Discovery of Surfactant-Like Peptides from a Phage-Displayed Peptide Library
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Screening of Surfactant-Like Peptides from a Phage-Displayed Peptide Library
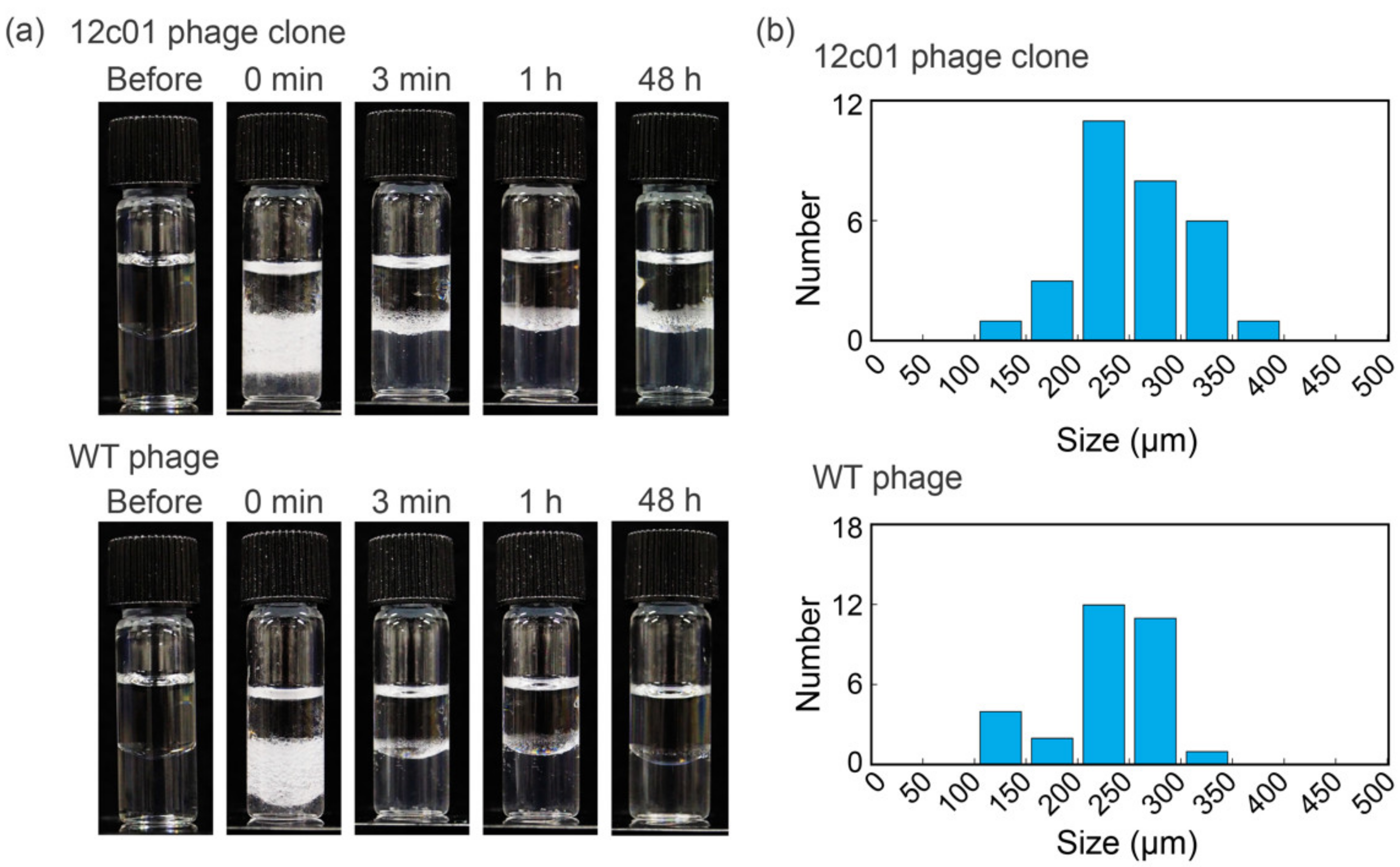
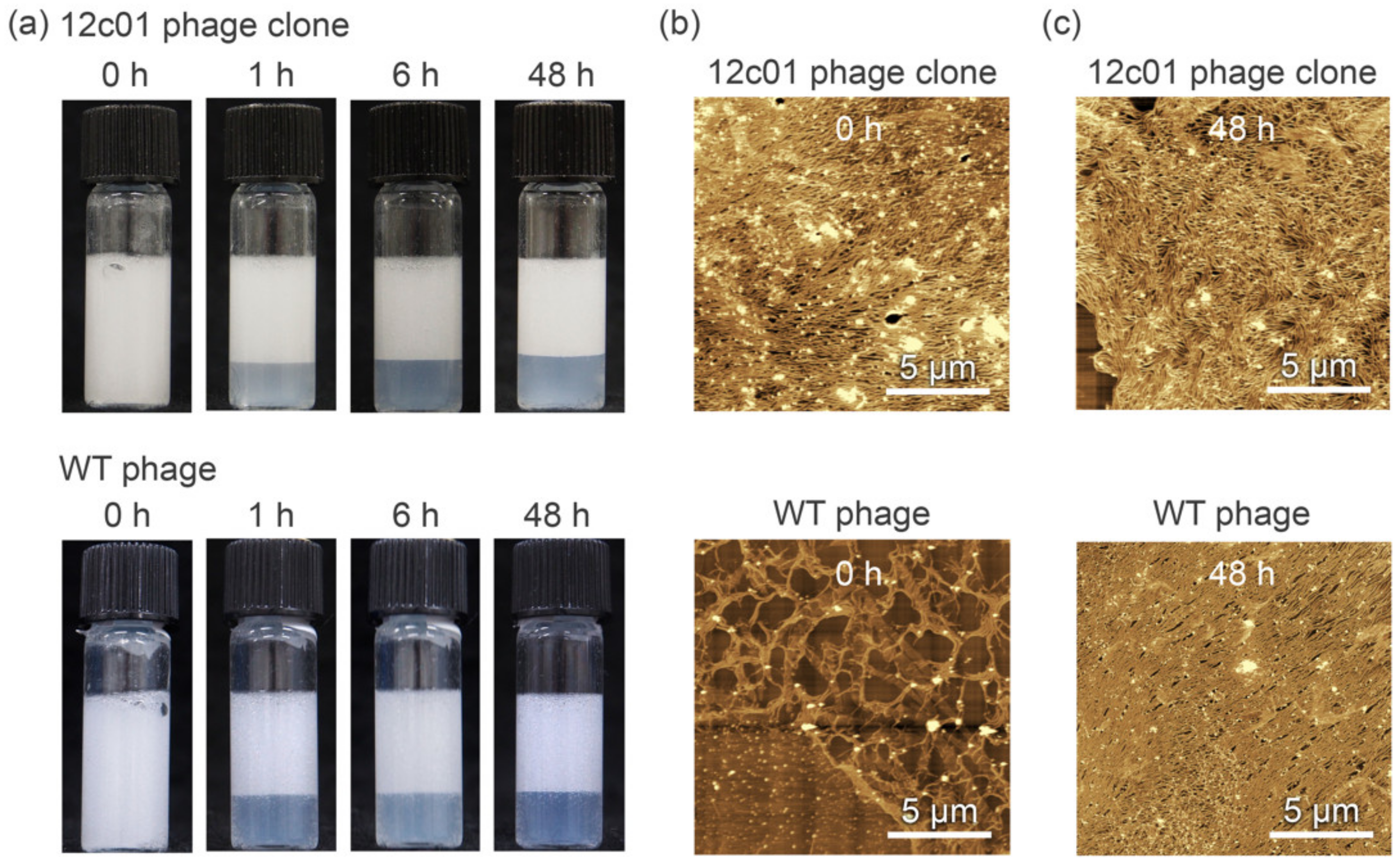
2.3. Preparation of Phage-Based Emulsions
2.4. Atomic Force Microscopy (AFM) Observations
2.5. Peptide Synthesis
2.6. Structural Characterization of Peptides
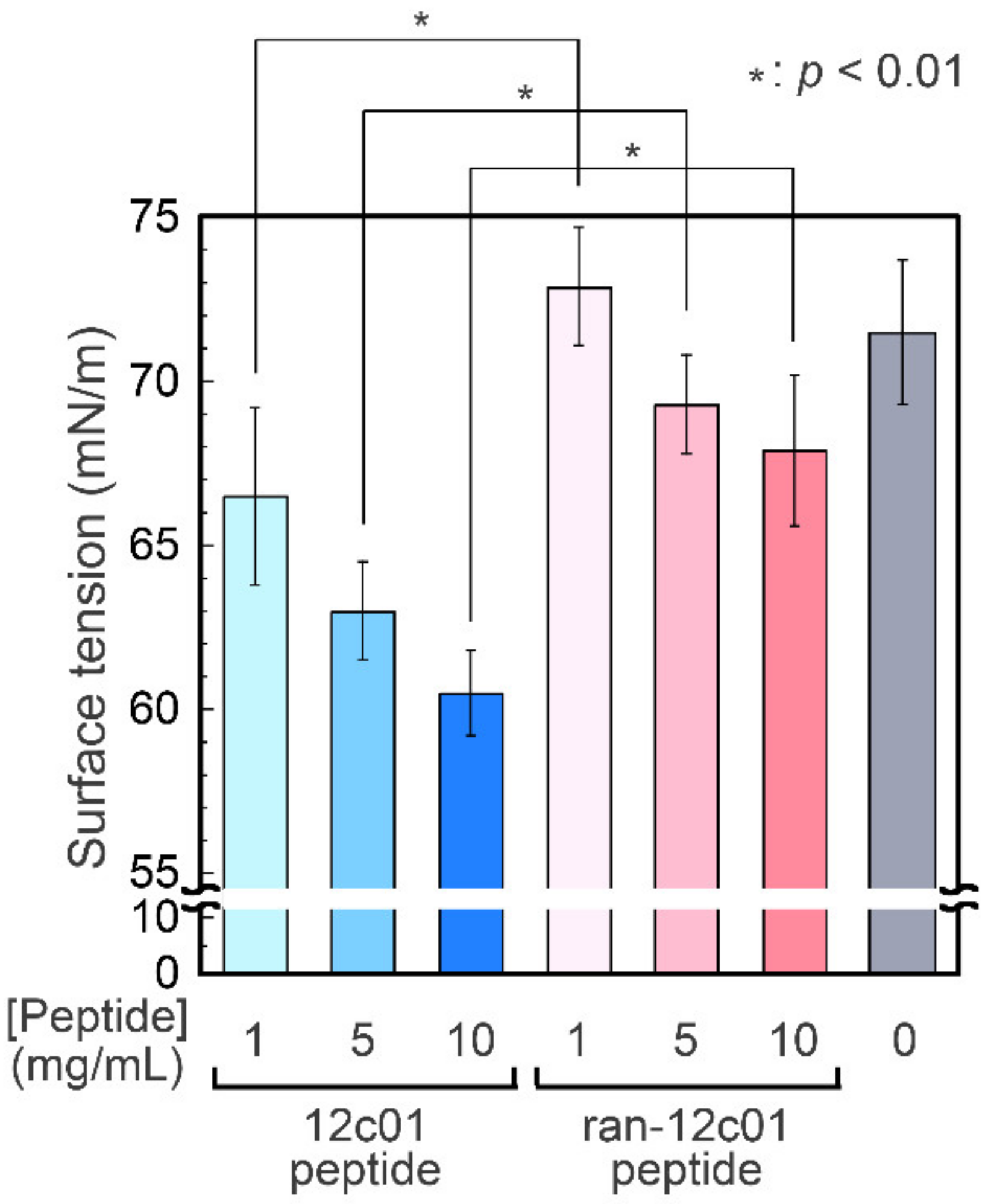
2.7. Surface Tension Measurements
2.8. Preparation of Peptide-Based Emulsions
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sarikaya, M.; Tamerler, C.; Jen, A.; Schulten, K.; Baneyx, F. Molecular Biomimetics: Nanotechnology through Biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar] [CrossRef]
- Baneyx, F.; Schwartz, D.T. Selection and Analysis of Solid-Binding Peptides. Curr. Opin. Biotechnol. 2007, 18, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P.; Petrenko, V.A. Phage Display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Günay, K.; Klok, H.-A. Identification of Soft Matter Binding Peptide Ligands Using Phage Display. Bioconjugate Chem. 2015, 26, 2002–2015. [Google Scholar] [CrossRef] [PubMed]
- Shiba, K. Exploitation of Peptide Motif Sequences and Their Use in Nanobiotechnology. Curr. Opin. Biotechnol. 2010, 21, 412–425. [Google Scholar] [CrossRef]
- Naik, R.; Stringer, S.; Agarwal, G.; Jones, S.; Stone, M. Biomimetic Synthesis and Patterning of Silver Nanoparticles. Nat. Mater. 2002, 1, 169–172. [Google Scholar] [CrossRef]
- Sano, K.-I.; Sasaki, H.; Shiba, K. Specificity and Biomineralization Activities of Ti-Binding Peptide-1 (Tbp-1). Langmuir 2005, 21, 3090–3095. [Google Scholar] [CrossRef]
- Umetsu, M.; Mizuta, M.; Tsumoto, K.; Ohara, S.; Takami, S.; Watanabe, H.; Kumagai, I.; Adschiri, T. Bioassisted Room-Temperature Immobilization and Mineralization of Zinc Oxide—The Structural Ordering of Zno Nanoparticles into a Flower-Type Morphology. Adv. Mater. 2005, 17, 2571–2575. [Google Scholar] [CrossRef]
- Naik, R.R.; Jones, S.E.; Murray, C.J.; McAuliffe, J.C.; Vaia, R.A.; Stone, M.O. Peptide Templates for Nanoparticle Synthesis Derived from Polymerase Chain Reaction-Driven Phage Display. Adv. Funct. Mater. 2004, 14, 25–30. [Google Scholar] [CrossRef]
- Wei, Z.; Maeda, Y.; Matsui, H. Discovery of Catalytic Peptides for Inorganic Nanocrystal Synthesis by a Combinatorial Phage Display Approach. Anegw. Chem. Int. Ed. 2011, 50, 10585–10588. [Google Scholar] [CrossRef]
- Maeda, Y.; Javid, N.; Duncan, K.; Birchall, L.; Gibson, K.F.; Cannon, D.; Kanetsuki, Y.; Knapp, C.; Tuttle, T.; Ulijn, R.V.; et al. Discovery of Catalytic Phages by Biocatalytic Self-Assembly. J. Am. Chem. Soc. 2014, 136, 15893–15896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, Y.; Makhlynets, O.V.; Matsui, H.; Korendovych, I.V. Design of Catalytic Peptides and Proteins through Rational and Combinatorial Approaches. Annu. Rev. Biomed. Eng. 2016, 18, 311–328. [Google Scholar] [CrossRef] [PubMed]
- Bibette, J.; Calderon, F.L.; Poulin, P. Emulsions: Basic Principles. Rep. Prog. Phys. 1999, 62, 969. [Google Scholar] [CrossRef]
- Zarzar, L.D.; Sresht, V.; Sletten, E.M.; Kalow, J.A.; Blankschtein, D.; Swager, T.M. Dynamically Reconfigurable Complex Emulsions Via Tunable Interfacial Tensions. Nature 2015, 518, 520–524. [Google Scholar] [CrossRef] [Green Version]
- Kislukhin, A.A.; Xu, H.; Adams, S.R.; Narsinh, K.H.; Tsien, R.Y.; Ahrens, E.T. Paramagnetic Fluorinated Nanoemulsions for Sensitive Cellular Fluorine-19 Magnetic Resonance Imaging. Nat. Mater. 2016, 15, 662–668. [Google Scholar] [CrossRef]
- Hanson, J.A.; Chang, C.B.; Graves, S.M.; Li, Z.; Mason, T.G.; Deming, T.J. Nanoscale Double Emulsions Stabilized by Single-Component Block Copolypeptides. Nature 2008, 455, 85–88. [Google Scholar] [CrossRef]
- Rebello, S.; Asok, A.K.; Mundayoor, S.; Jisha, M.S. Surfactants: Toxicity, Remediation and Green Surfactants. Environ. Chem. Lett. 2014, 12, 275–287. [Google Scholar] [CrossRef]
- Lam, R.S.H.; Nickerson, M.T. Food Proteins: A Review on Their Emulsifying Properties Using a Structure–Function Approach. Food Chem. 2013, 141, 975–984. [Google Scholar] [CrossRef]
- Dickinson, E. Colloids in Food: Ingredients, Structure, and Stability. Annu. Rev. Food Sci. Technol. 2015, 6, 1–23. [Google Scholar] [CrossRef]
- Kim, W.; Wang, Y.; Selomulya, C. Dairy and Plant Proteins as Natural Food Emulsifiers. Trends Food Sci. Technol. 2020, 105, 261–272. [Google Scholar] [CrossRef]
- McClements, D.J. Protein-Stabilized Emulsions. Curr. Opin. Colloid Interface Sci. 2004, 9, 305–313. [Google Scholar] [CrossRef]
- Dickinson, E. Exploring the Frontiers of Colloidal Behaviour Where Polymers and Particles Meet. Food Hydrocoll. 2016, 52, 497–509. [Google Scholar] [CrossRef]
- Raffaini, G.; Milani, R.; Ganazzoli, F.; Resnati, G.; Metrangolo, P. Atomistic Simulation of Hydrophobin Hfbii Conformation in Aqueous and Fluorous Media and at the Water/Vacuum Interface. J. Mol. Graph. Model. 2016, 63, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Cox, P.W.; Hooley, P. Hydrophobins: New Prospects for Biotechnology. Fungal Biol. Rev. 2009, 23, 40–47. [Google Scholar] [CrossRef]
- Sawada, T.; Mihara, H. Dense Surface Functionalization Using Peptides That Recognize Differences in Organized Structures of Self-Assembling Nanomaterials. Mol. BioSyst. 2012, 8, 1264–1274. [Google Scholar] [CrossRef]
- Sawada, T.; Takahashi, T.; Mihara, H. Affinity-Based Screening of Peptides Recognizing Assembly States of Self-Assembling Peptide Nanomaterials. J. Am. Chem. Soc. 2009, 131, 14434–14441. [Google Scholar] [CrossRef]
- Sawada, T.; Mihara, H.; Serizawa, T. Peptides as New Smart Bionanomaterials: Molecular Recognition and Self-Assembly Capabilities. Chem. Rec. 2013, 13, 172–186. [Google Scholar] [CrossRef]
- Date, T.; Sekine, J.; Matsuno, H.; Serizawa, T. Polymer-Binding Peptides for the Noncovalent Modification of Polymer Surfaces: Effects of Peptide Density on the Subsequent Immobilization of Functional Proteins. ACS Appl. Mater. Interfaces 2011, 3, 351–360. [Google Scholar] [CrossRef]
- Date, T.; Yoshino, S.; Matsuno, H.; Serizawa, T. Surface Modification of Stereoregular and Stereocomplex Poly(Methyl Methacrylate) Films with Biologically Identified Peptides. Polym. J. 2012, 44, 366–369. [Google Scholar] [CrossRef]
- Date, T.; Sawada, T.; Serizawa, T. Self-Assembled Peptides on Polymer Surfaces: Towards Morphology-Dependent Surface Functionalization. Soft Matter 2013, 9, 3469–3472. [Google Scholar] [CrossRef]
- Dexter, A.F.; Malcolm, A.S.; Middelberg, A.P.J. Reversible Active Switching of the Mechanical Properties of a Peptide Film at a Fluid–Fluid Interface. Nat. Mater. 2006, 5, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; He, L.; Middelberg, A.P.J.; Mark, A.E.; Poger, D. Determining the Structure of Interfacial Peptide Films: Comparing Neutron Reflectometry and Molecular Dynamics Simulations. Langmuir 2014, 30, 10080–10089. [Google Scholar] [CrossRef] [PubMed]
- McGregor, C.-L.; Chen, L.; Pomroy, N.C.; Hwang, P.; Go, S.; Chakrabartty, A.; Privé, G.G. Lipopeptide Detergents Designed for the Structural Study of Membrane Proteins. Nat. Biotechnol. 2003, 21, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Varenik, M.; Bloch, D.N.; Atsmon-Raz, Y.; Jacoby, G.; Adler-Abramovich, L.; Shimon, L.J.W.; Beck, R.; Miller, Y.; Regev, O.; et al. A Minimal Length Rigid Helical Peptide Motif Allows Rational Design of Modular Surfactants. Nat. Commun. 2017, 8, 14018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Sawada, T.; Li, X.; Serizawa, T. Controlled Assembly of Filamentous Viruses into Hierarchical Nano- to Microstructures at Liquid/Liquid Interfaces. RSC Adv. 2020, 10, 26313–26318. [Google Scholar] [CrossRef]
- Chan, W.C.; White, P.D. (Eds.) Fmoc Solid Phase Peptide Synthesis, 1st ed.; Oxford University: New York, NY, USA, 2000; pp. 41–76. [Google Scholar]
- Serizawa, T.; Sawada, T.; Matsuno, H. Highly Specific Affinities of Short Peptides against Synthetic Polymers. Langmuir 2007, 23, 11127–11133. [Google Scholar] [CrossRef]
- Sawada, T.; Okeya, Y.; Hashizume, M.; Serizawa, T. Screening of Peptides Recognizing Simple Polycyclic Aromatic Hydrocarbons. Chem. Commun. 2013, 49, 5088–5090. [Google Scholar] [CrossRef]
- Sawada, T.; Serizawa, T. Immobilization of Highly-Oriented Filamentous Viruses onto Polymer Substrates. J. Mater. Chem. B 2013, 1, 149–152. [Google Scholar] [CrossRef]
- Suzuki, S.; Sawada, T.; Ishizone, T.; Serizawa, T. Affinity-Based Thermoresponsive Precipitation of Proteins Modified with Polymer-Binding Peptides. Chem. Commun. 2016, 52, 5670–5673. [Google Scholar] [CrossRef] [Green Version]
- Sawada, T. Filamentous Virus-Based Soft Materials Based on Controlled Assembly through Liquid Crystalline Formation. Polym. J. 2017, 49, 639–647. [Google Scholar] [CrossRef]
- McClements, D.J.; Gumus, C.E. Natural Emulsifiers—Biosurfactants, Phospholipids, Biopolymers, and Colloidal Particles: Molecular and Physicochemical Basis of Functional Performance. Adv. Colloid Interface Sci. 2016, 234, 3–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, A.F.M. Solubility Parameters. Chem. Rev. 1975, 75, 731–753. [Google Scholar] [CrossRef]
- Marcus, Y. The Properties of Organic Liquids That Are Relevant to Their Use as Solvating Solvents. Chem. Soc. Rev. 1993, 22, 409–416. [Google Scholar] [CrossRef]
- Reichardt, C. Solvatochromic Dyes as Solvent Polarity Indicators. Chem. Rev. 1994, 94, 2319–2358. [Google Scholar] [CrossRef]
- Lee, J.N.; Park, C.; Whitesides, G.M. Solvent Compatibility of Poly(Dimethylsiloxane)-Based Microfluidic Devices. Anal. Chem. 2003, 75, 6544–6554. [Google Scholar] [CrossRef]


| Clone | Frequency | Sequence |
|---|---|---|
| 12c01 | 16/26 | SQDIRTWNGTRS |
| 12c02 | 1/26 | TFYNNPTHTPSH |
| 12c03 | 1/26 | SGLPASAAMPLN |
| 12c04 | 1/26 | SPSVRNGVSPYG |
| 12c05 | 1/26 | QLYHPMSRGAVK |
| 12c06 | 1/26 | YLNNDHFGQALT |
| n. i.1 | 5/26 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawada, T.; Oyama, R.; Tanaka, M.; Serizawa, T. Discovery of Surfactant-Like Peptides from a Phage-Displayed Peptide Library. Viruses 2020, 12, 1442. https://doi.org/10.3390/v12121442
Sawada T, Oyama R, Tanaka M, Serizawa T. Discovery of Surfactant-Like Peptides from a Phage-Displayed Peptide Library. Viruses. 2020; 12(12):1442. https://doi.org/10.3390/v12121442
Chicago/Turabian StyleSawada, Toshiki, Rina Oyama, Michihiro Tanaka, and Takeshi Serizawa. 2020. "Discovery of Surfactant-Like Peptides from a Phage-Displayed Peptide Library" Viruses 12, no. 12: 1442. https://doi.org/10.3390/v12121442
APA StyleSawada, T., Oyama, R., Tanaka, M., & Serizawa, T. (2020). Discovery of Surfactant-Like Peptides from a Phage-Displayed Peptide Library. Viruses, 12(12), 1442. https://doi.org/10.3390/v12121442






