, H.; Zhang, L.; Guo, Y.; He, W.; Ojha, A.; Parcells, M.S.; Luo, G.; Li, W.; Zhong, G.; et al. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv 2020. [Google Scholar] [CrossRef]
- Xiong, H.; Wu, Y.; Cao, J.; Yang, R.; Ma, J.; Qiao, X.; Yao, X.; Zhang, B.; Zhang, Y.; Hou, W.; et al. Robust neutralization assay based on SARS-CoV-2 S-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressed BHK21 cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 680–686. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-) 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.-C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science (80-) 2020. [Google Scholar] [CrossRef] [PubMed]
- Joyce, M.G.; Sankhala, R.S.; Chen, W.-H.; Choe, M.; Bai, H.; Hajduczki, A.; Yan, L.; Sterling, S.L.; Peterson, C.; Green, E.C.; et al. A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- McBride, C.E.; Li, J.; Machamer, C.E. The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds COPI and Promotes Interaction with Membrane Protein. J. Virol. 2007, 81, 2418–2428. [Google Scholar] [CrossRef]
- Sadasivan, J.; Singh, M.; Sarma, J. Das Cytoplasmic tail of coronavirus spike protein has intracellular targeting signals. J. Biosci. 2017, 42, 231–244. [Google Scholar] [CrossRef]
- Giroglou, T.; Cinatl, J.; Rabenau, H.; Drosten, C.; Schwalbe, H.; Doerr, H.W.; von Laer, D. Retroviral Vectors Pseudotyped with Severe Acute Respiratory Syndrome Coronavirus S Protein. J. Virol. 2004, 78, 9007–9015. [Google Scholar] [CrossRef]
- Schwegmann-Weßels, C.; Glende, J.; Ren, X.; Qu, X.; Deng, H.; Enjuanes, L.; Herrler, G. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J. Gen. Virol. 2009, 90, 1724–1729. [Google Scholar] [CrossRef]
- Moore, M.J.; Dorfman, T.; Li, W.; Wong, S.K.; Li, Y.; Kuhn, J.H.; Coderre, J.; Vasilieva, N.; Han, Z.; Greenough, T.C.; et al. Retroviruses Pseudotyped with the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Efficiently Infect Cells Expressing Angiotensin-Converting Enzyme 2. J. Virol. 2004, 78, 10628–10635. [Google Scholar] [CrossRef]
- Jiang, W.; Hua, R.; Wei, M.; Li, C.; Qiu, Z.; Yang, X.; Zhang, C. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Cribbs, A.P.; Kennedy, A.; Gregory, B.; Brennan, F.M. Simplified production and concentration of lentiviral vectors to achieve high transduction in primary human T cells. BMC Biotechnol. 2013, 13. [Google Scholar] [CrossRef]
- Lei, C.; Fu, W.; Qian, K.; Li, T.; Zhang, S.; Ding, M.; Hu, S. Potent neutralization of 2019 novel coronavirus by recombinant ACE2-Ig. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bloch, E.M.; Shoham, S.; Casadevall, A.; Sachais, B.S.; Shaz, B.; Winters, J.L.; van Buskirk, C.; Grossman, B.J.; Joyner, M.; Henderson, J.P.; et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Investig. 2020. [Google Scholar] [CrossRef]
- Denning, W.; Das, S.; Guo, S.; Xu, J.; Kappes, J.C.; Hel, Z. Optimization of the transductional efficiency of lentiviral vectors: Effect of sera and polycations. Mol. Biotechnol. 2013, 53, 308–314. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.A.; Hui, K.P.Y.; Yen, H.-L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, 5247, 30003. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. J. Am. Med. Assoc. 2020. [Google Scholar] [CrossRef]
- Dodd, R.Y.; Stramer, S.L. COVID-19 and Blood Safety: Help with a Dilemma. Transfus. Med. Rev. 2020. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crawford, K.H.D.; Eguia, R.; Dingens, A.S.; Loes, A.N.; Malone, K.D.; Wolf, C.R.; Chu, H.Y.; Tortorici, M.A.; Veesler, D.; Murphy, M.; et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 2020, 12, 513. https://doi.org/10.3390/v12050513
Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020; 12(5):513. https://doi.org/10.3390/v12050513
Chicago/Turabian StyleCrawford, Katharine H. D., Rachel Eguia, Adam S. Dingens, Andrea N. Loes, Keara D. Malone, Caitlin R. Wolf, Helen Y. Chu, M. Alejandra Tortorici, David Veesler, Michael Murphy, and et al. 2020. "Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays" Viruses 12, no. 5: 513. https://doi.org/10.3390/v12050513
APA StyleCrawford, K. H. D., Eguia, R., Dingens, A. S., Loes, A. N., Malone, K. D., Wolf, C. R., Chu, H. Y., Tortorici, M. A., Veesler, D., Murphy, M., Pettie, D., King, N. P., Balazs, A. B., & Bloom, J. D. (2020). Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses, 12(5), 513. https://doi.org/10.3390/v12050513




