1. Introduction
Bacterial biofilm is a complex and dynamically changing structure responding to the external environment. Microbial cell aggregates are surrounded by complex polymer substances (matrix) composed of proteins, polysaccharides, eDNA (extracellular DNA), glycolipids and fatty acids [
1]. Bacteria inside the biofilm are not synchronized in terms of metabolic activity, and biofilm structure is highly heterogeneous in this respect [
2,
3]. The protection against unfavorable environmental conditions can be implemented by the extracellular polymer structure (EPS) production, increasing significantly the resistance to chemical compounds and the immune system response [
4]. Biofilm is a long-term strategy for bacterial survival under stress conditions. Cells embedded in the consortia cause biomaterials dysfunction, chronic infections, are highly insensitive to treatment and pose a threat in industry or are responsible for biofouling [
5]. The high insensitivity of biofilm to antibiotics forces the search for alternative treatment solutions. Bacteriophages turn out to be a convenient weapon against both planktonic and sessile forms of bacteria [
3,
6,
7]. To evaluate the complex and sublime interactions of antibacterials and biofilm cells it was necessary to develop various measurement techniques.
Currently, there are several methods allowing the characterization of biofilm components. The colony count (CFU/mL) of sessile cells colonizing biofilm structure is commonly used. The crystal violet (CV) assay is used for biomass measurement because the dye is non-specific and interacts with negatively charged moieties of cells and biofilm matrix [
8]. Other staining techniques are more specific showing the dye interaction with different biofilm elements. The commonly used Live/Dead BacLight kit composed of SYTO
® dyes with orange acridine or propidium iodide (Merck Millipore, Burlington, MA, USA), distinguishes live and dead cells [
9]. Resazurin (Alamar Blue), XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) and TTC (2,3,5-triphenyl-2H-tetrazolium chloride) are another group of markers for determining metabolically active cells. Calcofluor white or FITC-labeled lectins are used to stain the glycoconjugate fraction of biofilm [
10]. FITC also interacts with the amino residues of proteins and amino sugars [
11,
12]. For fast determination, it is possible to use Nile Red staining protocol [
10]. These dyes can be detected spectrophotometrically, but also fluorometrically using microplate readers or CLSM microscopy [
13].
Visualizations of biofilm development and its eradication by antibiotics, phages or other combined therapies are an inseparable group of methods used in biofilm research. Microscopy techniques based on transmission electron microscopy (TEM), scanning electron microscopy (SEM), atomic force (AFM) or confocal microscopy are often used [
14,
15]. However, this group of methods is quite expensive and requires the use of sophisticated microscopy tools and material preparation techniques.
There is also an interesting method for biofilm measurement based on interferometry spectroscopy allowing us to determine the changes in the diffusion rate through the biofilm matrix [
3,
7,
16,
17].
Each method used for biofilm monitoring has both advantages and limitations. In most cases, they are based on mandatory material preparation techniques, and do not allow measurements of long-term biofilm growth dynamics under the culture conditions. Monitoring of changes in the biofilm structure (EPS and cells) in real-time is particularly difficult. In this paper, we present the application of the impedance spectroscopy (IS) based on the application of quartz tuning forks (QTFs) as an impedance sensor. We have already published the physical background of QTF system applications in biofilm mass measurements [
15,
18].
In this study, we present our impedance spectroscopy set up for the monitoring of electrical properties of biofilm structure and bacterial culture. Measuring conductance and, we were able to detect the changes in biofilm development as well as in planktonic cells propagation. The application of QTF impedance systems were tested and verified on a Pseudomonas aeruginosa PAO1 culture model treated with Pseudomonas lytic phage LUZ19.
2. Materials and Methods
2.1. Bacterial Strain and Phages
Pseudomonas aeruginosa PAO1 (ATCC 15692) was used as a model of biofilm-forming bacteria. Bacterial cells were stored at −70 °C in Trypticase Soy Broth (TSB, Becton Dickinson and Company, Cockeysville, MD, USA) supplemented with 20% glycerol (U.S. Merck Corporate Headquarters, Kenilworth, NJ, USA). For the experiments, strains were refreshed on Trypticase Soy Agar (TSA, Becton Dickinson and Company, Cockeysville, MD, USA) at 37 °C for 18 h. Pseudomonas LUZ19 podovirus equipped with polysaccharide depolymerase was kindly provided from the collection of the Laboratory of Gene Technology, KU, Leuven, Belgium.
2.2. Bacterial Growth Measured by the Colony Count
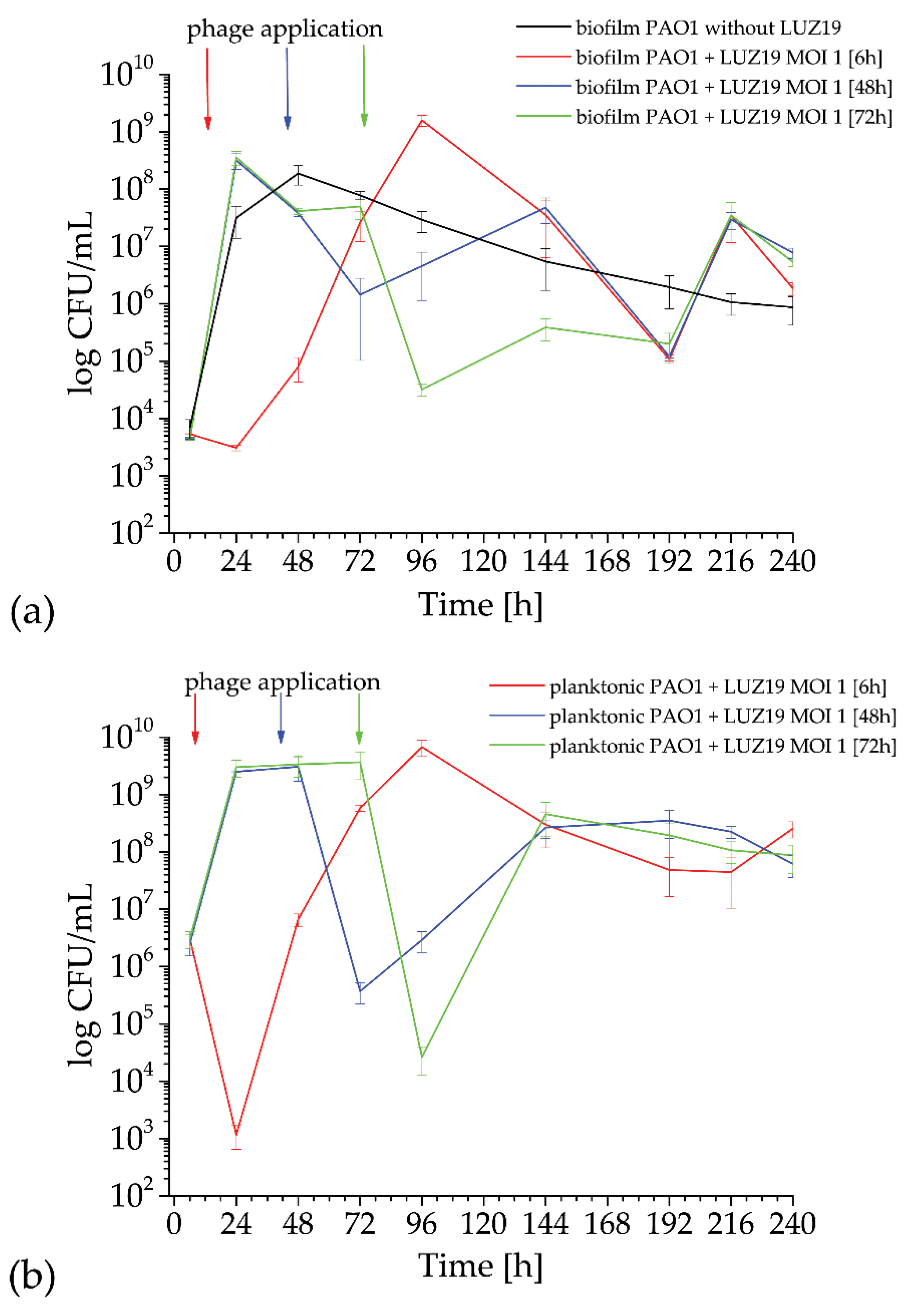
P. aeruginosa PAO1 culture was incubated at 37 °C in 1.5 mL of TSB medium with immersed quartz tuning forks (QTF). The biofilm and planktonic growth dynamics were measured by the classical colony forming units (CFU/mL) at specific time points: at 6, 12, 24, 48, 72, 96, 144, 192, 216 and 240 h of the experiment. QTFs covered with biofilm were transferred to eppendorf tubes containing 150 µL of PS (physiological saline, Avantor Performance Materials, Gliwice, Poland). To liberate cells from the biofilms and to measure the cell counts, biofilm was disintegrated using the ultrasonic cleaning bath USC300TH with 45 kHz frequency and 80 W power for 25 min for all sensors (VWR International Ltd., Lutterworth, Leicestershire, UK), released cells were plated on Trypticase Soy Agar plates (Becton Dickinson and Company, Cockeysville, MD, USA) and the colony count was evaluated after 18 h incubation at 37 °C. The colony count was done for phage treated and untreated samples.
2.3. Phage Treatment
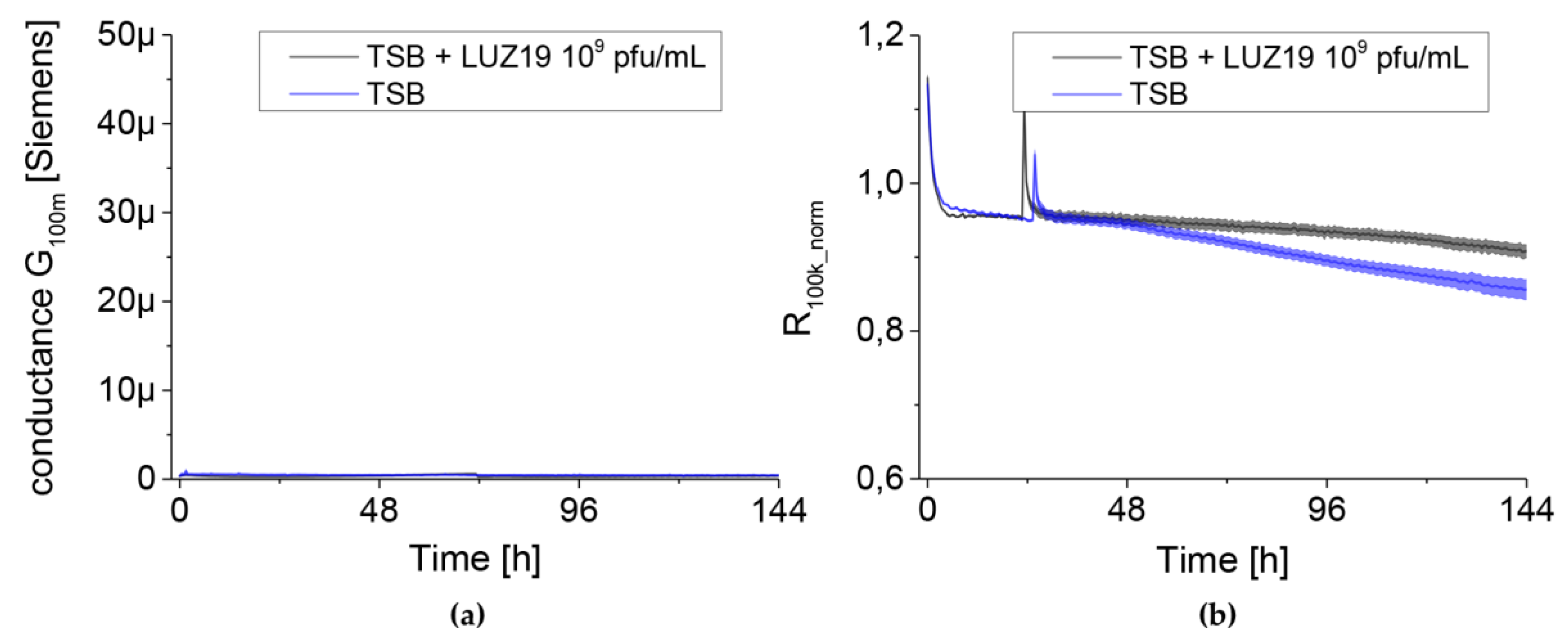
Pseudomonas phage LUZ19 filtrate at 109 pfu/mL (plaque forming units) was diluted to obtain a corresponding MOI 0.1, 1 and 10 used from biofilm treatment at different time points: 6, 48, 72 h. Phages were suspended in TSB medium in 1.5 mL into which sensors with adhered biofilm were transferred. Phage LUZ19 at a concentration of 109 pfu/mL and TSB alone were used as negative controls (no biofilm). All experiments were performed three times in at least 4 technical replicates.
2.4. Bacterial Biofilm Growth Measured by the Impedance Method
As was reported before [
18], the Quartz Tuning Forks (QTFs) may be used as the impedance sensors for biofilm growth monitoring. The measured object may be the impedance sensor which electric impedance change with the changes of the environment in which it is placed.
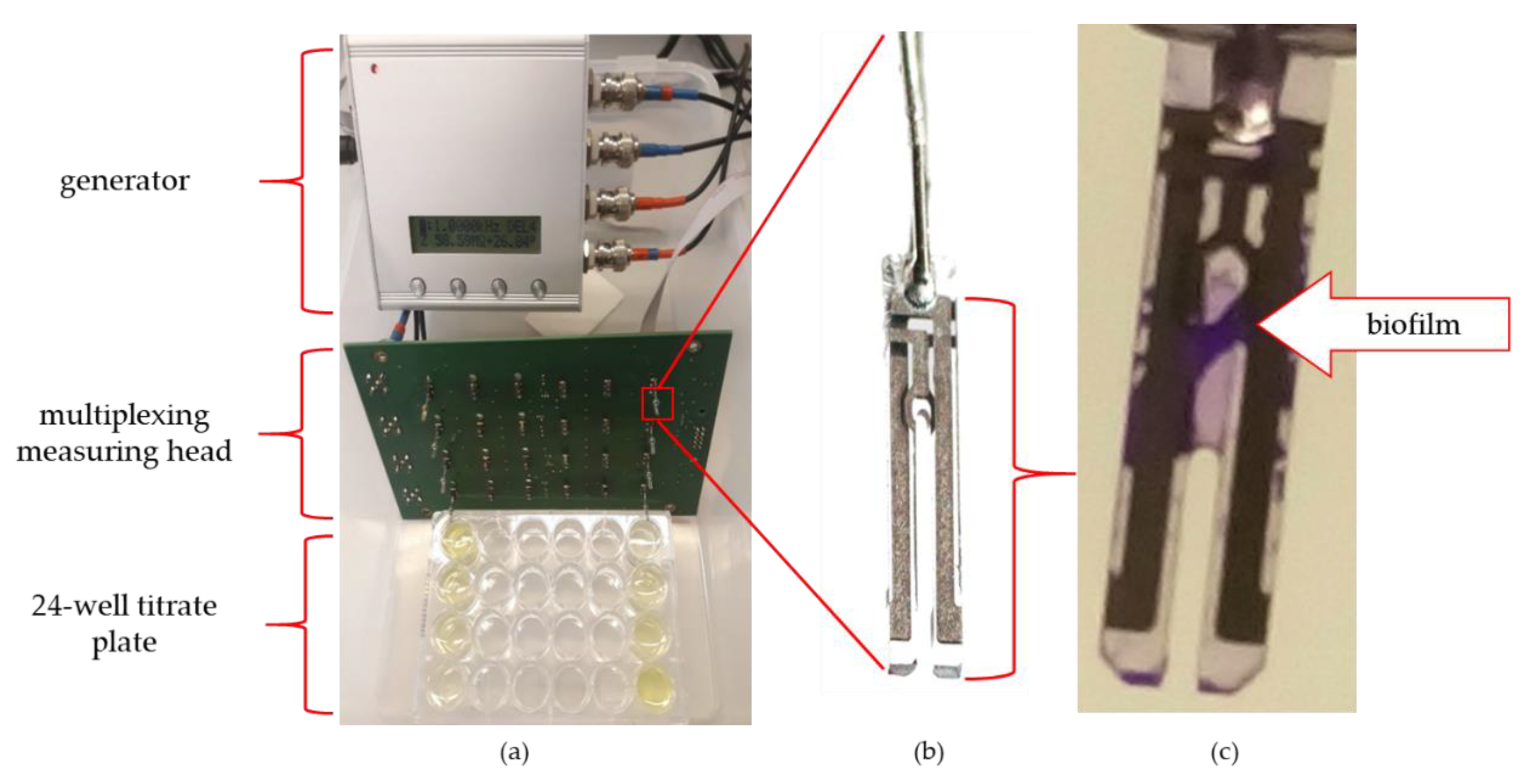
To perform the impedance spectra, the IMP-STM32 impedance analyser was used [
19] connected to a dedicated multiplexing measuring head with sockets for 24 QTF sensors (
Figure 1). Such a setup allowed for the quasi-simultaneous impedance spectra measurements of 24 sequentially switched sensors in the frequency range from 100 mHz to 100 kHz in approximately 30-min intervals.
Prior to the experiments, the measuring head was sterilized with alcohol-based chemicals approximately 24 h before performing the experiments. Immediately before installing the sensors, the system was sterilized in a laminar chamber using ultraviolet for about 10 min. The casings were removed from the QTF before the experiment and the sensors were rinsed with isopropanol (Avantor Performance Materials, Gliwice, Poland), which was evaporated at room temperature. A plate with bacterial cultures was prepared for the ready set of 24 sensors. Refreshed bacterial cultures were diluted in TSB to an OD550 (optical density) equal to 0.2. The culture of 106 CFU/mL was suspended in 1.5 mL in 24 wells culture plate (VWR International, LLC Radnor Corporate Centre, Radnor, PA, USA) and incubated at 37 °C for 240 h. Pseudomonas phages LUZ19 filtrate at MOI 0.1, 1 or 10 was added at 6, 48 and 72 h of incubation. Each experiment was performed in triplicate with four technical repetitions (N = 12).
2.5. Impedance Spectra Analysis
The electrical impedance
expressed in ohms (
) is the complex measure of the object’s electrical response to the alternating voltage excitation, where
is a resistance, X is a reactance and
is the imaginary unit. The technique in which impedance spectra, that is the representation of such responses measured over a wide range of frequencies, is called the impedance spectroscopy [
20]. Results of such a measurement may be also presented as the complex admittance
expressed in siemens (
), where real and imaginary parts are conductance G and susceptance B.
A typical impedance spectra analysis method is fitting them with the impedance of the electric equivalent circuit (EEC) using dedicated software (for example, Scribner ZView that was used in presented research). Two EECs were used (
Figure 2a,b).
These consisted of resistors and constant phase elements (CPE). CPE is the element frequently used in EEC modelling. Its impedance is equal , where Q and T are CPE parameters and f is frequency. Its components relate to: Rs, series resistance which corresponds mainly to the growth medium resistance; CPEdl, electric double layer capacitance between the growth medium and electrodes; Rct, charge transfer resistance; Rb and CPEb, resistance and capacitance of the objects adhered to the electrode surface (mostly cells and biofilm).
Most of the fits were done with primary EEC however, in some experiments, the influence of Rb and CPEb on the measurement spectrum was not measurable. In these cases the simplified EEC gave better-fitting results. The optimal model was chosen based on the fit quality (χ
2) and the fitting error of EEC components. In simplified analyses after EEC modelling, the use of Rs and Qb parameters was proposed. Biologically, the changes in Rs parameter should be understood as an increase in the number of planktonic forms in the medium surrounding the QTF sensor [
18]. The second important parameter of the EEC model is Q
b (electrical conductivity) measured at 100 mHz. The biological equivalent of changes for the conductance (Q
b) is the adhesion of bacterial cells to the sensor surface as well as the formation or degradation of the biofilm matrix. The analysis of the frequency ranges at which the most important EEC components influence the impedance spectrum shape allowed a simplification of the biofilm state assessment. The plot shown in
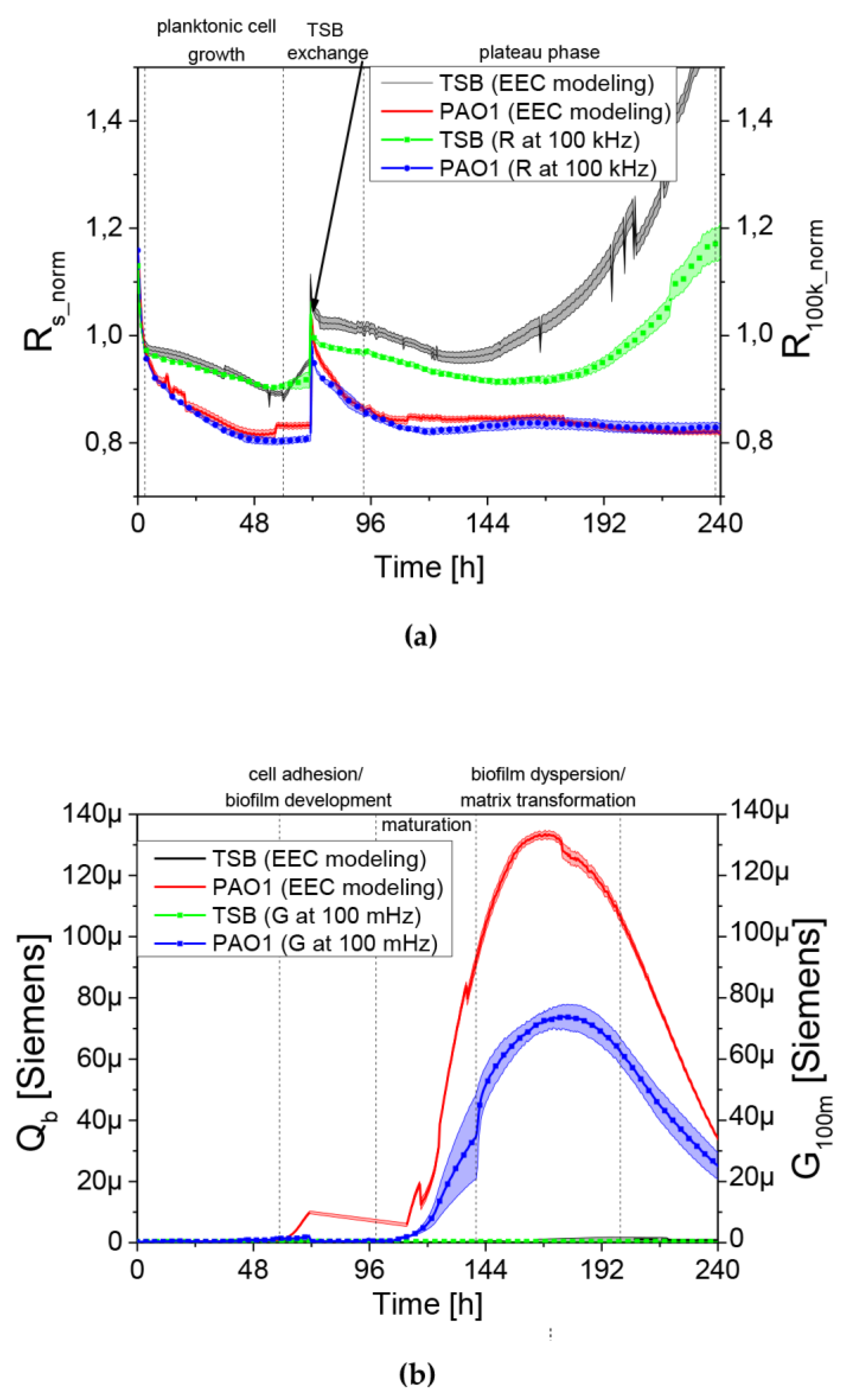
Figure 2d was created by simulating the conductance spectrum of the EEC fitted to one of the preliminary results (dots) and with R
s, R
ct and Q
b changed twofold. As may be observed, a change in R
s altered the high-frequency part of the spectrum while Q
b influences the low-frequency part. Therefore the Rs variations may be estimated by the real part of the impedance measured at 100 kHz. As the initial value of such a parameter varied between sensors it was normalized by its value at the 3rd h of the experiment yielding R
100k_norm presented in the Results section. Similarly, the Q
b variations were estimated by the variations of the conductance (that is the real part of the inverse of the impedance) G
100m measured at 100 mHz. The similarity between parameters obtained using the EEC and simplified analysis is shown in Figure 6a,b.
2.6. Biofilm Monitoring by Scanning Electron Microscopy (SEM)
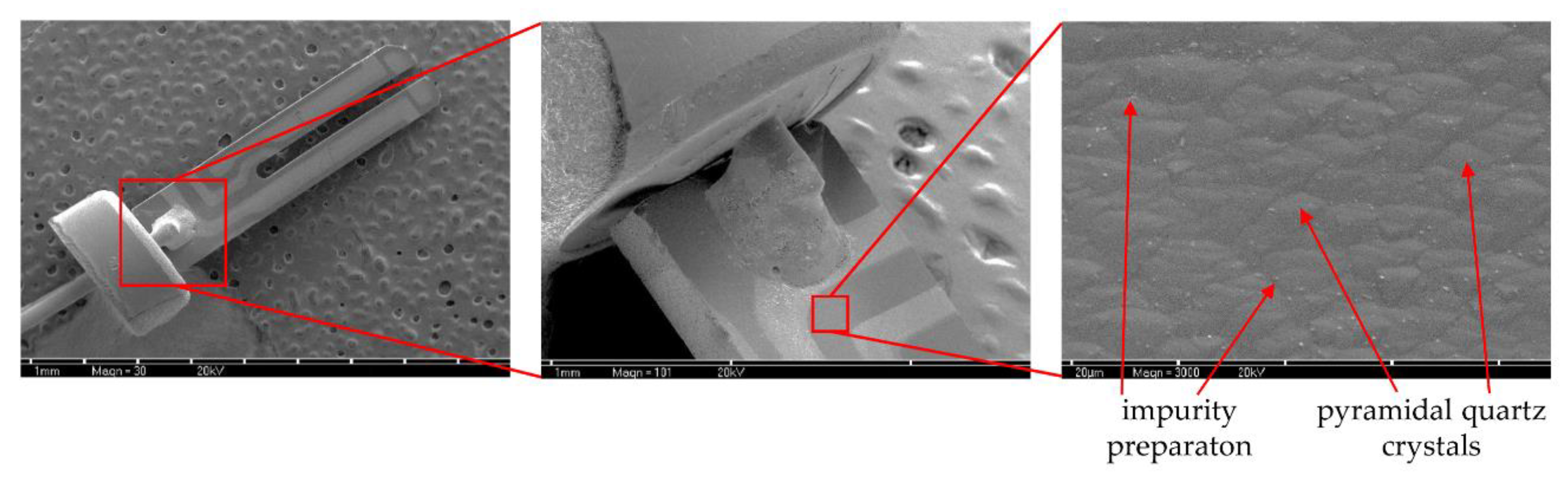
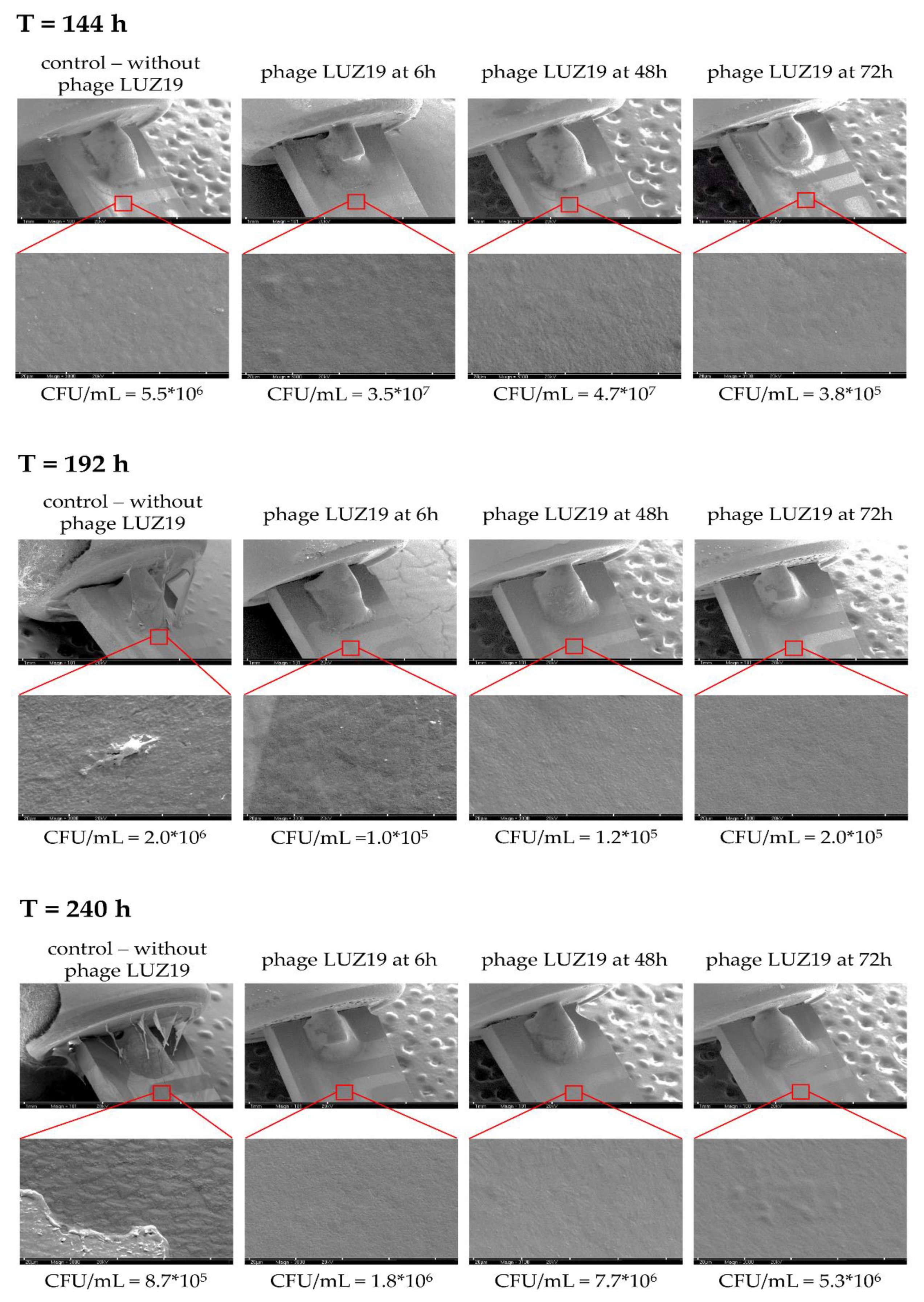
Sensors covered with bacterial biofilm were transferred to eppendorf tubes with PS to remove unadhered cells. Next, the sensors were placed in a series of increasing ethanol concentrations: 60%, 70%, 80%, 96% and the remaining water content was evaporated in vacuum conditions. After 24 h the preparations were transferred to a vacuum evaporator and the silver conductive layer spraying process was carried out. Biofilm presence on QTF surface was visualized by SEM at 30×, 100× and 3000× magnifications (Tesla BS 300, Brno, Czechoslovakia).
2.7. Statistical Analyses
The statistical analysis was performed for three independent experiments and four technical repetitions (N = 12) using a one-way ANOVA. To compare the differences between variances, Levene’s statistical method was applied. Results were considered significant at p < 0.05 value.
4. Discussion
Our previous experience with the application QTF sensor platform regarded biofilm biomass measurements when
P. aeruginosa sessile cells were treated with antibiotics [
15,
21]. The changes in the resonant frequency of QTFs covered with biofilm structure were measured, which were then converted into the mass of bacterial biofilm at selected culture time points. That technology was limited to particular incubation times and it required complex material preparation procedures. In the presented study, we were looking for real-time measurement techniques remotely monitoring the biofilm formation and degradation. We have shown that the construction and arrangement of electrical covers of QTF allow for its application as an impedance spectroscopy sensor [
18]. The applied impedance spectroscopy (IS) technique using QTF sensors is a convenient technology for biofilm growth monitoring in a real-time manner. IS was commonly used to determine the behavior of materials in chemical systems. It has become a tool for chemical and electrochemical analysis of materials with different levels of conductivity [
21]. It has also found an application in the broadly understood biological sciences. Measurements of electrical parameters of biological objects have been monitored since around 1970, mainly to determine food contamination by bacteria [
22]. Impedance spectroscopy is becoming widely used in experiments with bacterial biofilms as described by the Paredes group or van Duuren team [
23,
24]. Usually, these experiments base their measurements on a commercial instrument, the xCELLigence Real-Time Cell Analyzer (RTCA) from Acea Biosciences that measures impedance in 96 well plates equipped with gold microelectrodes by measuring the so-called cell index for
Pseudomonas and
Streptococcal biofilms [
24,
25]. Our previous experience also showed that IS using QTF is an effective technique for non-invasive analysis of biofilm culture parameters [
18] extending now the aspect of the phage application model. The sensors used are planar and often the electrodes are finger-shaped. QTF sensor design is very convenient for spectra characterization and the creation of an electric equivalent model. In our technology, QTF sensors are immersed in bacterial culture enabling bacterial cells adhesion and an increase of biofilm monitoring. We also use a neutral metallization for tested objects, because QTF is mainly made of quartz and aluminum. The materials that make up the sensor are also relatively inexpensive, which is why the technology we propose is a relatively cheap tool for the monitoring of bacterial biofilm physiology.
The results obtained in our study are presented as simplified analyses measured in the form of conductance and resistance. Due to differences in the quality of QTF sensors, their surface functionalization in the culture medium and stabilization of temperature conditions (heating the TSB medium to 37 °C), the results for series resistance (R
100k) were normalized to three h of experiment and presented as R
100k_norm. Compared to commonly used methods, the undoubted advantage of IS based on QTF is the ability to measure parameters characterizing simultaneously the changes in planktonic forms in the culture medium and the biofilm formed on the sensor surface (parameters R
100k_norm and G
100m, respectively). The dynamics of both parameters resulted from the equivalent electrical circuit (EEC) model we had previously set up and verified. The impedance spectroscopy measurements are highly repeatable at a high resolution. The spectra collection took place for the same sensor every 30 min. This allowed us to obtain reliable results, without the need for complex sample manipulation, as is the case with many recommended biofilm monitoring methods that have so far been described in
Crit Rev Microbiol [
13].
Most of the laboratory methods used to determine biofilm growth rely on the measurements at a given culture point. This applies to methods based on CFU/mL calculation, as well as staining techniques [
26]. Moreover, these experiments are usually limited to several hours of incubation, and in many cases, we are not able to characterize the phenomenon of biofilm dynamic changes. In the case of IS using QTF, the experiments were carried out after up to 10 days of incubation. Our measurements were carried out semi-parallel to each sensor. The built platform allowed 24 independent experiments to be performed simultaneously, while the entire measurement took place in a titration plate in batch culture.
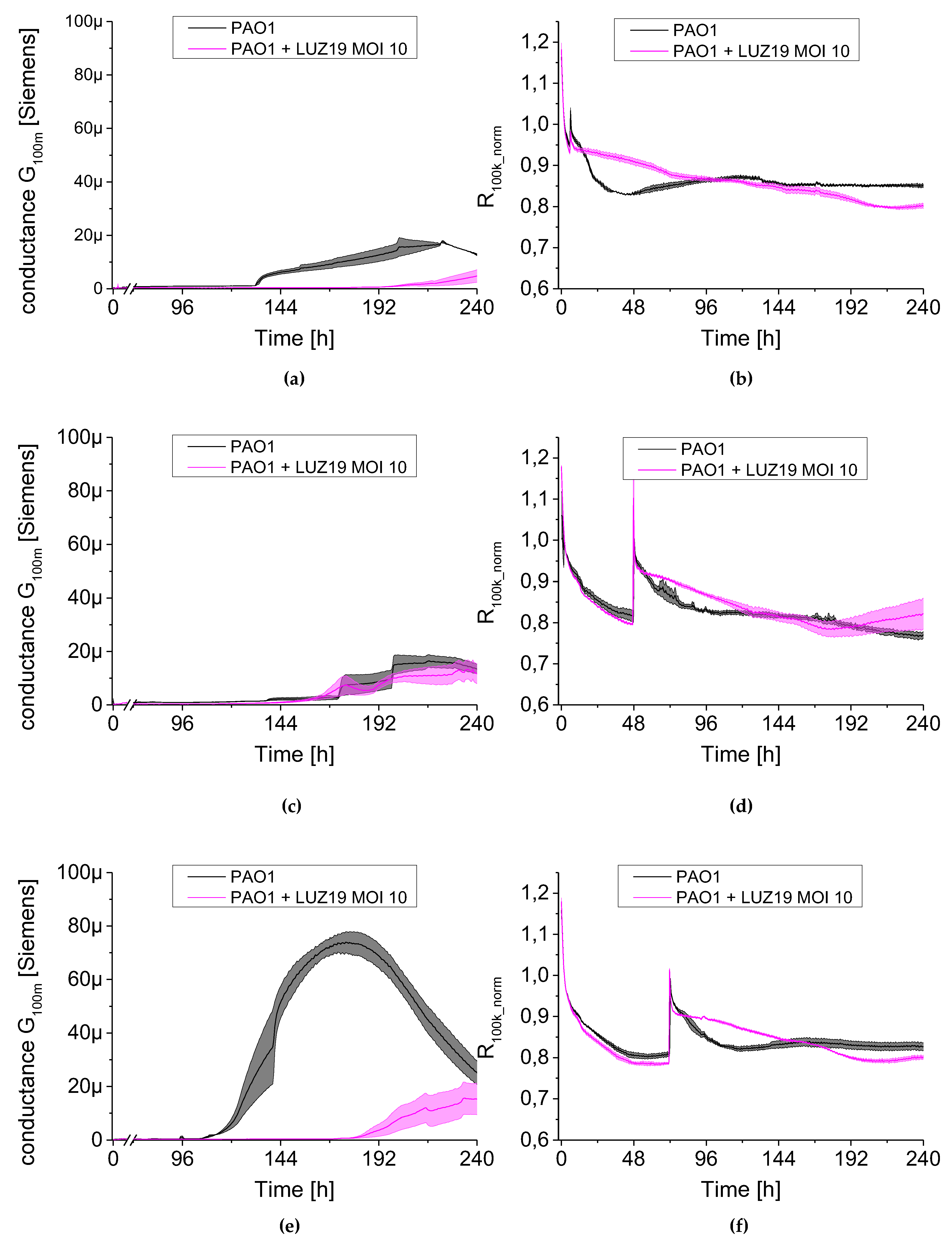
We observed a clear relationship between the series resistance parameter (R
100k_norm) and conductance (G
100m). When R
100k_norm resistance reached the plateau phase, G
100m conductance increased significantly. The lowering resistance is associated with an increase in planktonic cell number in the medium (described earlier in [
18]). The increase in the conductance is identified with the appearance and level of surface coverage by a conductive EPS matrix. Based on the G
100m parameter, we measured the QTF coverage with an EPS matrix that changes the charge transfer across the barrier between the electrode and electrolyte. However, the drop in the conductance parameter for G
100m is associated with a reduction of EPS or a change in the physicochemical properties of the biofilm matrix. The culture was carried out in stationary conditions, without medium exchange, therefore, the conductance reduction scenario was associated with the reuse of matrix components as an energy source of bacterial culture. The SEM visualization of QTF surface on the 6th, 8th and 10th day of biofilm formation combined with the colony count results provide the proof that the above conclusions were correct.
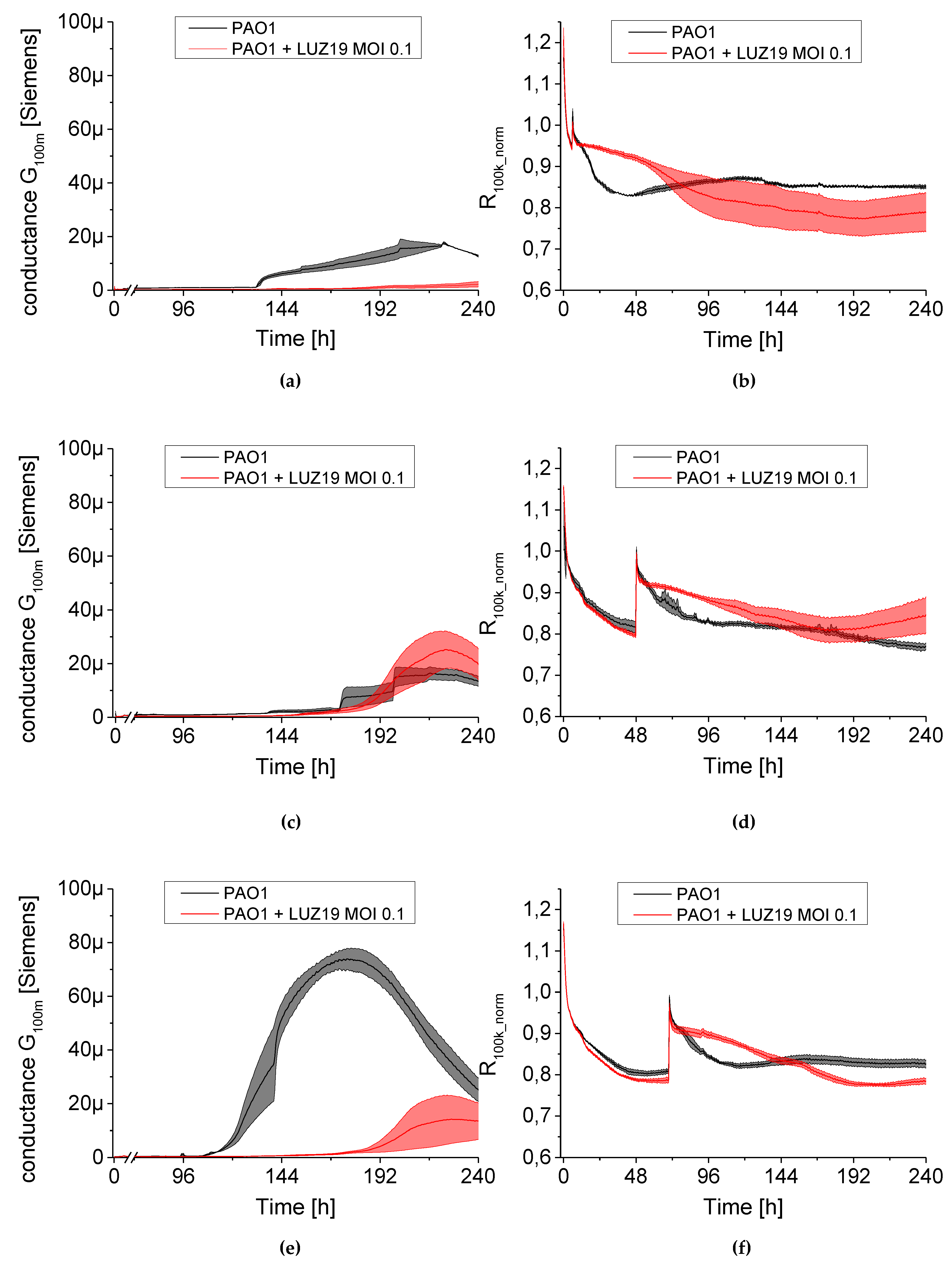
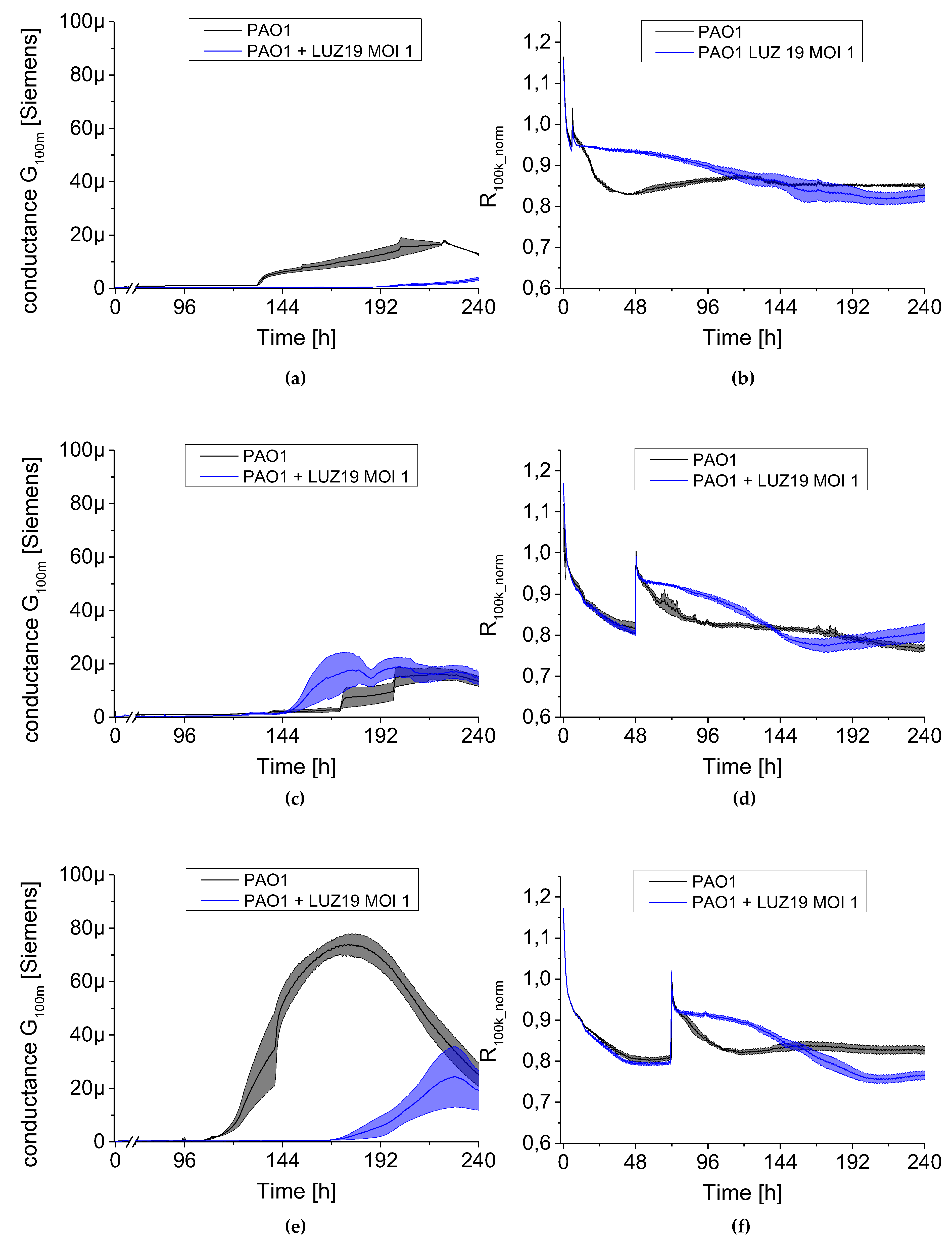
We used a QTF as an impedance sensor to measure electrical changes of PAO1 culture during phage LUZ19 infection at three different MOIs corresponding to 0.1, 1 and 10. An inhibitory effect on biofilm formation was seen when the infection took place at the 6th and 72nd h of incubation, regardless of MOI applied. It has also been shown that generally, the biofilm dynamics were independent of phage LUZ19 infecting dose. Interestingly, 48-h biofilm turned out to be a breakthrough moment, because at this time phage administration did not affect the biofilm formation, although the killing influence on both sessile and planktonic cells were recorded. Comparing the results from three different biofilm monitoring techniques (colony count, SEM, and impedance), we may conclude that G100m parameter shows, in fact, the amount of the biofilm EPS accumulating in the sensor surface.
The impedance spectroscopy model we designed is able to monitor the behavior of planktonic culture surrounding the QTF sensor. The R100k_norm parameter indicating the culture propagation dynamics in the case of phage infection means the inhibition of phage-sensitive cell growth and secondary regrowth events of selected phage resistant bacteria. That population regrowth and dynamic phage–bacteria balance in the biofilm structure was confirmed in the colony count detection.
The application of
Pseudomonas phages in order to eradicate bacterial biofilm is widely described in the literature showing both successes and failures. Some phages like phage E27 or M4, are not able to penetrate the biofilm matrix, thus, they are ineffective in biofilm elimination [
27,
28]. The strong restriction of phage access to cells embedded in the dense biofilm EPS matrix can be partially overcome by virion-associated depolymerases produced by some phages such as LUZ19, tested in the presented study [
29]. Phage infection usually led to a significant cell number reduction (3 logs) on catheter surfaces but the incomplete eradication resulted in a fast population regrowth within the next 24 h [
28]. Because of the presence of phage insensitive portions in the sessile cells, the idea of combined phage-antibiotic treatment was also studied [
30]. The synergy effect of podovirus phage LUZ7 and streptomycin was noticed against
P. aeruginosa PAO1 biofilm compared to a single preparation [
30]. A similar synergy observation was done by the Knezevic group [
31]. Nevertheless, the heterogenic character of the sessile population protects the biofilm from a complete elimination regardless of an applied anti-biofilm agent. The limitation in the biofilm eradication is also caused by rapid emergence of phage-resistant phenotypes [
32,
33].
As phages are co-evolving with its host, there are some contrary mechanisms of phage influence on biofilm formation and maintenance. This specifically concerns prophages having a huge impact on bacterial phenotypes. It is already proven that the remodeling of a biofilm EPS matrix and the development of small-colony phenotypic variants are associated with the presence of filamentous prophage Pf4 (Pf) in
P. aeruginosa genome [
34]. It turns out that Pf participates in the organization of biofilm matrix, while being induced, release and transform into crystal lattices. This modification of matrix improves the adhesion process and increases drying and antibiotics tolerance [
34]. Tobramycin applied with the biofilm treatment accumulates inside liquid crystals of Pf phage particles, whereas a strong packaging of negatively polarized phage polymers interact with positively charged antibiotics [
35]. Pf phage activity is not only limited to the modification of a specific host EPS matrix, but may affect the biofilm growth of distant accompanying organisms (
Aspergillus fumigatus), by direct capture of Fe
3+ ions (and other multivalent metals) [
36,
37]. Above prophage activity is a newly discovered mechanism of restrictive regulation of the biofilms especially in
P. aeruginosa strains associated with cystic fibrosis patient infection.
In conclusion, the technology we presented in this study gives a reproducible and precise measurement of biofilm structure and planktonic culture dynamics analyzed simultaneously and in a real-time manner without specific sample preparation. The impedance parameters such as the conductance and resistance of biological samples give more insight into the bacterial culture changes happening during the natural growth and the fluctuation of culture characteristics when treated with antibacterial agents. Moreover, the impedance parameters turned out to be specific for EPS amount measurement. Therefore this system seems to be very useful in the monitoring of antibacterial effects in biofilm prevention and eradication, and especially for the study on phage influence on biofilm matrix development.