RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane
Abstract
1. Introduction
2. Materials and Methods
2.1. Production of Isolated Tsg101 UEV Domain
2.2. NMR Spectroscopy (Binding Studies)
2.3. Plasmids and Reagents
2.4. Transfection and Assays for Particle Budding
2.5. Fluorescence Microscopy
2.6. Yeast Two-Hybrid Assay
3. Results
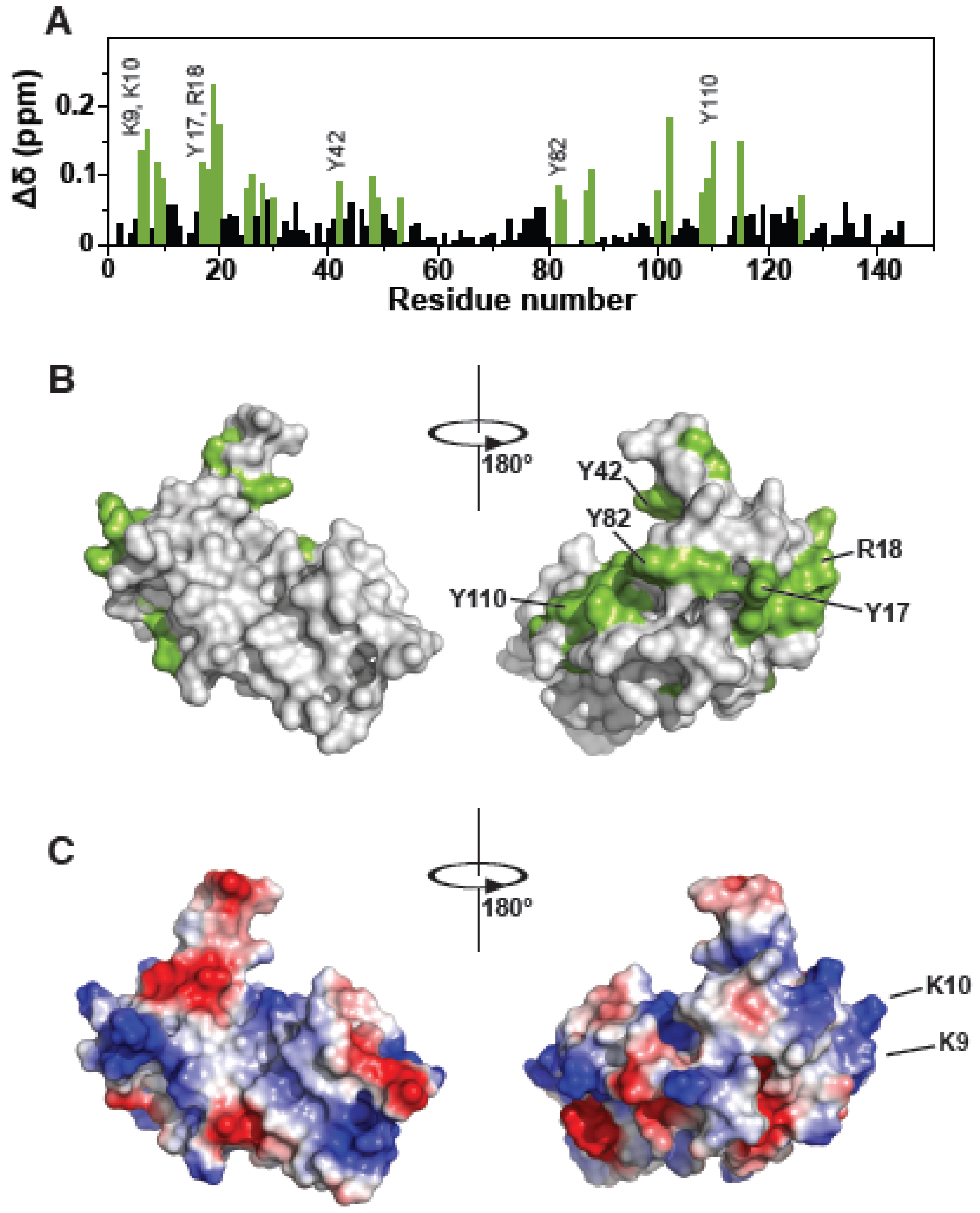
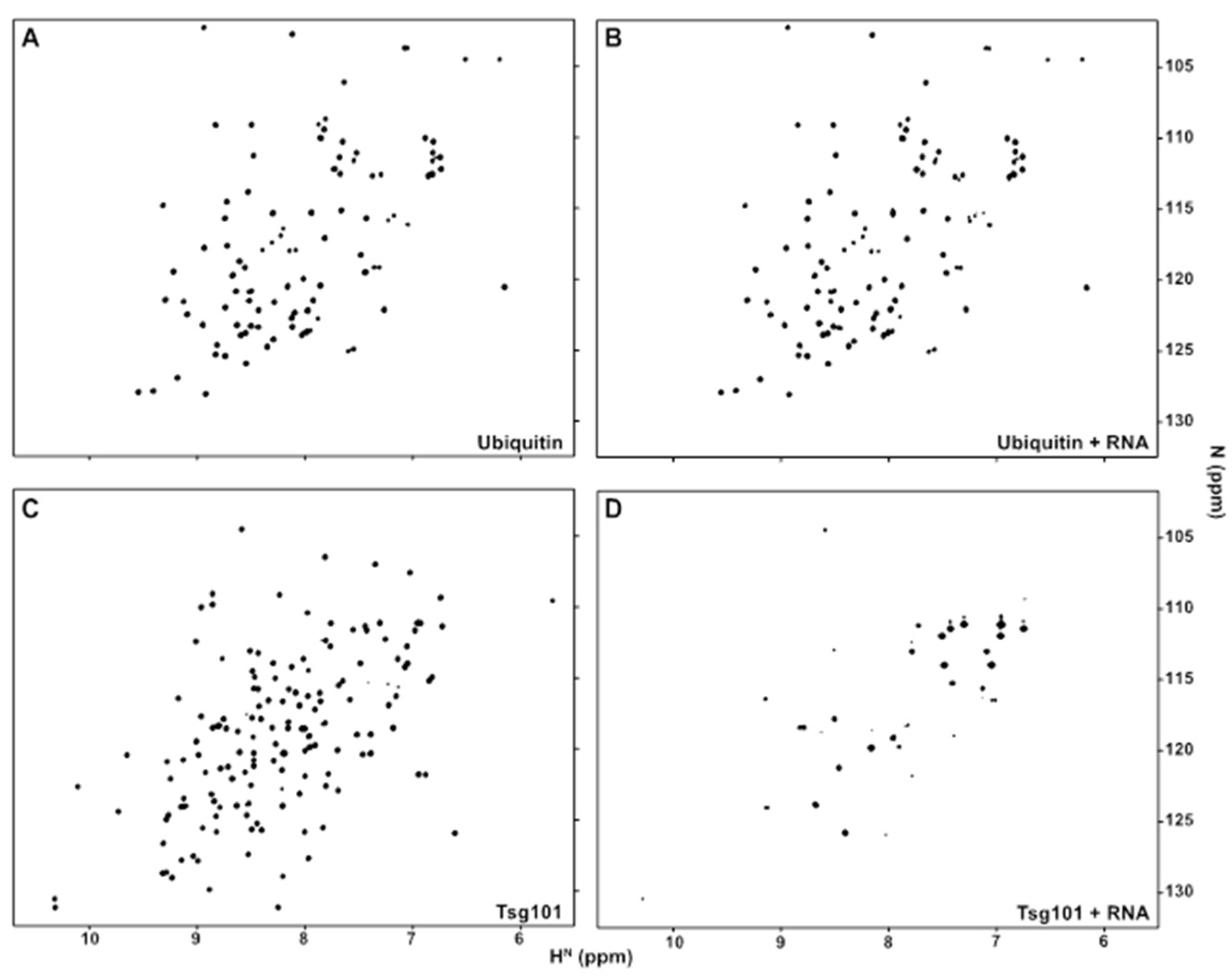
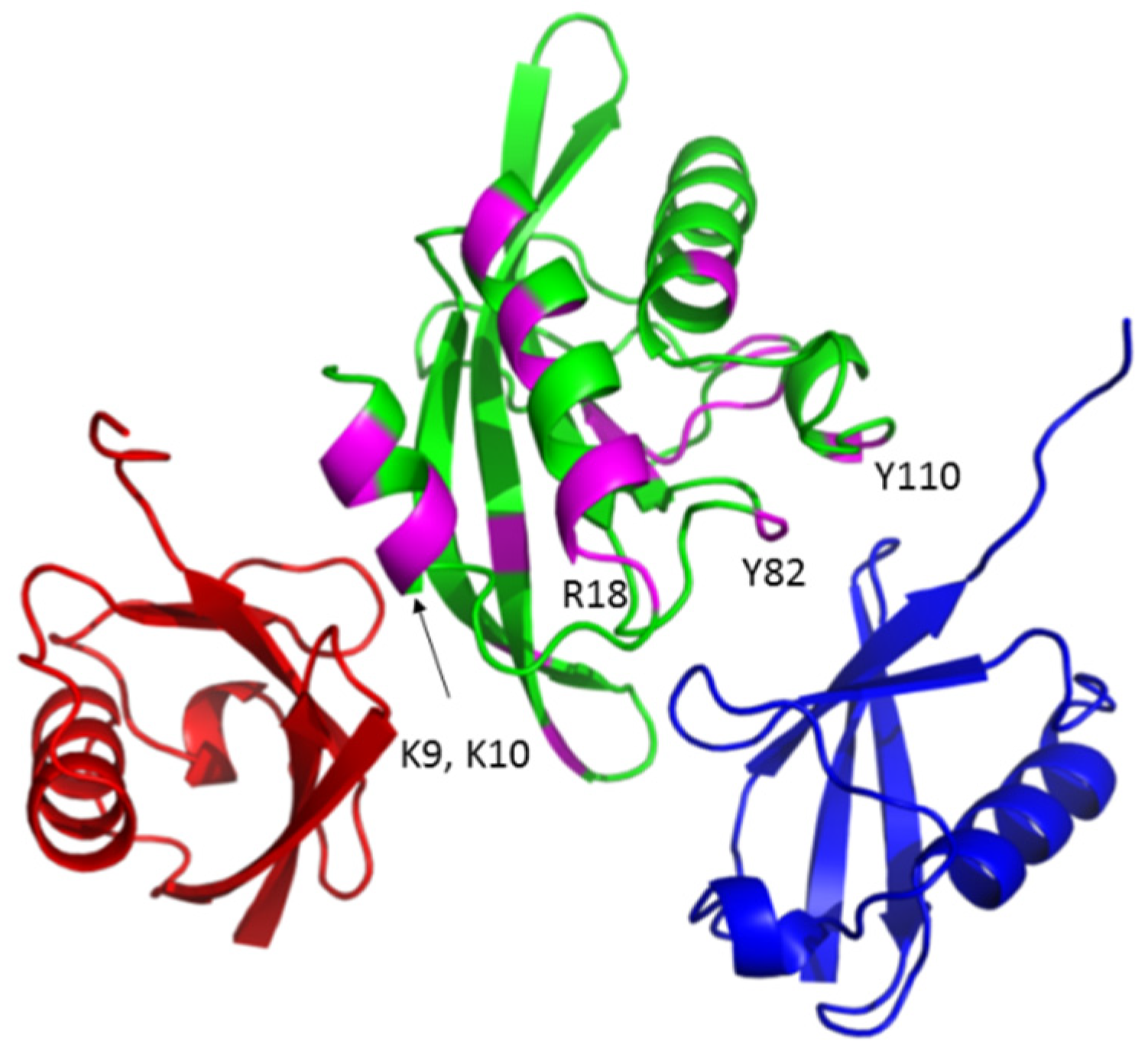
3.1. The Tsg101 UEV Domain Binds tRNA
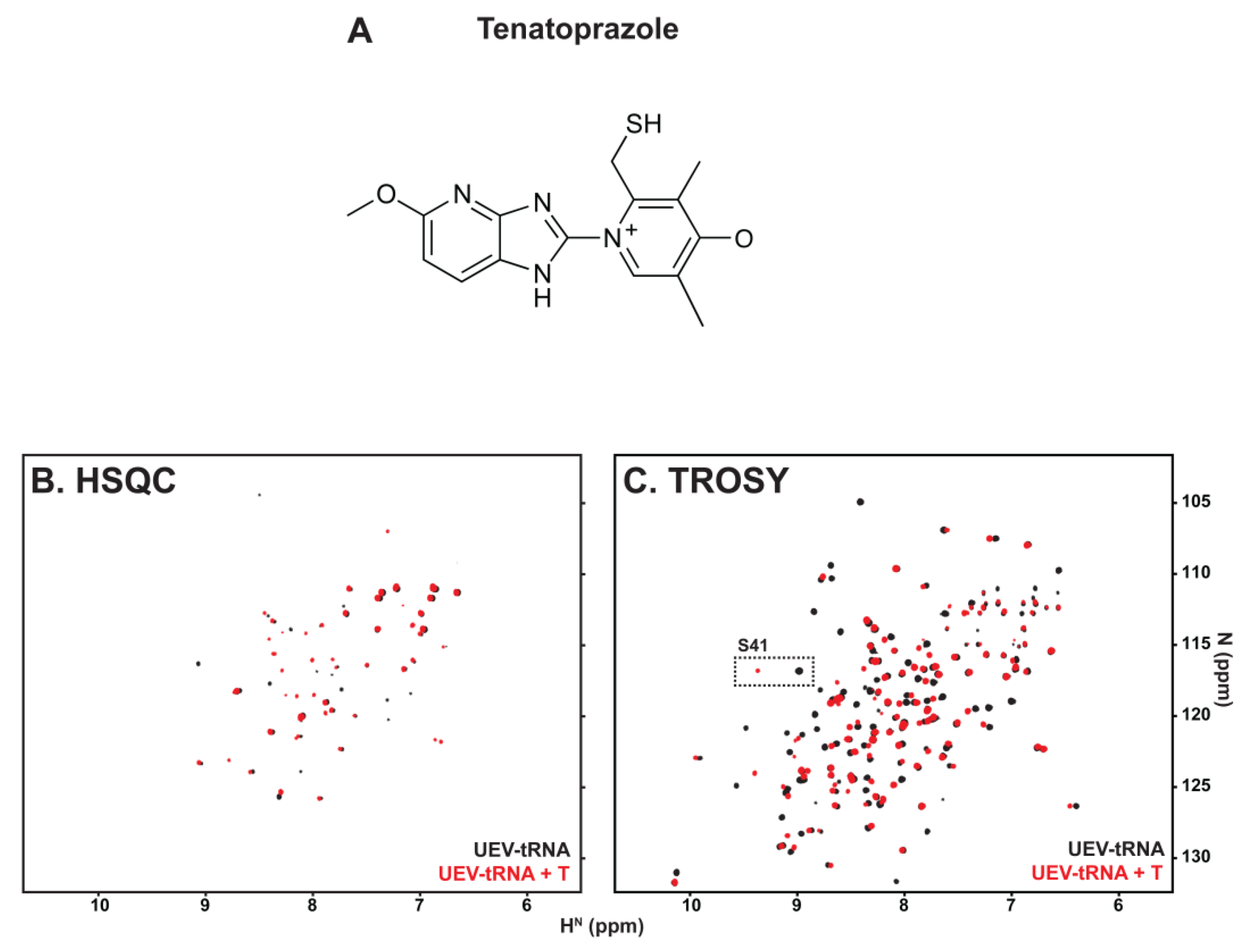
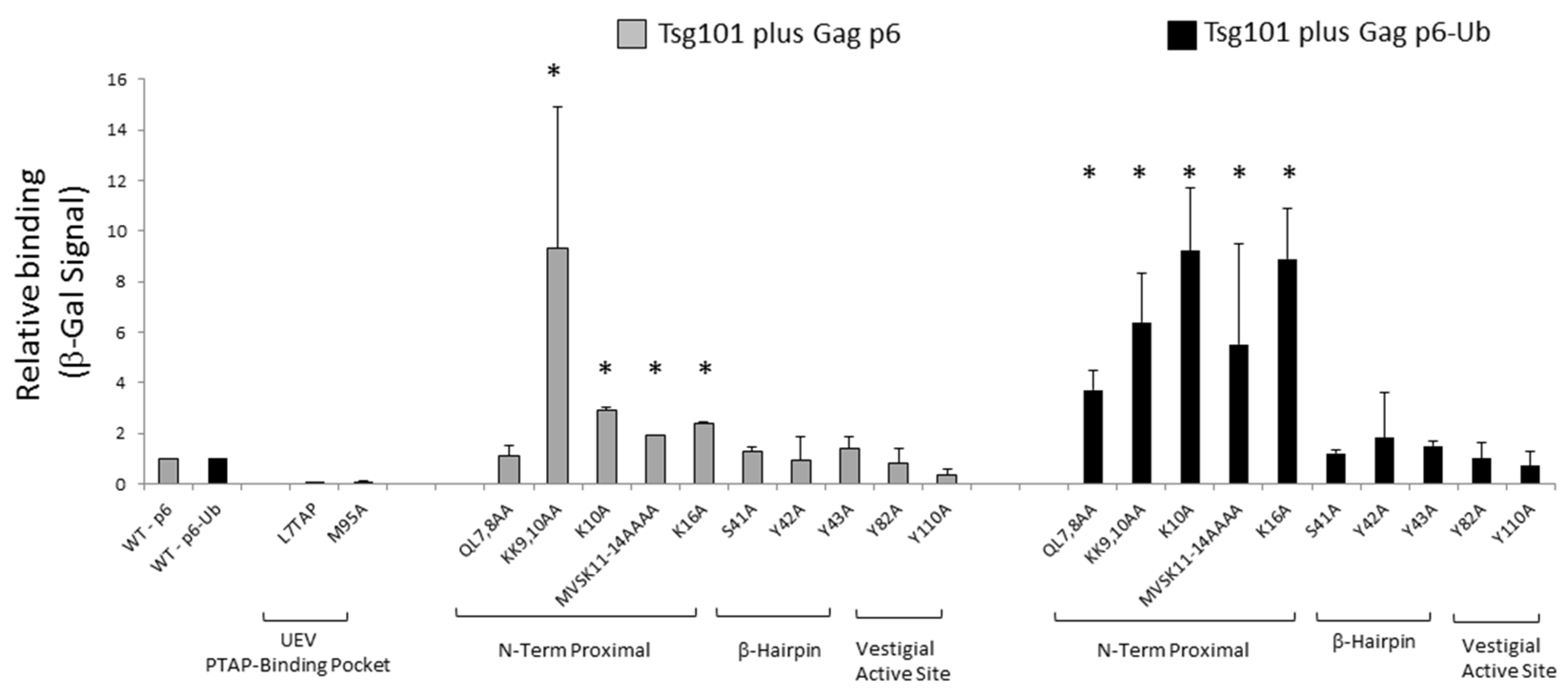
3.2. tRNA Binding Does Not Prevent Access to the Site Necessary for Mono-Ub Binding
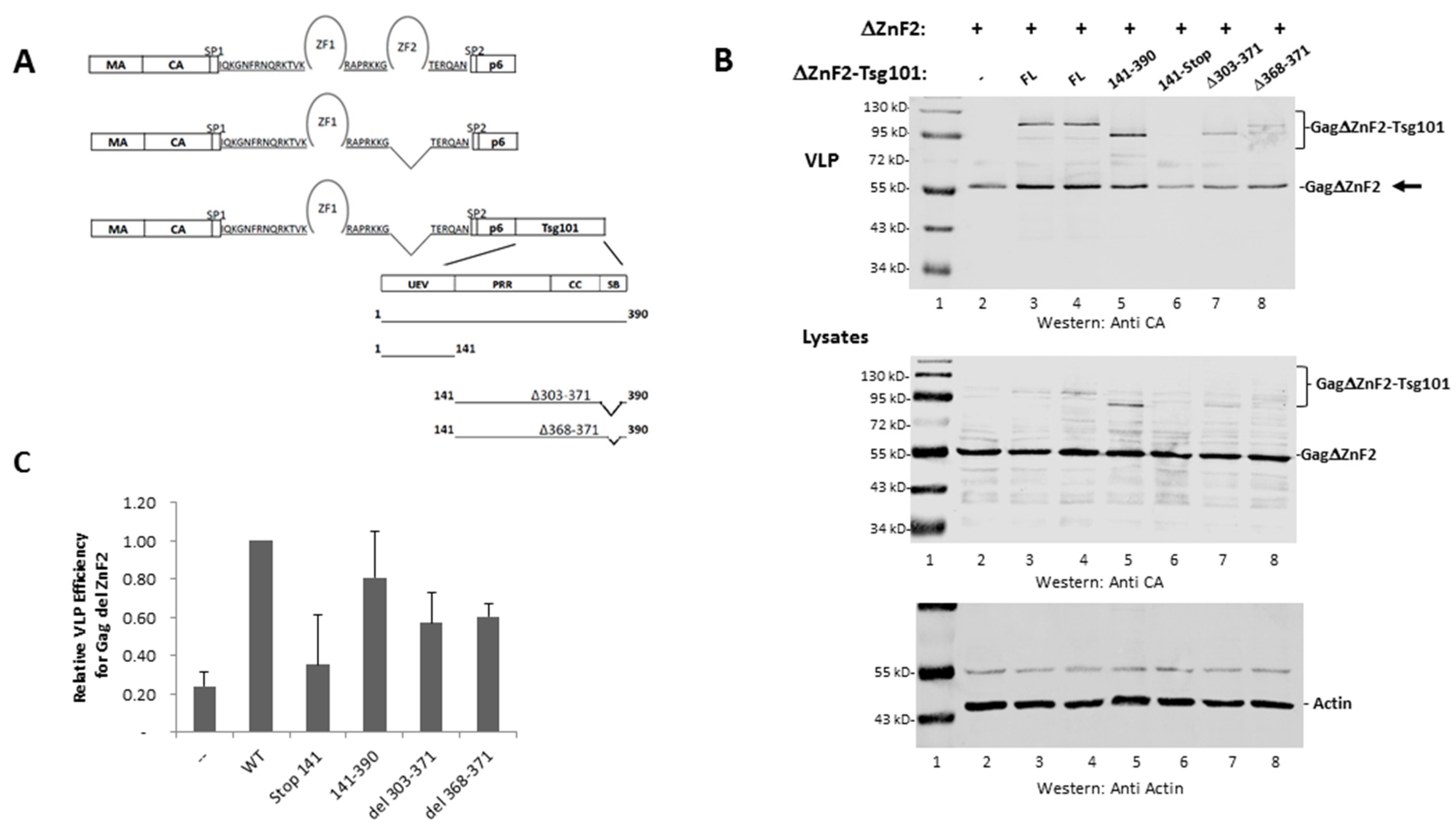
3.3. The UEV Domain Is Not Required when Tsg101 Is at the Budding Site
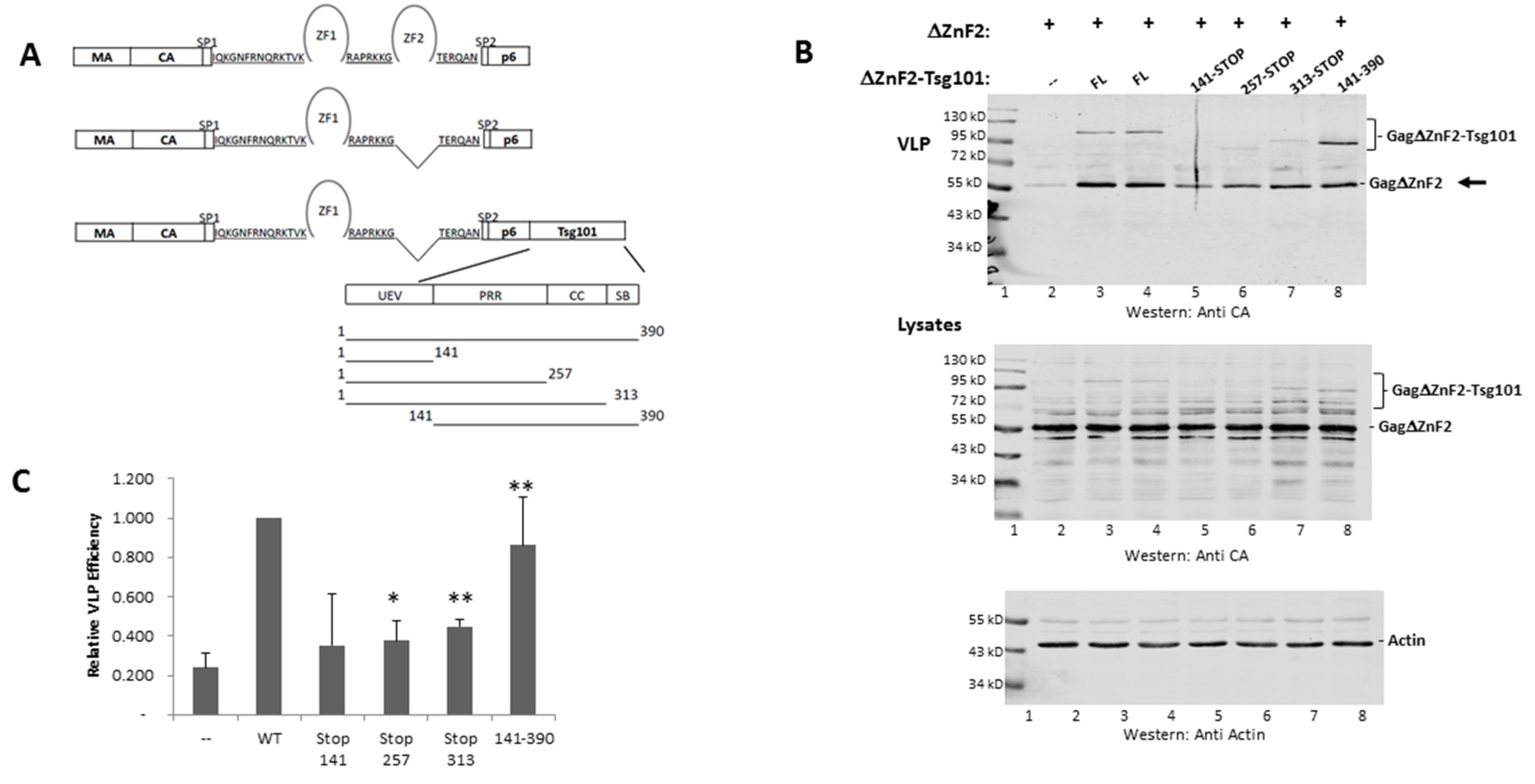
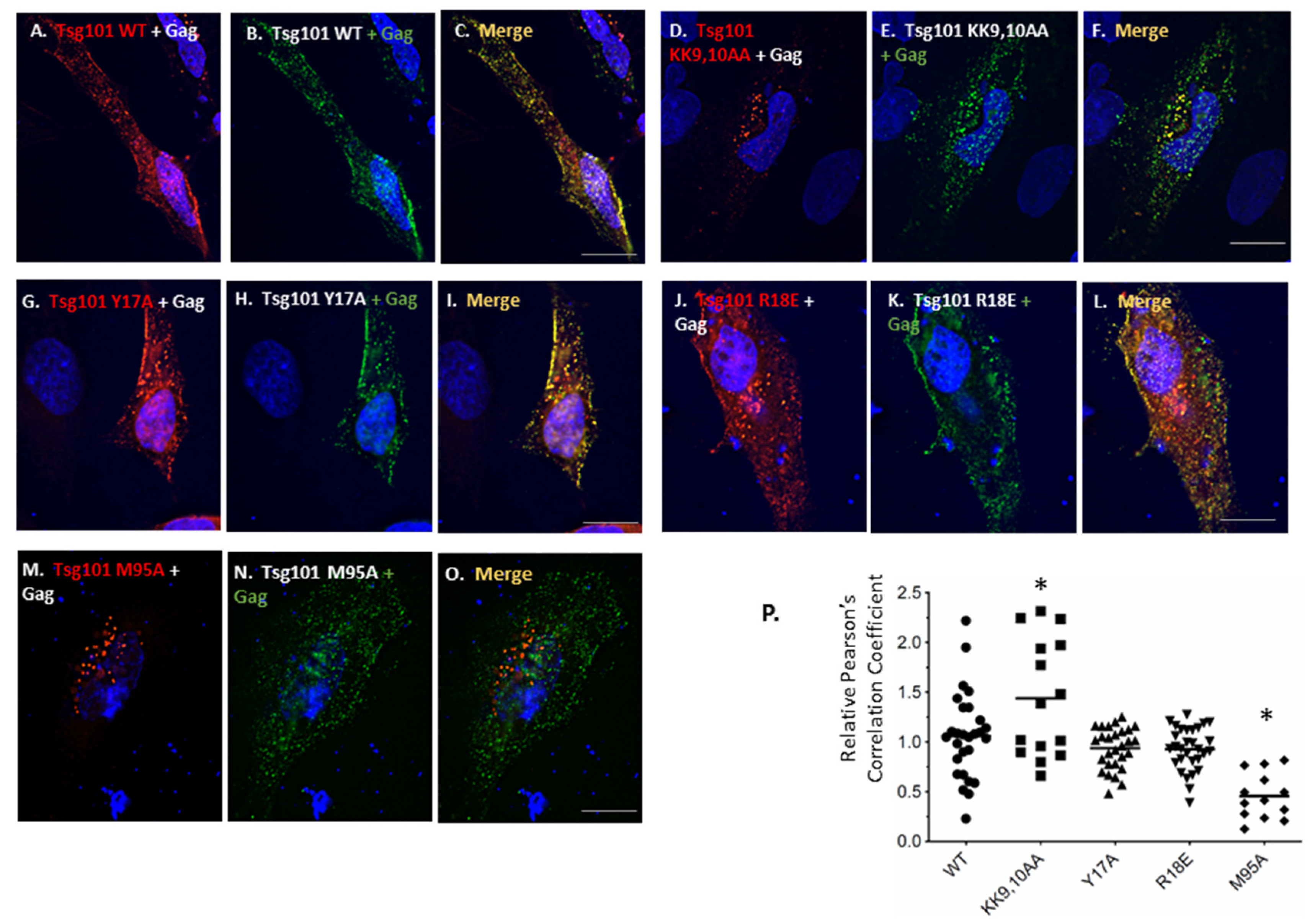
3.4. Recruitment from the Cell Interior Requires the tRNA Binding Determinants
3.5. Mutation of tRNA Binding Determinants Promotes Recognition of Ub-Modified Gag
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sette, P.; Dussupt, V.; Bouamr, F. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J. Virol. 2012, 86, 11608–11615. [Google Scholar] [CrossRef]
- Katoh, K.; Shibata, H.; Hatta, K.; Maki, M. CHMP4b is a major binding partner of the ALG-2-interacting protein Alix among the three CHMP4 isoforms. Arch. Biochem. Biophys. 2004, 421, 159–165. [Google Scholar] [CrossRef]
- Martin-Serrano, J.; Yarovoy, A.; Perez-Caballero, D.; Bieniasz, P.D. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 12414–12419. [Google Scholar] [CrossRef] [PubMed]
- Strack, B.; Calistri, A.; Craig, S.; Popova, E.; Gottlinger, H.G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 2003, 114, 689–699. [Google Scholar] [CrossRef]
- von Schwedler, U.K.; Stuchell, M.; Muller, B.; Ward, D.M.; Chung, H.Y.; Morita, E.; Wang, H.E.; Davis, T.; He, G.P.; Cimbora, D.M.; et al. The protein network of HIV budding. Cell 2003, 114, 701–713. [Google Scholar] [CrossRef]
- Fisher, R.D.; Chung, H.Y.; Zhai, Q.; Robinson, H.; Sundquist, W.I.; Hill, C.P. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 2007, 128, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Usami, Y.; Popov, S.; Gottlinger, H.G. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 2007, 81, 6614–6622. [Google Scholar] [CrossRef] [PubMed]
- Dussupt, V.; Javid, M.P.; Abou-Jaoude, G.; Jadwin, J.A.; de La Cruz, J.; Nagashima, K.; Bouamr, F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009, 5, e1000339. [Google Scholar] [CrossRef]
- Popov, S.; Popova, E.; Inoue, M.; Gottlinger, H.G. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 2008, 82, 1389–1398. [Google Scholar] [CrossRef]
- McCullough, J.; Fisher, R.D.; Whitby, F.G.; Sundquist, W.I.; Hill, C.P. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 7687–7691. [Google Scholar] [CrossRef]
- VerPlank, L.; Bouamr, F.; LaGrassa, T.J.; Agresta, B.; Kikonyogo, A.; Leis, J.; Carter, C.A. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 2001, 98, 7724–7729. [Google Scholar] [CrossRef] [PubMed]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Cote, M.; Rich, R.L.; et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001, 7, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Dussupt, V.; Sette, P.; Bello, N.F.; Javid, M.P.; Nagashima, K.; Bouamr, F. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J. Virol. 2011, 85, 2304–2315. [Google Scholar] [CrossRef]
- El Meshri, S.E.; Boutant, E.; Mouhand, A.; Thomas, A.; Larue, V.; Richert, L.; Vivet-Boudou, V.; Mely, Y.; Tisne, C.; Muriaux, D.; et al. The NC domain of HIV-1 Gag contributes to the interaction of Gag with TSG101. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1421–1431. [Google Scholar] [CrossRef]
- Wang, W.; Naiyer, N.; Mitra, M.; Li, J.; Williams, M.C.; Rouzina, I.; Gorelick, R.J.; Wu, Z.; Musier-Forsyth, K. Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity. Nucleic Acids Res. 2014, 42, 7145–7159. [Google Scholar] [CrossRef]
- Chamontin, C.; Rassam, P.; Ferrer, M.; Racine, P.J.; Neyret, A.; Laine, S.; Milhiet, P.E.; Mougel, M. HIV-1 nucleocapsid and ESCRT-component Tsg101 interplay prevents HIV from turning into a DNA-containing virus. Nucleic Acids Res. 2015, 43, 336–347. [Google Scholar] [CrossRef]
- Patnaik, A.; Chau, V.; Wills, J.W. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 2000, 97, 13069–13074. [Google Scholar] [CrossRef]
- Strack, B.; Calistri, A.; Accola, M.A.; Palu, G.; Gottlinger, H.G. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 2000, 97, 13063–13068. [Google Scholar] [CrossRef]
- Joshi, A.; Munshi, U.; Ablan, S.D.; Nagashima, K.; Freed, E.O. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic 2008, 9, 1972–1983. [Google Scholar] [CrossRef]
- Tanzi, G.O.; Piefer, A.J.; Bates, P. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J. Virol. 2003, 77, 8440–8447. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Higginson, D.S.; Stray, K.M.; Fisher, R.D.; Garrus, J.E.; Payne, M.; He, G.P.; Wang, H.E.; Morham, S.G.; Sundquist, W.I. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 2003, 162, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Pincetic, A.; Medina, G.; Carter, C.; Leis, J. Avian sarcoma virus and human immunodeficiency virus, type 1 use different subsets of ESCRT proteins to facilitate the budding process. J. Biol. Chem. 2008, 283, 29822–29830. [Google Scholar] [CrossRef] [PubMed]
- Medina, G.; Pincetic, A.; Ehrlich, L.S.; Zhang, Y.; Tang, Y.; Leis, J.; Carter, C.A. Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting. Virology 2008, 377, 30–38. [Google Scholar] [CrossRef]
- Gorelick, R.J.; Chabot, D.J.; Rein, A.; Henderson, L.E.; Arthur, L.O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 1993, 67, 4027–4036. [Google Scholar] [CrossRef]
- Zargarian, L.; Tisne, C.; Barraud, P.; Xu, X.; Morellet, N.; Rene, B.; Mely, Y.; Fosse, P.; Mauffret, O. Dynamics of linker residues modulate the nucleic acid binding properties of the HIV-1 nucleocapsid protein zinc fingers. PLoS ONE 2014, 9, e102150. [Google Scholar] [CrossRef]
- Mizuno, A.; Ido, E.; Goto, T.; Kuwata, T.; Nakai, M.; Hayami, M. Mutational analysis of two zinc finger motifs in HIV type 1 nucleocapsid proteins: Effects on proteolytic processing of Gag precursors and particle formation. AIDS Res. Hum. Retrovir. 1996, 12, 793–800. [Google Scholar] [CrossRef]
- Tanchou, V.; Decimo, D.; Pechoux, C.; Lener, D.; Rogemond, V.; Berthoux, L.; Ottmann, M.; Darlix, J.L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 1998, 72, 4442–4447. [Google Scholar] [CrossRef]
- Grigorov, B.; Decimo, D.; Smagulova, F.; Pechoux, C.; Mougel, M.; Muriaux, D.; Darlix, J.L. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 2007, 4, 54. [Google Scholar] [CrossRef]
- El Meshri, S.E.; Dujardin, D.; Godet, J.; Richert, L.; Boudier, C.; Darlix, J.L.; Didier, P.; Mely, Y.; de Rocquigny, H. Role of the nucleocapsid domain in HIV-1 Gag oligomerization and trafficking to the plasma membrane: A fluorescence lifetime imaging microscopy investigation. J. Mol. Biol. 2015, 427 Pt B, 1480–1494. [Google Scholar] [CrossRef]
- de Rocquigny, H.; El Meshri, S.E.; Richert, L.; Didier, P.; Darlix, J.L.; Mely, Y. Role of the nucleocapsid region in HIV-1 Gag assembly as investigated by quantitative fluorescence-based microscopy. Virus Res. 2014, 193, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.; Ehrlich, L.S.; Watanabe, S.; Khan, M.; Strub, M.P.; Luan, C.H.; Powell, M.D.; Leis, J.; Tjandra, N.; Carter, C.A. Tsg101 chaperone function revealed by HIV-1 assembly inhibitors. Nat. Commun. 2017, 8, 1391. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.M.; Ehrlich, L.S.; Strickland, M.; Li, X.; Soloveva, V.; Goff, A.J.; Stauft, C.B.; Bhaduri-McIntosh, S.; Tjandra, N.; Carter, C. Selective Targeting of Virus Replication by Proton Pump Inhibitors. Sci. Rep. 2020, 10, 4003. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. Nmrpipe-a Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, P.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Pervushin, K.; Riek, R.; Wider, G.; Wuthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 1997, 94, 12366–12371. [Google Scholar] [CrossRef]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc 2013, 73, 1–16. [Google Scholar] [CrossRef]
- Goff, A.; Ehrlich, L.S.; Cohen, S.N.; Carter, C.A. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 2003, 77, 9173–9182. [Google Scholar] [CrossRef]
- Sette, P.; Jadwin, J.A.; Dussupt, V.; Bello, N.F.; Bouamr, F. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J. Virol. 2010, 84, 8181–8192. [Google Scholar] [CrossRef]
- Sette, P.; O’Connor, S.K.; Yerramilli, V.S.; Dussupt, V.; Nagashima, K.; Chutiraka, K.; Lingappa, J.; Scarlata, S.; Bouamr, F. HIV-1 Nucleocapsid Mimics the Membrane Adaptor Syntenin PDZ to Gain Access to ESCRTs and Promote Virus Budding. Cell Host Microbe 2016, 19, 336–348. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Alam, S.L.; Rich, R.L.; Myszka, D.G.; Davis, D.R.; Sundquist, W.I. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002, 21, 2397–2406. [Google Scholar] [CrossRef] [PubMed]
- Lambert, N.; Robertson, A.; Jangi, M.; McGeary, S.; Sharp, P.A.; Burge, C.B. RNA Bind-n-Seq: Quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell 2014, 54, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Alam, S.L.; Davis, D.R.; Sundquist, W.I. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 2002, 9, 812–817. [Google Scholar] [CrossRef]
- Teo, H.; Veprintsev, D.B.; Williams, R.L. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J. Biol. Chem. 2004, 279, 28689–28696. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Schubert, H.L.; Kelly, B.N.; Hill, G.C.; Holton, J.M.; Hill, C.P. Ubiquitin recognition by the human TSG101 protein. Mol. Cell 2004, 13, 783–789. [Google Scholar] [CrossRef]
- Feng, G.H.; Lih, C.J.; Cohen, S.N. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res. 2000, 60, 1736–1741. [Google Scholar]
- Bishop, N.; Woodman, P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 2001, 276, 11735–11742. [Google Scholar] [CrossRef]
- McDonald, B.; Martin-Serrano, J. Regulation of Tsg101 expression by the steadiness box: A role of Tsg101-associated ligase. Mol. Biol. Cell 2008, 19, 754–763. [Google Scholar] [CrossRef]
- Stuchell, M.D.; Garrus, J.E.; Muller, B.; Stray, K.M.; Ghaffarian, S.; McKinnon, R.; Krausslich, H.G.; Morham, S.G.; Sundquist, W.I. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 2004, 279, 36059–36071. [Google Scholar] [CrossRef]
- Ono, A.; Ablan, S.D.; Lockett, S.J.; Nagashima, K.; Freed, E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA 2004, 101, 14889–14894. [Google Scholar] [CrossRef] [PubMed]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef] [PubMed]
- Vlach, J.; Eastep, G.N.; Ghanam, R.H.; Watanabe, S.M.; Carter, C.A.; Saad, J.S. Structural basis for targeting avian sarcoma virus Gag polyprotein to the plasma membrane for virus assembly. J. Biol. Chem. 2018, 293, 18828–18840. [Google Scholar] [CrossRef]
- Todd, G.C.; Duchon, A.; Inlora, J.; Olson, E.D.; Musier-Forsyth, K.; Ono, A. Inhibition of HIV-1 Gag-membrane interactions by specific RNAs. RNA 2017, 23, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Saad, J.S.; Muriaux, D.M. Editorial: Role of lipids in virus assembly. Front. Microbiol. 2015, 6, 410. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.; Setz, C.; Wild, J.; Schubert, U. The PTAP sequence within the p6 domain of human immunodeficiency virus type 1 Gag regulates its ubiquitination and MHC class I antigen presentation. J. Immunol. 2011, 186, 5706–5718. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef]
- Amit, I.; Yakir, L.; Katz, M.; Zwang, Y.; Marmor, M.D.; Citri, A.; Shtiegman, K.; Alroy, I.; Tuvia, S.; Reiss, Y.; et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004, 18, 1737–1752. [Google Scholar] [CrossRef]
- Sette, P.; Nagashima, K.; Piper, R.C.; Bouamr, F. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 2013, 10, 79. [Google Scholar] [CrossRef]
- Ciechanover, A.; Wolin, S.L.; Steitz, J.A.; Lodish, H.F. Transfer RNA is an essential component of the ubiquitin- and ATP-dependent proteolytic system. Proc. Natl. Acad. Sci. USA 1985, 82, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Ferber, S.; Ciechanover, A. Transfer RNA is required for conjugation of ubiquitin to selective substrates of the ubiquitin- and ATP-dependent proteolytic system. J. Biol. Chem. 1986, 261, 3128–3134. [Google Scholar] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, S.M.; Strickland, M.; Tjandra, N.; Carter, C.A. RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane. Viruses 2020, 12, 447. https://doi.org/10.3390/v12040447
Watanabe SM, Strickland M, Tjandra N, Carter CA. RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane. Viruses. 2020; 12(4):447. https://doi.org/10.3390/v12040447
Chicago/Turabian StyleWatanabe, Susan M., Madeleine Strickland, Nico Tjandra, and Carol A. Carter. 2020. "RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane" Viruses 12, no. 4: 447. https://doi.org/10.3390/v12040447
APA StyleWatanabe, S. M., Strickland, M., Tjandra, N., & Carter, C. A. (2020). RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane. Viruses, 12(4), 447. https://doi.org/10.3390/v12040447



