Risk Mapping of Influenza D Virus Occurrence in Ruminants and Swine in Togo Using a Spatial Multicriteria Decision Analysis Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Surveillance Data
2.2. Identification of Risk Factors and Experts Survey
2.3. Spatial Data Collection and Geoprocessing
2.4. Generation of the Final Maps
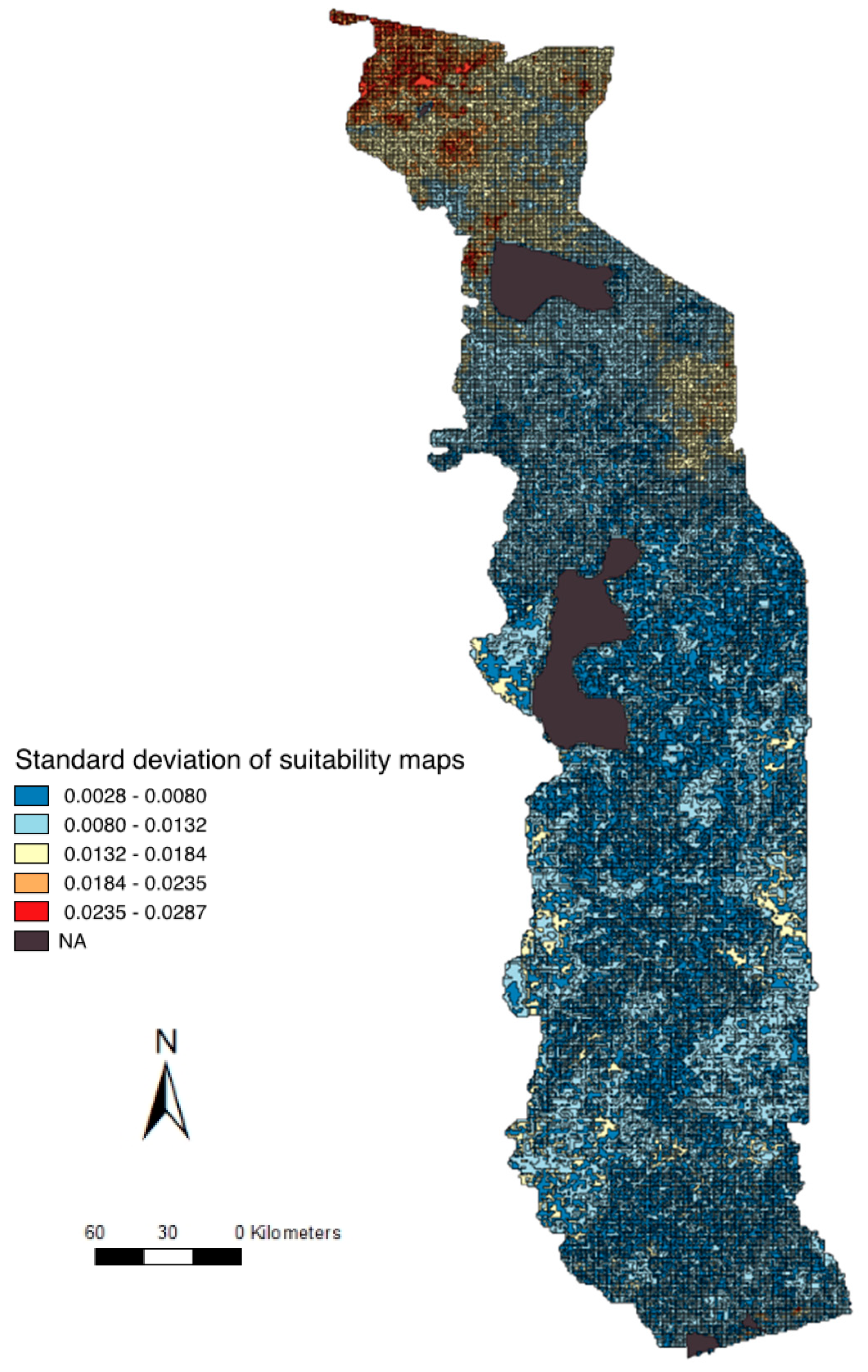
2.5. Uncertainty Analysis and Validation
2.6. Risk Maps Comparison with Serological Results
3. Results
3.1. Surveillance Data
3.2. Risk Mapping
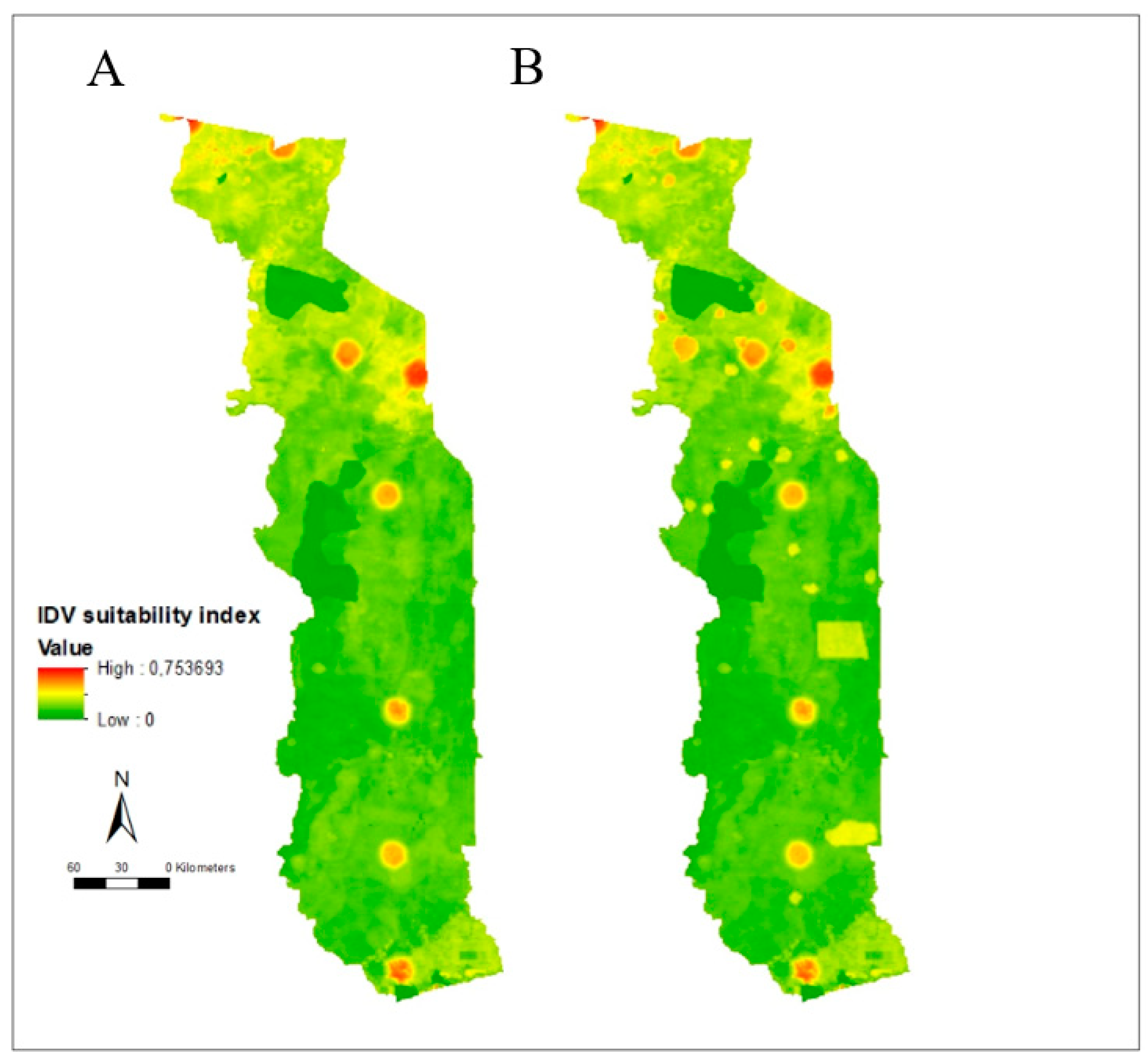
3.3. Suitability Map, Uncertainty Analysis, and Serological Comparison
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a Novel Swine Influenza Virus from Oklahoma in 2011 Which Is Distantly Related to Human Influenza C Viruses. PLOS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef]
- Salem, E.; Cook, E.; Lbacha, H.; Oliva, J.; Awoume, F.; Aplogan, G.; Couacy Hymann, E.; Muloi, D.; Deem, S.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017, 23, 1556–1559. [Google Scholar] [CrossRef]
- Quast, M.; Sreenivasan, C.; Sexton, G.; Nedland, H.; Singrey, A.; Fawcett, L.; Miller, G.; Lauer, D.; Voss, S.; Pollock, S.; et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015, 180, 281–285. [Google Scholar] [CrossRef]
- Nedland, H.; Wollman, J.; Sreenivasan, C.; Quast, M.; Singrey, A.; Fawcett, L.; Christopher-Hennings, J.; Nelson, E.; Kaushik, R.S.; Wang, D.; et al. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 2018, 65, e148–e154. [Google Scholar] [CrossRef]
- Ferguson, L.; Luo, K.; Olivier, A.; Cunningham, F.; Blackman, S.; Hanson-Dorr, K.; Sun, H.; Baroch, J.; Lutman, M.; Quade, B.; et al. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg. Infect. Dis. 2018, 24, 1020–1028. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. mBio. 2014, 5, e00031-14. [Google Scholar] [CrossRef]
- Collin, E.A.; Sheng, Z.; Lang, Y.; Ma, W.; Hause, B.M.; Li, F. Cocirculation of Two Distinct Genetic and Antigenic Lineages of Proposed Influenza D Virus in Cattle. J. Virol. 2015, 89, 1036–1042. [Google Scholar] [CrossRef]
- Ferguson, L.; Eckard, L.; Epperson, W.B.; Long, L.-P.; Smith, D.; Huston, C.; Genova, S.; Webby, R.; Wan, X.-F. Influenza D virus infection in Mississippi beef cattle. Virology. 2015, 486, 28–34. [Google Scholar] [CrossRef]
- Silveira, S.; Falkenberg, S.M.; Kaplan, B.S.; Crossley, B.; Ridpath, J.F.; Bauermann, F.B.; Fossler, C.P.; Dargatz, D.A.; Dassanayake, R.P.; Vincent, A.L.; et al. Serosurvey for Influenza D Virus Exposure in Cattle, United States, 2014–2015. Emerg. Infect. Dis. 2019, 25, 2074–2080. [Google Scholar] [CrossRef]
- Foni, E.; Chiapponi, C.; Baioni, L.; Zanni, I.; Merenda, M.; Rosignoli, C.; Kyriakis, C.S.; Luini, M.V.; Mandola, M.L.; Bolzoni, L.; et al. Influenza D in Italy: Towards a better understanding of an emerging viral infection in swine. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Flynn, O.; Gallagher, C.; Mooney, J.; Irvine, C.; Ducatez, M.; Hause, B.M.; McGrath, G.; Ryan, E. Influenza D Virus in Cattle, Ireland. Emerg. Infect. Dis. 2018, 24, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Pelletier, C.; Meyer, G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, C.; Oliva, J.; Pauly, M.; Losch, S.; Wildschutz, F.; Muller, C.; Hübschen, J.; Ducatez, M. Influenza D Virus Circulation in Cattle and Swine, Luxembourg, 2012–2016. Emerg. Infect. Dis. 2018, 24, 1388–1389. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; Zhang, H.; Chen, S.N.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.H.; Wen, X.H.; Wei, W.K.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef]
- Horimoto, T.; Hiono, T.; Mekata, H.; Odagiri, T.; Lei, Z.; Kobayashi, T.; Norimine, J.; Inoshima, Y.; Hikono, H.; Murakami, K.; et al. Nationwide Distribution of Bovine Influenza D Virus Infection in Japan. PLoS ONE 2016, 11, e0163828. [Google Scholar] [CrossRef]
- Salem, E.; Hägglund, S.; Cassard, H.; Corre, T.; Näslund, K.; Foret, C.; Gauthier, D.; Pinard, A.; Delverdier, M.; Zohari, S.; et al. Pathogenesis, Host Innate Immune Response, and Aerosol Transmission of Influenza D Virus in Cattle. J. Virol. 2019, 93, e01853-18. [Google Scholar] [CrossRef]
- Mitra, N.; Cernicchiaro, N.; Torres, S.; Li, F.; Hause, B.M. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 2016, 97, 1771–1784. [Google Scholar] [CrossRef]
- Ng, T.F.F.; Kondov, N.O.; Deng, X.; Van Eenennaam, A.; Neibergs, H.L.; Delwart, E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J. Virol. 2015, 89, 5340–5349. [Google Scholar] [CrossRef]
- Couacy-Hymann, E.; Kouakou, V.A.; Aplogan, G.L.; Awoume, F.; Kouakou, C.K.; Kakpo, L.; Sharp, B.R.; McClenaghan, L.; McKensie, P.; Webster, R.G.; et al. Surveillance for influenza viruses in poultry and swine, west Africa, 2006–2008. Emerg. Infect. Dis. 2012, 18, 1446–1452. [Google Scholar] [CrossRef]
- Fusade-Boyer, M.; Pato, P.S.; Komlan, M.; Dogno, K.; Trushar, J.; Rubrum, A.; Kouakou, C.K.; Couacy-Hymann, E.; Batawui, D.; Go-Maro, E.; et al. Evolution of Highly Pathogenic Avian Influenza A(H5N1) Virus in Poultry, Togo, 2018. Emerg. Infect. Dis. 2019, 25, 2287–2289. [Google Scholar] [CrossRef]
- Stevens, K.B.; Pfeiffer, D.U. Spatial modelling of disease using data- and knowledge-driven approaches. Spat. Spatio-Temporal Epidemiol. 2011, 2, 125–133. [Google Scholar] [CrossRef]
- Tran, A.; Trevennec, K.; Lutwama, J.; Sserugga, J.; Gély, M.; Pittiglio, C.; Pinto, J.; Chevalier, V. Development and Assessment of a Geographic Knowledge-Based Model for Mapping Suitable Areas for Rift Valley Fever Transmission in Eastern Africa. PLoS Negl. Trop. Dis. 2016, 10, e0004999. [Google Scholar] [CrossRef]
- De Glanville, W.A.; Vial, L.; Costard, S.; Wieland, B.; Pfeiffer, D.U. Spatial multi-criteria decision analysis to predict suitability for African swine fever endemicity in Africa. BMC Vet. Res. 2014, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Stevens, K.; de Glanville, W.; Costard, S.; Metras, R.; Theuri, W.; Kruska, R.; Randolph, T.; Grace, D.; Hendrickx, S.; Pfeiffer, D. Mapping the Likelihood of Introduction and Spread of Highly Pathogenic Avian Influenza Virus H5N1 in Africa and Indonesia using Multicriteria Decision Modelling. HPAI Research Brief. 2009, 7. Available online: https://cgspace.cgiar.org/bitstream/handle/10568/489/hpai_rbr07.pdf?sequence=1 (accessed on 19 January 2020).
- Lee, J.; Wang, L.; Palinski, R.; Walsh, T.; He, D.; Li, Y.; Wu, R.; Lang, Y.; Sunwoo, S.Y.; Richt, J.A.; et al. Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs. Viruses 2019, 11, 905. [Google Scholar] [CrossRef] [PubMed]
- Oliva, J.; Eichenbaum, A.; Belin, J.; Gaudino, M.; Guillotin, J.; Alzieu, J.P.; Nicollet, P.; Brugidou, R.; Gueneau, E.; Michel, E.; et al. Serological Evidence of Influenza D Virus Circulation Among Cattle and Small Ruminants in France. Viruses 2019, 11, 516. [Google Scholar] [CrossRef]
- Luo, J.; Ferguson, L.; Smith, D.R.; Woolums, A.R.; Epperson, W.B.; Wan, X.F. Serological evidence for high prevalence of Influenza D Viruses in Cattle, Nebraska, United States, 2003–2004. Virology 2017, 501, 88–91. [Google Scholar] [CrossRef]
- Saaty, R.W. The analytic hierarchy process—What it is and how it is used. Math. Model. 1987, 9, 161–176. [Google Scholar] [CrossRef]
- Zadeh, L.A. Fuzzy sets. Inf. Control. 1965, 8, 338–353. [Google Scholar] [CrossRef]
- Ligmann-Zielinska, A.; Jankowski, P. Spatially-explicit integrated uncertainty and sensitivity analysis of criteria weights in multicriteria land suitability evaluation. Environ. Model. Softw. 2014, 57, 235–247. [Google Scholar] [CrossRef]
- Moreno, A.; Lelli, D.; Lavazza, A.; Sozzi, E.; Zanni, I.; Chiapponi, C.; Foni, E.; Capucci, L.; Brocchi, E. MAb-based competitive ELISA for the detection of antibodies against influenza D virus. Transbound Emerg. Dis. 2019, 66, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Knol, A.; Slottje, P.; Sluijs, J.P.; Lebret, E. The use of expert elicitation in environmental health impact assessment: A seven step procedure. Environ. Health 2010, 9, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Krueger, T.; Page, T.; Hubacek, K.; Smith, L.; Hiscock, K. The role of expert opinion in environmental modelling. Environ. Model. Softw. 2012, 36, 4–18. [Google Scholar] [CrossRef]
- Paul, M.C.; Goutard, F.L.; Roulleau, F.; Holl, D.; Thanapongtharm, W.; Roger, F.L.; Tran, A. Quantitative assessment of a spatial multicriteria model for highly pathogenic avian influenza H5N1 in Thailand, and application in Cambodia. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Vial, L.; Ducheyne, E.; Filatoc, S.; Gerilovych, A.; McVey, D.S.; Sindryakova, I.; Morgunov, S.; Pérez de Leon, A.A.; Kolbasov, D.; De Clercq, E.M. Spatial multi-criteria decision analysis for modelling suitable habitats of Ornithodoros soft ticks in the Western Palearctic region. Vet. Parasitol. 2018, 249, 2–16. [Google Scholar] [CrossRef]
- Stevens, K.B.; Gilbert, M.; Pfeiffer, D.U. Modeling habitat suitability for occurrence of highly pathogenic avian influenza virus H5N1 in domestic poultry in Asia: A spatial multicriteria decision analysis approach. Spat. Spatio-Temporal Epidemiol. 2013, 4, 1–14. [Google Scholar] [CrossRef]

| Risk Factor | Explanation | References |
|---|---|---|
| Swine density | IDV was discovered in swine and it is efficiently transmissible in this species. | [1,25] |
| Cattle density | Cattle are susceptible to IDV and harbor the highest seropositivity rates. Cattle are considered as the main host of the virus. IDV is also transmissible by aerosol between cattle. | [8,12,13,16,26,27] |
| Small ruminants density | Specific antibodies against IDV were detected in small ruminants, justifying their density as a risk factor. | [3,26] |
| Presence of respiratory clinical signs in cattle | Several studies report that IDV is more commonly isolated from cattle with respiratory clinical signs and can be airborne transmitted among cattle. | [8,16,17,18] |
| Cattle age | Calves appear more susceptible to IDV infection than adults. | [8,27] |
| Proximity to cattle market | Some cities in Togo receive cattle from all over the country and sometimes from neighboring countries. Cattle can stay in fields around the city waiting to be transferred to the slaughterhouse or to be sold to other farmers. Cattle markets represent focus points where cattle of different sanitary status and from different origins are parked, likely leading to an easier circulation of the virus. | Local expert opinion |
| Transhumance areas | Transhumance occurs each year in Togo between January and May. During this period, about 50,000 cattle come from Sahelian countries and are parked on dedicated fields, with the possibility of contact with local cattle. Trade with local farmers occurs during this period. Transhumance areas and periods were therefore considered a risk factor for IDV occurrence. | Local expert opinion |
| Proximity to wildlife | In wildlife, IDV has been detected only in feral swine but because of the wide range of hosts susceptible to infection, wild ruminants and other species from wildlife could play a role in transmission. | [5] |
| Proximity to water | Water areas can represent focus points where cattle from different farms can have close contact between each other and with wildlife, extensive breeding being the main breeding system for cattle and small ruminants in Togo. | Local expert opinion |
| Species | Nb. Sera Samples | Nb. IDV Seropositive Samples | Positive Sera (%) [Median HI Positive Titer; HI Titers Range] | Nb. Nasal Swabs | Nb. IDV Positive Swabs |
|---|---|---|---|---|---|
| Cattle | 399 | 18 | 4.5 [20; 10–320] | 10 | 0 |
| Small ruminants | 737 | 28 | 3.8 [40; 10–160] | 840 | 0 |
| Swine | 80 | 0 | 0 | 346 | 0 |
| Risk Factor | Mean Weight | Risk Relationships * | Thresholds |
|---|---|---|---|
| Cattle density | 0.38 | Linear increasing | a = minimum raster layer value b = maximum raster layer value |
| Small ruminants density | 0.08 | Linear increasing | a = minimum raster layer value b = maximum raster layer value |
| Swine density | 0.11 | Linear increasing | a = minimum raster layer value b = maximum raster layer value |
| Proximity to water | 0.01 | Sigmoid decreasing | a = 2.5 km b = 5 km |
| Proximity to cattle market | 0.24 | Sigmoid decreasing | a = 5 km b = 10 km |
| Proximity to wildlife | 0.02 | Sigmoid decreasing | a = 2 km b = 4 km |
| Proximity to transhumance areas | 0.16 | Sigmoid decreasing | a = 0.5 km b = 2.5 km |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fusade-Boyer, M.; Pato, P.S.; Komlan, M.; Dogno, K.; Batawui, K.; Go-Maro, E.; McKenzie, P.; Guinat, C.; Secula, A.; Paul, M.; et al. Risk Mapping of Influenza D Virus Occurrence in Ruminants and Swine in Togo Using a Spatial Multicriteria Decision Analysis Approach. Viruses 2020, 12, 128. https://doi.org/10.3390/v12020128
Fusade-Boyer M, Pato PS, Komlan M, Dogno K, Batawui K, Go-Maro E, McKenzie P, Guinat C, Secula A, Paul M, et al. Risk Mapping of Influenza D Virus Occurrence in Ruminants and Swine in Togo Using a Spatial Multicriteria Decision Analysis Approach. Viruses. 2020; 12(2):128. https://doi.org/10.3390/v12020128
Chicago/Turabian StyleFusade-Boyer, Maxime, Pidemnéwé S. Pato, Mathias Komlan, Koffi Dogno, Komla Batawui, Emilie Go-Maro, Pamela McKenzie, Claire Guinat, Aurélie Secula, Mathilde Paul, and et al. 2020. "Risk Mapping of Influenza D Virus Occurrence in Ruminants and Swine in Togo Using a Spatial Multicriteria Decision Analysis Approach" Viruses 12, no. 2: 128. https://doi.org/10.3390/v12020128
APA StyleFusade-Boyer, M., Pato, P. S., Komlan, M., Dogno, K., Batawui, K., Go-Maro, E., McKenzie, P., Guinat, C., Secula, A., Paul, M., Webby, R. J., Tran, A., Waret-Szkuta, A., & Ducatez, M. F. (2020). Risk Mapping of Influenza D Virus Occurrence in Ruminants and Swine in Togo Using a Spatial Multicriteria Decision Analysis Approach. Viruses, 12(2), 128. https://doi.org/10.3390/v12020128





