Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Phages, and Culture Conditions
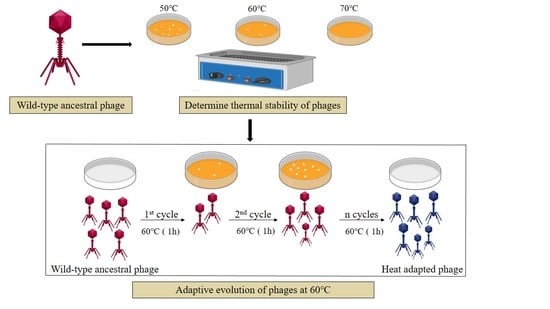
2.2. Phage Adaptive Evolution
2.3. Stability of the Ancestral and Heat-Adapted Phages
2.4. Determination of the Host Range and Lytic Curve Pattern of the Phages
2.5. Genome Extraction, Sequencing, and Analysis
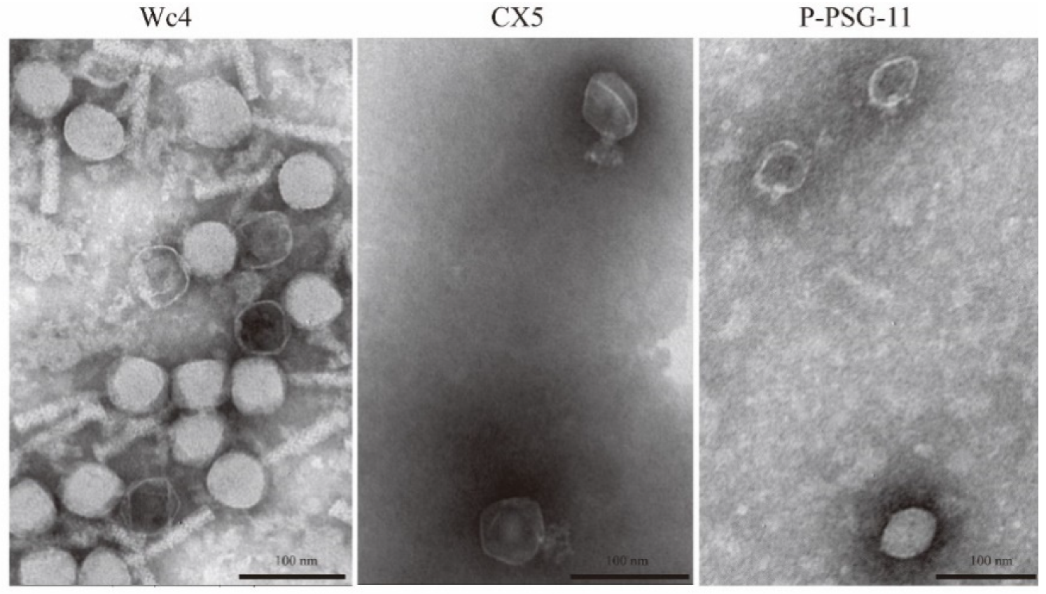
2.6. Electron Microscopy
3. Results
3.1. Thermal Stability of the Phages
3.2. Phage Evolution
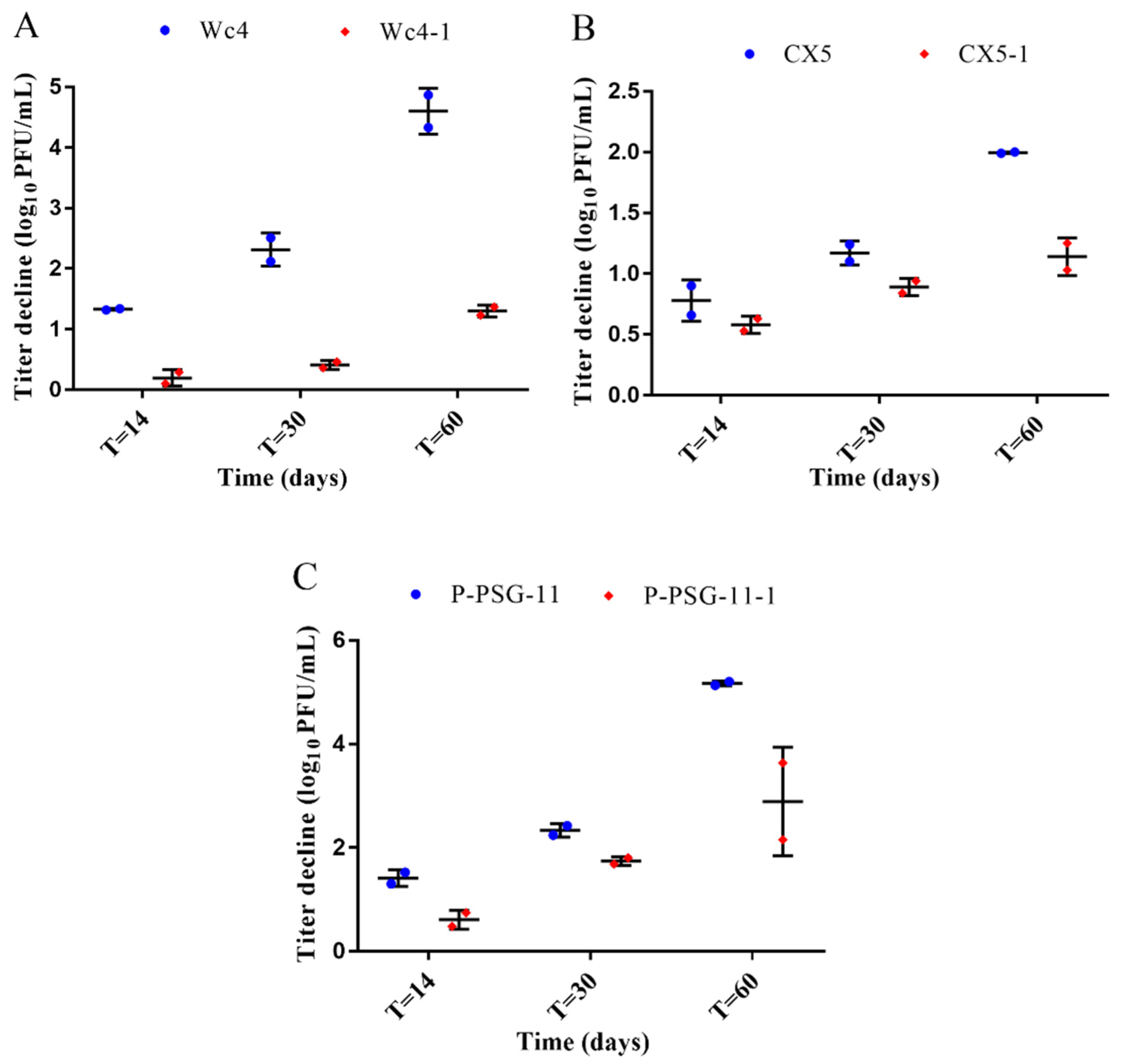
3.3. Stability of the Ancestral and Adapted Phages at 60 °C
3.4. Stability of the Ancestral and Adapted Phages at 37 °C
3.5. Determination of Infectivity and Lytic Curve Patterns
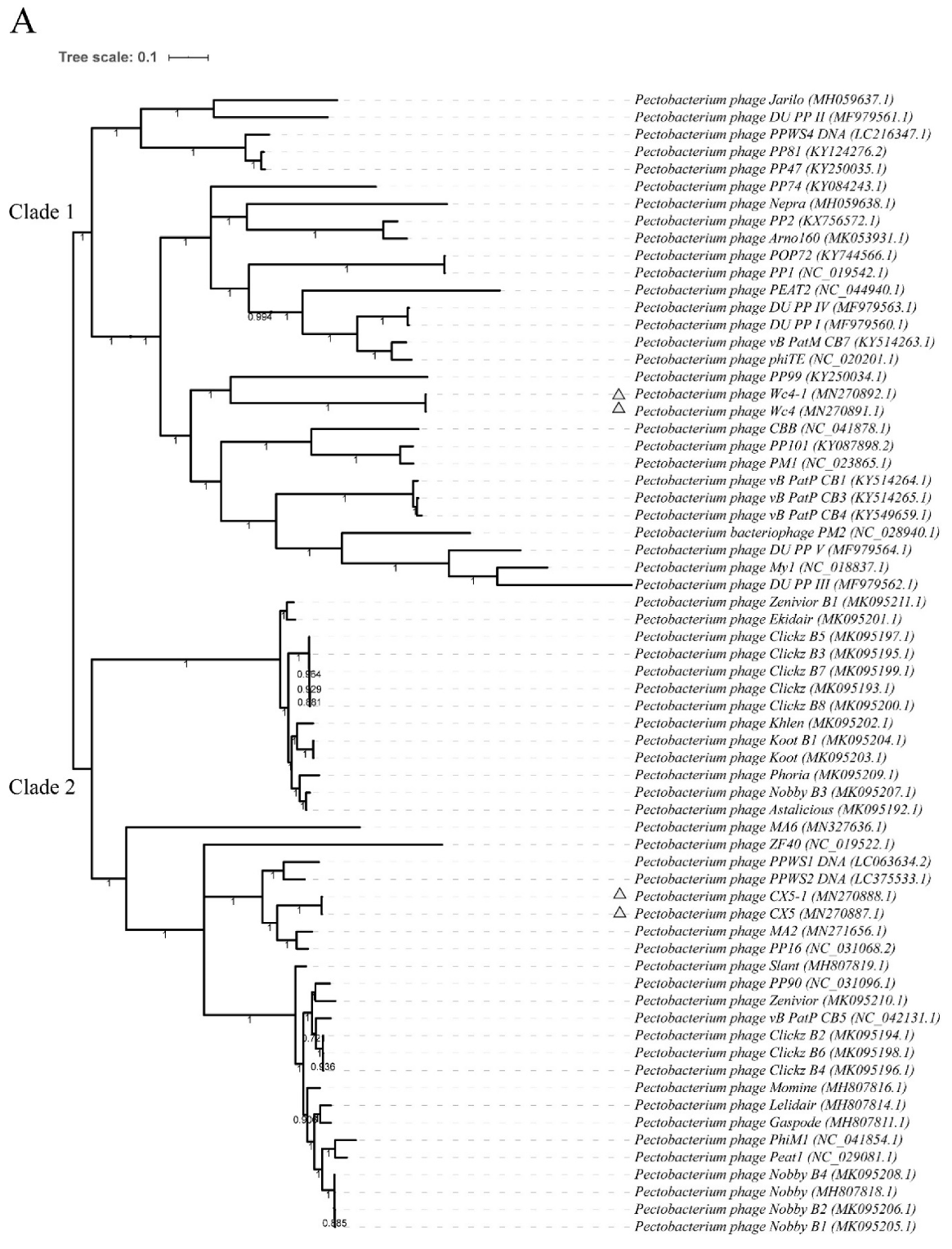
3.6. Genomic Sequence Analysis
3.7. Genomic Variation in the Ancestral Phages and Adapted Phages
3.8. Stability of the Mutations during Growth under Non-Selective Conditions
3.9. Morphology of the Phages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [PubMed]
- Hatfull, G.F. Dark matter of the biosphere: The amazing world of bacteriophage diversity. J. Virol. 2015, 89, 8107. [Google Scholar] [PubMed]
- Navarro, F.; Muniesa, M. Phages in the human body. Front. Microbiol. 2017, 8, 566. [Google Scholar] [PubMed]
- Letarov, A.; Kulikov, E. The bacteriophages in human- and animal body-associated microbial communities. J. Appl. Microbiol. 2009, 107, 1–13. [Google Scholar]
- Muniesa, M.; Imamovic, L.; Jofre, J. Bacteriophages and genetic mobilization in sewage and faecally polluted environments. Microb. Biotechnol. 2011, 4, 725–734. [Google Scholar]
- Suttle, C.A. Marine viruses — major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar]
- Ashelford, K.E.; Day, M.J.; Fry, J.C. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 2003, 69, 285. [Google Scholar]
- Prigent, M.; Leroy, M.; Confalonieri, F.; Dutertre, M.; DuBow, M.S. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the sahara desert. Extremophiles 2005, 9, 289–296. [Google Scholar]
- Prestel, E.; Salamitou, S.; DuBow, M.S. An examination of the bacteriophages and bacteria of the namib desert. J. Microbiol 2008, 46, 364–372. [Google Scholar]
- Zablocki, O.; Adriaenssens, E.M.; Cowan, D. Diversity and ecology of viruses in hyperarid desert soils. Appl. Environ. Microbiol. 2016, 82, 770. [Google Scholar]
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2019, 53, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- LeLièvre, V.; Besnard, A.; Schlusselhuber, M.; Desmasures, N.; Dalmasso, M. Phages for biocontrol in foods: What opportunities for salmonella sp. Control along the dairy food chain? Food Microbiol. 2019, 78, 89–98. [Google Scholar] [CrossRef] [PubMed]
- D’Accolti, M.; Soffritti, I.; Piffanelli, M.; Bisi, M.; Mazzacane, S.; Caselli, E. Efficient removal of hospital pathogens from hard surfaces by a combined use of bacteriophages and probiotics: Potential as sanitizing agents. Infect. Drug Resist. 2018, 11, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.M.; Gorman, S.P.; Donnelly, R.F.; Gilmore, B.F. Recent advances in bacteriophage therapy: How delivery routes, formulation, concentration and timing influence the success of phage therapy. J. Pharm Pharm. 2011, 63, 1253–1264. [Google Scholar] [CrossRef]
- Vandenheuvel, D.; Lavigne, R.; Brussow, H. Bacteriophage therapy: Advances in formulation strategies and human clinical trials. Annu. Rev. Virol. 2015, 2, 599–618. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, X.; Zhang, H.; Watts, A.B.; Ghosh, D. Manufacturing and ambient stability of shelf freeze dried bacteriophage powder formulations. Int. J. Pharm. 2018, 542, 1–7. [Google Scholar] [CrossRef]
- Chang, R.Y.; Wong, J.; Mathai, A.; Morales, S.; Kutter, E.; Britton, W.; Li, J.; Chan, H.K. Production of highly stable spray dried phage formulations for treatment of pseudomonas aeruginosa lung infection. Eur. J. Pharm. Biopharm. 2017, 121, 1–13. [Google Scholar] [CrossRef]
- Merabishvili, M.; Vervaet, C.; Pirnay, J.P.; De Vos, D.; Verbeken, G.; Mast, J.; Chanishvili, N.; Vaneechoutte, M. Stability of staphylococcus aureus phage isp after freeze-drying (lyophilization). PLoS ONE 2013, 8, e68797. [Google Scholar] [CrossRef]
- Leung, S.S.Y.; Parumasivam, T.; Nguyen, A.; Gengenbach, T.; Carter, E.A.; Carrigy, N.B.; Wang, H.; Vehring, R.; Finlay, W.H.; Morales, S.; et al. Effect of storage temperature on the stability of spray dried bacteriophage powders. Eur. J. Pharm. Biopharm. 2018, 127, 213–222. [Google Scholar] [CrossRef]
- Leung, S.S.; Parumasivam, T.; Gao, F.G.; Carrigy, N.B.; Vehring, R.; Finlay, W.H.; Morales, S.; Britton, W.J.; Kutter, E.; Chan, H.K. Production of inhalation phage powders using spray freeze drying and spray drying techniques for treatment of respiratory infections. Pharm. Res. 2016, 33, 1486–1496. [Google Scholar] [CrossRef] [PubMed]
- Jonczyk, E.; Klak, M.; Miedzybrodzki, R.; Gorski, A. The influence of external factors on bacteriophages--review. Folia Microbiol. (Praha) 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, E.; Arribas, M.; Cabanillas, L.; Román, I.; Acosta, E. Evolutionary adaptation of an rna bacteriophage to the simultaneous increase in the within-host and extracellular temperatures. Sci. Rep. 2018, 8, 8080. [Google Scholar] [CrossRef]
- Kashiwagi, A.; Kadoya, T.; Kumasaka, N.; Kumagai, T.; Tsushima, F.S.; Yomo, T. Influence of adaptive mutations, from thermal adaptation experiments, on the infection cycle of rna bacteriophage qbeta. Arch. Virol. 2018, 163, 2655–2662. [Google Scholar] [CrossRef]
- Kashiwagi, A.; Sugawara, R.; Sano Tsushima, F.; Kumagai, T.; Yomo, T. Contribution of silent mutations to thermal adaptation of rna bacteriophage qbeta. J. Virol. 2014, 88, 11459–11468. [Google Scholar] [CrossRef]
- Cox, J.; Schubert, A.M.; Travisano, M.; Putonti, C. Adaptive evolution and inherent tolerance to extreme thermal environments. BMC Evol. Biol. 2010, 10, 75. [Google Scholar] [CrossRef]
- Holder, K.K.; Bull, J.J. Profiles of adaptation in two similar viruses. Genetics 2001, 159, 1393–1404. [Google Scholar]
- Tom, E.F.; Molineux, I.J.; Paff, M.L.; Bull, J.J. Experimental evolution of uv resistance in a phage. Peer J. 2018, 6, e5190. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef]
- Czajkowski, R.; Pérombelon, M.; Jafra, S.; Lojkowska, E.; Potrykus, M.; van der Wolf, J.; Sledz, W. Detection, identification and differentiation of pectobacterium and dickeya species causing potato blackleg and tuber soft rot: A review. Ann. Appl. Biol. 2015, 166, 18–38. [Google Scholar] [CrossRef]
- Wei, C.; Liu, J.; Maina, A.N.; Mwaura, F.B.; Yu, J.; Yan, C.; Zhang, R.; Wei, H. Developing a bacteriophage cocktail for biocontrol of potato bacterial wilt. Virol. Sin. 2017, 32, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Muturi, P.; Yu, J.; Maina, A.N.; Kariuki, S.; Mwaura, F.B.; Wei, H. Bacteriophages isolated in china for the control of pectobacterium carotovorum causing potato soft rot in kenya. Virol. Sin. 2019, 34, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; Hendrix, H.; Lucid, A.; Neve, H.; Noben, J.P.; Franz, C.; O’Mahony, J.; Lavigne, R.; Coffey, A. Novel n4-like bacteriophages of pectobacterium atrosepticum. Pharmaceuticals 2018, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. Megahit: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Kasif, S.; Fleischmann, R.D.; Peterson, J.; White, O.; Salzberg, S.L. Alignment of whole genomes. Nucleic Acids Res. 1999, 27, 2369–2376. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Tavares, P.; Petit, M.A.; Guerois, R.; Zinn-Justin, S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genom. 2014, 15, 1027. [Google Scholar] [CrossRef]
- Ahmad, A.A.; Elhalag, K.M.; Addy, H.S.; Nasr-Eldin, M.A.; Hussien, A.S.; Huang, Q. Sequencing, genome analysis and host range of a novel ralstonia phage, rsop1egy, isolated in egypt. Arch. Virol. 2018, 163, 2271–2274. [Google Scholar] [CrossRef]
- González-Menéndez, E.; Fernández, L.; Gutiérrez, D.; Rodríguez, A.; Martínez, B.; García, P. Comparative analysis of different preservation techniques for the storage of staphylococcus phages aimed for the industrial development of phage-based antimicrobial products. PLoS ONE 2018, 13, e0205728. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by pseudomonas aeruginosa (phagoburn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Gonzalez-Garcia, V.A.; Bocanegra, R.; Pulido-Cid, M.; Martin-Benito, J.; Cuervo, A.; Carrascosa, J.L. Characterization of the initial steps in the t7 DNA ejection process. Bacteriophage 2015, 5, e1056904. [Google Scholar] [CrossRef]
- Kemp, P.; Garcia, L.R.; Molineux, I.J. Changes in bacteriophage t7 virion structure at the initiation of infection. Virology 2005, 340, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Garcia, V.A.; Pulido-Cid, M.; Garcia-Doval, C.; Bocanegra, R.; van Raaij, M.J.; Martin-Benito, J.; Cuervo, A.; Carrascosa, J.L. Conformational changes leading to t7 DNA delivery upon interaction with the bacterial receptor. J. Biol. Chem. 2015, 290, 10038–10044. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.; Chagoyen, M.; Pulido-Cid, M.; Camacho, A.; Carrascosa, J.L. Structural characterization of t7 tail machinery reveals a conserved tubular structure among other podoviridae family members and suggests a common mechanism for DNA delivery. Bacteriophage 2013, 3, e27011. [Google Scholar] [CrossRef]


| Bacteria | Phage | Source | Area and Year of Phage Isolation |
|---|---|---|---|
| Pectobacterium carotovorum subsp carotovorum (Pcc) strain KPM17 | Wc4 [32] | Soil | Wuhan, China (2016) |
| Pectobacterium atrosepticum (Pba) strain WHG10001 | CX5 | Fresh Water | Wuhan, China (2017) |
| Ralstonia solanacearum strain GIM1.74. | P-PSG-11 [31] | Fresh Water | Migori, Kenya (2014) |
| Adapted Phage | Genome Size (bp) | Genome Position | Protein Encoded by the Mutant Gene | Protein Length (aa) | Amino Acid Change and Position |
|---|---|---|---|---|---|
| Wc4-1 | 92,039 | 56238 | Putative protein | 261 | Asp → Tyr (96) |
| CX5-1 | 43,885 | 17228 | Tail tubular protein (gp12) | 776 | Tyr → Cys (555) |
| P-PSG-11-1 | 40,313 | 7037 | Tail tubular protein (gp11) | 195 | Val → Ala (81) |
| Phage | Nucleotide | Position in Genome | Percentage or Average Percentage of Reads Mapped to the Nucleotides (% of All Reads) | |||
|---|---|---|---|---|---|---|
| A | T | C | G | |||
| Wc4 | C | 56238 | 0.21 | 0 | 99.78 | 0.01 |
| All positions | 0.29 | 0.01 | 99.68 | 0.02 | ||
| Wc4-1 | A | 56238 | 99.61 | 0.05 | 0.33 | 0.01 |
| All positions | 99.69 | 0.05 | 0.25 | 0.01 | ||
| CX5 | A | 17228 | 98.61 | 0.15 | 0.93 | 0.31 |
| All positions | 99.71 | 0.10 | 0.18 | 0.01 | ||
| CX5-1 | G | 17228 | 0.01 | 0.15 | 0.02 | 99.82 |
| All positions | 0.01 | 0.16 | 0.02 | 99.81 | ||
| P-PSG-11 | T | 7037 | 0.13 | 99.74 | 0.01 | 0.12 |
| All positions | 0.09 | 99.77 | 0.01 | 0.13 | ||
| P-PSG-11-1 | C | 7037 | 0.12 | 0.01 | 99.87 | 0 |
| All positions | 0.14 | 0.01 | 99.83 | 0.02 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage. Viruses 2020, 12, 423. https://doi.org/10.3390/v12040423
Kering KK, Zhang X, Nyaruaba R, Yu J, Wei H. Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage. Viruses. 2020; 12(4):423. https://doi.org/10.3390/v12040423
Chicago/Turabian StyleKering, Kelvin K., Xiaoxu Zhang, Raphael Nyaruaba, Junping Yu, and Hongping Wei. 2020. "Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage" Viruses 12, no. 4: 423. https://doi.org/10.3390/v12040423
APA StyleKering, K. K., Zhang, X., Nyaruaba, R., Yu, J., & Wei, H. (2020). Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage. Viruses, 12(4), 423. https://doi.org/10.3390/v12040423