Leveraging 3D Model Systems to Understand Viral Interactions with the Respiratory Mucosa
Abstract
1. Introduction
2. In Vitro Airway Models with Emergent Properties
2.1. Air–Liquid Interface Transwell Systems
2.2. Organoids
2.3. Incorporation of Airway Mechanics, Heterotypic Cell–Cell Interactions, and On-Chip Systems
3. Application of In Vitro Airway Models with Emergent Properties in Virology
4. Components of the Extracellular Barrier in the Respiratory Tract
4.1. Mucus and Mucins
4.2. Secreted (Non-Mucin) Components
4.2.1. Defensins
4.2.2. Proteases
4.2.3. Other Secreted Components
4.3. Microbiota
5. Utility of In Vitro Airway Models with Emergent Properties in Probing Virus Interactions in the Extracellular Space
6. Assaying Virus–Host Interactions in the Extracellular Space In Vitro
6.1. Host-Specific Barrier Properties and How to Assess Them
6.2. Viral Particle Tracking, Host–Virus Interactions, and Specific Barrier Component Contributions
7. Conclusions and Future Perspectives
Funding
Conflicts of Interest
Abbreviations
| AAV1 | adeno-associated virus serotype 1 |
| AAV6 | adeno-associated virus serotype 6 |
| ALI | air–liquid interface |
| ASL | airway surface liquid |
| CF | cystic fibrosis |
| CDHR3 | cadherin related family member 3 |
| COPD | chronic obstructive pulmonary disorder |
| HAT | human airway trypsin-like protease |
| hPSC | human pluripotent stem cell |
| IAV | influenza A virus |
| LRT | lower respiratory tract |
| MCC | mucociliary clearance |
| MSD | mean squared displacement |
| PCL | periciliary layer |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| TMPRSS2 | transmembrane protease serine 2 |
| URT | upper respiratory tract |
References
- Lechner, J.F.; Haugen, A.; McClendon, I.A.; Pettis, E.W. Clonal growth of normal adult human bronchial epithelial cells in a serum-free medium. In Vitro 1982, 18, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Jorissen, M.; Van der Schueren, B.; Van den Berghe, H.; Cassiman, J.J. The preservation and regeneration of cilia on human nasal epithelial cells cultured in vitro. Arch. Otorhinolaryngol. 1989, 246, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Yankaskas, J.; Cheng, E.; Knowles, M.R.; Boucher, R. Growth and differentiation of human nasal epithelial cells in culture. Serum-free, hormone-supplemented medium and proteoglycan synthesis. Am. Rev. Respir. Dis. 1985, 132, 311–320. [Google Scholar] [PubMed]
- Benali, R.; Tournier, J.M.; Chevillard, M.; Zahm, J.M.; Klossek, J.M.; Hinnrasky, J.; Gaillard, D.; Maquart, F.X.; Puchelle, E. Tubule formation by human surface respiratory epithelial cells cultured in a three-dimensional collagen lattice. Am. J. Physiol. 1993, 264, L183–L192. [Google Scholar] [CrossRef]
- Whitcutt, M.J.; Adler, K.B.; Wu, R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell. Dev. Biol. 1988, 24, 420–428. [Google Scholar] [CrossRef]
- Wheeler, A.H.; Nungester, W.J. Effect of mucin on influenza virus infection in hamsters. Science 1942, 96, 92–93. [Google Scholar] [CrossRef]
- Fazekas de S Groth, S. Nasal mucus and influenza viruses. I. The haemagglutinin inhibitor in nasal secretions. J. Hyg. 1952, 50, 471–490. [Google Scholar]
- Pannu, J.S.; Sigel, M.M. Inhibition of viruses by secretions from the female genital tract. Proc. Soc. Exp. Biol. Med. 1963, 114, 763–766. [Google Scholar] [CrossRef]
- Kesimer, M.; Ehre, C.; Burns, K.A.; Davis, C.W.; Sheehan, J.K.; Pickles, R.J. Molecular organization of the mucins and glycocalyx underlying mucus transport over mucosal surfaces of the airways. Mucosal Immunol. 2013, 6, 379–392. [Google Scholar] [CrossRef]
- Davis, A.S.; Chertow, D.S.; Moyer, J.E.; Suzich, J.; Sandouk, A.; Dorward, D.W.; Logun, C.; Shelhamer, J.H.; Taubenberger, J.K. Validation of normal human bronchial epithelial cells as a model for influenza A infections in human distal trachea. J. Histochem. Cytochem. 2015, 63, 312–328. [Google Scholar] [CrossRef]
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Kesimer, M.; Kirkham, S.; Pickles, R.J.; Henderson, A.G.; Alexis, N.E.; Demaria, G.; Knight, D.; Thornton, D.J.; Sheehan, J.K. Tracheobronchial air-liquid interface cell culture: A model for innate mucosal defense of the upper airways? Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 296, L92–L100. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Goldsmith, C.S.; Maines, T.R.; Belser, J.A.; Gustin, K.M.; Pekosz, A.; Zaki, S.R.; Katz, J.M.; Tumpey, T.M. Tropism and infectivity of influenza virus, including highly pathogenic avian H5N1 virus, in ferret tracheal differentiated primary epithelial cell cultures. J. Virol. 2013, 87, 2597–2607. [Google Scholar] [CrossRef] [PubMed]
- Newby, C.M.; Rowe, R.K.; Pekosz, A. Influenza A virus infection of primary differentiated airway epithelial cell cultures derived from Syrian golden hamsters. Virology 2006, 354, 80–90. [Google Scholar] [CrossRef]
- You, Y.; Richer, E.J.; Huang, T.; Brody, S.L. Growth and differentiation of mouse tracheal epithelial cells: Selection of a proliferative population. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 283, L1315–L1321. [Google Scholar] [CrossRef]
- Kondo, M.; Tamaoki, J.; Takeyama, K.; Nakata, J.; Nagai, A. Interleukin-13 induces goblet cell differentiation in primary cell culture from Guinea pig tracheal epithelium. Am. J. Respir. Cell Mol. Biol. 2002, 27, 536–541. [Google Scholar] [CrossRef]
- Fulcher, M.L.; Gabriel, S.; Burns, K.A.; Yankaskas, J.R.; Randell, S.H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005, 107, 183–206. [Google Scholar]
- Karp, P.H.; Moninger, T.O.; Weber, S.P.; Nesselhauf, T.S.; Launspach, J.L.; Zabner, J.; Welsh, M.J. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol. Biol. 2002, 188, 115–137. [Google Scholar]
- Firth, A.L.; Dargitz, C.T.; Qualls, S.J.; Menon, T.; Wright, R.; Singer, O.; Gage, F.H.; Khanna, A.; Verma, I.M. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, E1723–E1730. [Google Scholar] [CrossRef]
- Firth, A.L.; Menon, T.; Parker, G.S.; Qualls, S.J.; Lewis, B.M.; Ke, E.; Dargitz, C.T.; Wright, R.; Khanna, A.; Gage, F.H.; et al. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep. 2015, 12, 1385–1390. [Google Scholar] [CrossRef]
- Wong, A.P.; Bear, C.E.; Chin, S.; Pasceri, P.; Thompson, T.O.; Huan, L.-J.; Ratjen, F.; Ellis, J.; Rossant, J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012, 30, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, F.J.; Suzuki, S.; Beermann, M.L.; Barillà, C.; Wang, R.; Villacorta-Martin, C.; Berical, A.; Jean, J.C.; Le Suer, J.; Matte, T.; et al. Derivation of airway basal stem cells from human pluripotent stem cells. Cell Stem Cell 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Bluhmki, T.; Bitzer, S.; Gindele, J.A.; Schruf, E.; Kiechle, T.; Webster, M.; Schymeinsky, J.; Ries, R.; Gantner, F.; Bischoff, D.; et al. Development of a miniaturized 96-Transwell air-liquid interface human small airway epithelial model. Sci. Rep. 2020, 10, 13022. [Google Scholar] [CrossRef] [PubMed]
- Tarran, R.; Button, B.; Picher, M.; Paradiso, A.M.; Ribeiro, C.M.; Lazarowski, E.R.; Zhang, L.; Collins, P.L.; Pickles, R.J.; Fredberg, J.J.; et al. Normal and cystic fibrosis airway surface liquid homeostasis. The effects of phasic shear stress and viral infections. J. Biol. Chem. 2005, 280, 35751–35759. [Google Scholar] [CrossRef]
- Derichs, N.; Jin, B.J.; Song, Y.; Finkbeiner, W.E.; Verkman, A.S. Hyperviscous airway periciliary and mucous liquid layers in cystic fibrosis measured by confocal fluorescence photobleaching. FASEB J. 2011, 25, 2325–2332. [Google Scholar] [CrossRef]
- Henderson, A.G.; Ehre, C.; Button, B.; Abdullah, L.H.; Cai, L.-H.; Leigh, M.W.; DeMaria, G.C.; Matsui, H.; Donaldson, S.H.; Davis, C.W.; et al. Cystic fibrosis airway secretions exhibit mucin hyperconcentration and increased osmotic pressure. J. Clin. Investig. 2014, 124, 3047–3060. [Google Scholar] [CrossRef]
- Farberman, M.M.; Ibricevic, A.; Joseph, T.D.; Akers, K.T.; Garcia-Medina, R.; Crosby, S.; Clarke, L.L.; Brody, S.L.; Ferkol, T.W. Effect of polarized release of CXC-chemokines from wild-type and cystic fibrosis murine airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2011, 45, 221–228. [Google Scholar] [CrossRef]
- Tarran, R.; Trout, L.; Donaldson, S.H.; Boucher, R.C. Soluble mediators, not cilia, determine airway surface liquid volume in normal and cystic fibrosis superficial airway epithelia. J. Gen. Physiol. 2006, 127, 591–604. [Google Scholar] [CrossRef]
- Lakshmi, S.P.; Reddy, A.T.; Banno, A.; Reddy, R.C. Airway epithelial cell peroxisome proliferator-activated receptor γ regulates inflammation and mucin expression in allergic airway disease. J. Immunol. 2018, 201, 1775–1783. [Google Scholar] [CrossRef]
- Sotty, J.; Garçon, G.; Denayer, F.O.; Alleman, L.Y.; Saleh, Y.; Perdrix, E.; Riffault, V.; Dubot, P.; Lo-Guidice, J.M.; Canivet, L. Toxicological effects of ambient fine (PM) and ultrafine (PM) particles in healthy and diseased 3D organo-typic mucocilary-phenotype models. Environ. Res. 2019, 176, 108538. [Google Scholar] [CrossRef]
- Stewart, C.E.; Torr, E.E.; Mohd Jamili, N.H.; Bosquillon, C.; Sayers, I. Evaluation of Differentiated Human Bronchial Epithelial Cell Culture Systems for Asthma Research. J. Allergy 2012, 2012, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.; Sarlang, S.; Thavagnanam, S.; Williamson, G.; O’donoghue, D.; Villenave, R.; Power, U.; Shields, M.; Heaney, L.; Skibinski, G. A 3-D well-differentiated model of pediatric bronchial epithelium demonstrates unstimulated morphological differences between asthmatic and nonasthmatic cells. Pediatr. Res. 2010, 67, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Hackett, T.-L.; Singhera, G.K.; Shaheen, F.; Hayden, P.; Jackson, G.R.; Hegele, R.G.; Van Eeden, S.; Bai, T.R.; Dorscheid, D.R.; Knight, D.A. Intrinsic phenotypic differences of asthmatic epithelium and its inflammatory responses to respiratory syncytial virus and air pollution. Am. J. Respir. Cell Mol. Biol. 2011, 45, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, B.; Happillon, M.; Antherieu, S.; Hardy, E.M.; Alleman, L.Y.; Grova, N.; Perdrix, E.; Appenzeller, B.M.; Lo Guidice, J.M.; Coddeville, P.; et al. Differential responses of healthy and chronic obstructive pulmonary diseased human bronchial epithelial cells repeatedly exposed to air pollution-derived PM. Environ. Pollut. 2016, 218, 1074–1088. [Google Scholar] [CrossRef]
- Mertens, T.C.J.; Karmouty-Quintana, H.; Taube, C.; Hiemstra, P.S. Use of airway epithelial cell culture to unravel the pathogenesis and study treatment in obstructive airway diseases. Pulm. Pharmacol. Ther. 2017, 45, 101–113. [Google Scholar] [CrossRef]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef]
- Suprynowicz, F.A.; Upadhyay, G.; Krawczyk, E.; Kramer, S.C.; Hebert, J.D.; Liu, X.; Yuan, H.; Cheluvaraju, C.; Clapp, P.W.; Boucher, R.C., Jr.; et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20035–20040. [Google Scholar] [CrossRef]
- Walters, M.S.; Gomi, K.; Ashbridge, B.; Moore, M.A.S.; Arbelaez, V.; Heldrich, J.; Ding, B.S.; Rafii, S.; Staudt, M.R.; Crystal, R.G. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir. Res. 2013, 14, 135. [Google Scholar] [CrossRef]
- Kreft, M.E.; Jerman, U.D.; Lasič, E.; Hevir-Kene, N.; Rižner, T.L.; Peternel, L.; Kristan, K. The characterization of the human cell line Calu-3 under different culture conditions and its use as an optimized in vitro model to investigate bronchial epithelial function. Eur. J. Pharm. Sci. 2015, 69, 1–9. [Google Scholar] [CrossRef]
- Chu, H.W.; Rios, C.; Huang, C.; Wesolowska-Andersen, A.; Burchard, E.G.; O’Connor, B.P.; Fingerlin, T.E.; Nichols, D.; Reynolds, S.D.; Seibold, M.A. CRISPR-Cas9-mediated gene knockout in primary human airway epithelial cells reveals a proinflammatory role for MUC18. Gene Ther. 2015, 22, 822–829. [Google Scholar] [CrossRef]
- Everman, J.L.; Sajuthi, S.; Saef, B.; Rios, C.; Stoner, A.M.; Numata, M.; Hu, D.; Eng, C.; Oh, S.; Rodriguez-Santana, J.; et al. Functional genomics of CDHR3 confirms its role in HRV-C infection and childhood asthma exacerbations. J. Clin. Immunol. 2019, 144, 962–971. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.D.; Siddiqui, S.; Cheng, D.; Bonser, L.R.; Sun, D.I.; Zlock, L.T.; Finkbeiner, W.E.; Woodruff, P.G.; Erle, D.J. Efficient RNP-directed human gene targeting reveals SPDEF is required for IL-13-induced mucostasis. Am. J. Respir. Cell Mol. Biol. 2020, 62, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Dye, B.R.; Dedhia, P.H.; Miller, A.J.; Nagy, M.S.; White, E.S.; Shea, L.D.; Spence, J.R. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife 2016, 5, e19732. [Google Scholar] [CrossRef] [PubMed]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.H.; Tsai, Y.H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, 4, e05098. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Huang, S.X.L.; Islam, M.N.; O’Neill, J.; Hu, Z.; Yang, Y.-G.; Chen, Y.-W.; Mumau, M.; Green, M.D.; Vunjak-Novakovic, G.; Bhattacharya, J.; et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 84–91. [Google Scholar] [CrossRef]
- Konishi, S.; Gotoh, S.; Tateishi, K.; Yamamoto, Y.; Korogi, Y.; Nagasaki, T.; Matsumoto, H.; Muro, S.; Hirai, T.; Ito, I.; et al. Directed Induction of Functional Multi-ciliated Cells in Proximal Airway Epithelial Spheroids from Human Pluripotent Stem Cells. Stem Cell Rep. 2016, 6, 18–25. [Google Scholar] [CrossRef]
- McCauley, K.B.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Ching, R.H.H.; Chan, S.K.H.; Nicholls, J.M.; Sachs, N.; Clevers, H.; Peiris, J.S.M.; Chan, M.C.W. Tropism, replication competence, and innate immune responses of influenza virus: An analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med. 2018, 6, 846–854. [Google Scholar] [CrossRef]
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.Y.; Chu, H.; Poon, V.K.M.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827. [Google Scholar] [CrossRef]
- Tan, Q.; Choi, K.M.; Sicard, D.; Tschumperlin, D.J. Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 2017, 113, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Huang, S.X.; de Carvalho, A.L.R.T.; Ho, S.H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.L.; Bhattacharya, J.; et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Porotto, M.; Ferren, M.; Chen, Y.W.; Siu, Y.; Makhsous, N.; Rima, B.; Briese, T.; Greninger, A.L.; Snoeck, H.W.; Moscona, A. Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids. MBio 2019, 10, e00723-19. [Google Scholar] [CrossRef] [PubMed]
- Evans, K.V.; Lee, J.H. Alveolar wars: The rise of in vitro models to understand human lung alveolar maintenance, regeneration, and disease. Stem Cells Transl. Med. 2020, 9, 867–881. [Google Scholar] [CrossRef]
- Liao, D.; Li, H. Dissecting the niche for alveolar type II cells with alveolar organoids. Front. Cell Dev. Biol. 2020, 8, 419. [Google Scholar] [CrossRef]
- Nikolić, M.Z.; Caritg, O.; Jeng, Q.; Johnson, J.A.; Sun, D.; Howell, K.J.; Brady, J.L.; Laresgoiti, U.; Allen, G.; Butler, R.; et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife 2017, 6, e26575. [Google Scholar] [CrossRef]
- Beers, M.F.; Moodley, Y. When is an alveolar type 2 cell an alveolar type 2 cell? A conundrum for lung stem cell biology and regenerative medicine. Am. J. Respir. Cell Mol. Biol. 2017, 57, 18–27. [Google Scholar] [CrossRef]
- Katsura, H.; Sontake, V.; Tata, A.; Kobayashi, Y.; Edwards, C.E.; Heaton, B.E.; Konkimalla, A.; Asakura, T.; Mikami, Y.; Fritch, E.J.; et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2 mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 2020, in press. [Google Scholar] [CrossRef]
- Youk, J.; Kim, T.; Evans, K.V.; Jeong, Y.I.; Hur, Y.; Hong, S.P.; Kim, J.H.; Yi, K.; Kim, S.Y.; Na, K.J.; et al. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell 2020, in press. [Google Scholar] [CrossRef]
- Yang, Q.; Oost, K.C.; Liberali, P. Engineering human knock-in organoids. Nat. Cell Biol. 2020, 22, 261–263. [Google Scholar] [CrossRef]
- Barkauskas, C.E.; Chung, M.I.; Fioret, B.; Gao, X.; Katsura, H.; Hogan, B.L.M. Lung organoids: Current uses and future promise. Development 2017, 144, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Danahay, H.; Pessotti, A.D.; Coote, J.; Montgomery, B.E.; Xia, D.; Wilson, A.; Yang, H.; Wang, Z.; Bevan, L.; Thomas, C.; et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 2015, 10, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.S.N.B.; Prota, L.F.M.; Morales, M.M.; Romero, P.V.; Zin, W.A.; Rocco, P.R.M. Understanding the mechanisms of lung mechanical stress. Braz. J. Med. Biol. Res. 2006, 39, 697–706. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dimova, S.; Vlaeminck, V.; Brewster, M.E.; Noppe, M.; Jorissen, M.; Augustijns, P. Stable ciliary activity in human nasal epithelial cells grown in a perfusion system. Int. J. Pharm. 2005, 292, 157–168. [Google Scholar] [CrossRef]
- Lee, J.H.; Bhang, D.H.; Beede, A.; Huang, T.L.; Stripp, B.R.; Bloch, K.D.; Wagers, A.J.; Tseng, Y.H.; Ryeom, S.; Kim, C.F. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 2014, 156, 440–455. [Google Scholar] [CrossRef]
- Barkauskas, C.E.; Cronce, M.J.; Rackley, C.R.; Bowie, E.J.; Keene, D.R.; Stripp, B.R.; Randell, S.H.; Noble, P.W.; Hogan, B.L.M. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investig. 2013, 123, 3025–3036. [Google Scholar] [CrossRef]
- Lechner, A.J.; Driver, I.H.; Lee, J.; Conroy, C.M.; Nagle, A.; Locksley, R.M.; Rock, J.R. Recruited monocytes and type 2 immunity promote lung regeneration following pneumonectomy. Cell Stem Cell 2017, 21, 120–134. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef]
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012, 4, 159ra147. [Google Scholar] [CrossRef]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.H.; Alves, S.E.; Salmon, M.; Ferrante, T.C.; Weaver, J.C.; et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef]
- Deinhardt-Emmer, S.; Rennert, K.; Schicke, E.; Cseresnyés, Z.; Windolph, M.; Nietzsche, S.; Heller, R.; Siwczak, F.; Haupt, K.F.; Carlstedt, S.; et al. Co-infection with Staphylococcus aureus after primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication 2020, 12, 025012. [Google Scholar] [CrossRef] [PubMed]
- Nawroth, J.C.; Lucchesi, C.; Cheng, D.; Shukla, A.; Ngyuen, J.; Shroff, T.; Varone, A.; Karalis, K.; Lee, H.-H.; Alves, S.; et al. A micro-engineered airway lung-chip models key features of viral-induced exacerbation of asthma. Am. J. Respir. Cell Mol. Biol. 2020, 63, 591–600. [Google Scholar] [CrossRef]
- Hao, W.; Bernard, K.; Patel, N.; Ulbrandt, N.; Feng, H.; Svabek, C.; Wilson, S.; Stracener, C.; Wang, K.; Suzich, J.; et al. Infection and propagation of human rhinovirus C in human airway epithelial cells. J. Virol. 2012, 86, 13524–13532. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Brockman-Schneider, R.; Bochkov, Y.A.; Pasic, T.R.; Gern, J.E. Biological characteristics and propagation of human rhinovirus-C in differentiated sinus epithelial cells. Virology 2013, 436, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Tapparel, C.; Sobo, K.; Constant, S.; Huang, S.; Van Belle, S.; Kaiser, L. Growth and characterization of different human rhinovirus C types in three-dimensional human airway epithelia reconstituted in vitro. Virology 2013, 446, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pyrc, K.; Sims, A.C.; Dijkman, R.; Jebbink, M.; Long, C.; Deming, D.; Donaldson, E.; Vabret, A.; Baric, R.; van der Hoek, L.; et al. Culturing the unculturable: Human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 2010, 84, 11255–11263. [Google Scholar] [CrossRef] [PubMed]
- Dijkman, R.; Koekkoek, S.M.; Molenkamp, R.; Schildgen, O.; van der Hoek, L. Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 2009, 83, 7739–7748. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Matrosovich, M.; Matrosovich, T.; Carr, J.; Roberts, N.A.; Klenk, H.D. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J. Virol. 2003, 77, 8418–8425. [Google Scholar] [CrossRef]
- Oh, D.Y.; Barr, I.G.; Mosse, J.A.; Laurie, K.L. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J. Clin. Microbiol. 2008, 46, 2189–2194. [Google Scholar] [CrossRef]
- Takada, K.; Kawakami, C.; Fan, S.; Chiba, S.; Zhong, G.; Gu, C.; Shimizu, K.; Takasaki, S.; Sakai-Tagawa, Y.; Lopes, T.J.S.; et al. A humanized MDCK cell line for the efficient isolation and propagation of human influenza viruses. Nat. Microbiol. 2019, 4, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wharton, S.A.; Whittaker, L.; Dai, M.; Ermetal, B.; Lo, J.; Pontoriero, A.; Baumeister, E.; Daniels, R.S.; McCauley, J.W. The characteristics and antigenic properties of recently emerged subclade 3C.3a and 3C.2a human influenza A(H3N2) viruses passaged in MDCK cells. Influenza Other Respir. Viruses 2017, 11, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.C.; Barclay, W.S.; Galiano, M.; Harvey, R. Passage of influenza A/H3N2 viruses in human airway cells removes artefactual variants associated with neuraminidase-mediated binding. J. Gen. Virol. 2020, 101, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Enkirch, T.; von Messling, V. Ferret models of viral pathogenesis. Virology 2015, 479, 259–270. [Google Scholar] [CrossRef]
- Chan, R.W.Y.; Chan, L.L.Y.; Mok, C.K.P.; Lai, J.; Tao, K.P.; Obadan, A.; Chan, M.C.W.; Perez, D.R.; Peiris, J.S.M.; Nicholls, J.M. Replication of H9 influenza viruses in the human ex vivo respiratory tract, and the influence of neuraminidase on virus release. Sci. Rep. 2017, 7, 6208. [Google Scholar] [CrossRef]
- Nicholls, J.M.; Chan, M.C.W.; Chan, W.Y.; Wong, H.K.; Cheung, C.Y.; Kwong, D.L.W.; Wong, M.P.; Chui, W.H.; Poon, L.L.M.; Tsao, S.W.; et al. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat. Med. 2007, 13, 147–149. [Google Scholar] [CrossRef]
- Weinheimer, V.K.; Becher, A.; Tönnies, M.; Holland, G.; Knepper, J.; Bauer, T.T.; Schneider, P.; Neudecker, J.; Rückert, J.C.; Szymanski, K.; et al. Influenza A viruses target type II pneumocytes in the human lung. J. Infect. Dis. 2012, 206, 1685–1694. [Google Scholar] [CrossRef]
- Hocke, A.C.; Becher, A.; Knepper, J.; Peter, A.; Holland, G.; Tönnies, M.; Bauer, T.T.; Schneider, P.; Neudecker, J.; Muth, D.; et al. Emerging human middle East respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am. J. Respir. Crit. Care Med. 2013, 188, 882–886. [Google Scholar] [CrossRef]
- Fischer, W.A., 2nd; King, L.S.; Lane, A.P.; Pekosz, A. Restricted replication of the live attenuated influenza A virus vaccine during infection of primary differentiated human nasal epithelial cells. Vaccine 2015, 33, 4495–4504. [Google Scholar] [CrossRef]
- Fischer, W.A., 2nd; Chason, K.D.; Brighton, M.; Jaspers, I. Live attenuated influenza vaccine strains elicit a greater innate immune response than antigenically-matched seasonal influenza viruses during infection of human nasal epithelial cell cultures. Vaccine 2014, 32, 1761–1767. [Google Scholar] [CrossRef]
- Wohlgemuth, N.; Ye, Y.; Fenstermacher, K.J.; Liu, H.; Lane, A.P.; Pekosz, A. The M2 protein of live, attenuated influenza vaccine encodes a mutation that reduces replication in human nasal epithelial cells. Vaccine 2017, 35, 6691–6699. [Google Scholar] [CrossRef] [PubMed]
- Schaap-Nutt, A.; Scull, M.A.; Schmidt, A.C.; Murphy, B.R.; Pickles, R.J. Growth restriction of an experimental live attenuated human parainfluenza virus type 2 vaccine in human ciliated airway epithelium in vitro parallels attenuation in African green monkeys. Vaccine 2010, 28, 2788–2798. [Google Scholar] [CrossRef] [PubMed]
- Rostad, C.A.; Stobart, C.C.; Gilbert, B.E.; Pickles, R.J.; Hotard, A.L.; Meng, J.; Blanco, J.C.G.; Moin, S.M.; Graham, B.S.; Piedra, P.A.; et al. A recombinant respiratory syncytial virus vaccine candidate attenuated by a low-fusion F protein is immunogenic and protective against challenge in cotton rats. J. Virol. 2016, 90, 7508–7518. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.F.; Ikizler, M.R.; Gonzales, R.A.; Carroll, K.N.; Johnson, J.E.; Werkhaven, J.A. Growth of respiratory syncytial virus in primary epithelial cells from the human respiratory tract. J. Virol. 2005, 79, 8651–8654. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.R.; Flannery, B.; Thompson, M.G.; Gaglani, M.; Jackson, M.L.; Monto, A.S.; Nowalk, M.P.; Talbot, H.K.; Treanor, J.J.; Belongia, E.A.; et al. Seasonal effectiveness of live attenuated and inactivated influenza vaccine. Pediatrics 2016, 137, e20153279. [Google Scholar] [CrossRef]
- Gaglani, M.; Pruszynski, J.; Murthy, K.; Clipper, L.; Robertson, A.; Reis, M.; Chung, J.R.; Piedra, P.A.; Avadhanula, V.; Nowalk, M.P.; et al. Influenza vaccine effectiveness against 2009 pandemic influenza A(H1N1) virus differed by vaccine type during 2013–2014 in the United States. J. Infect. Dis. 2016, 213, 1546–1556. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Sokolow, L.Z.; Olsen, S.J.; Bresee, J.S.; Broder, K.R.; Karron, R.A. Prevention and control of influenza with vaccines: Recommendations of the advisory committee on immunization practices, United States, 2015–2016 influenza season. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 818–825. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Sokolow, L.Z.; Broder, K.R.; Olsen, S.J.; Karron, R.A.; Jernigan, D.B.; Bresee, J.S. Prevention and control of seasonal influenza with vaccines. MMWR Recomm. Rep. 2016, 65, 1–54. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Sokolow, L.Z.; Broder, K.R.; Walter, E.B.; Bresee, J.S.; Fry, A.M.; Jernigan, D.B. Prevention and control of seasonal influenza with vaccines: Recommendations of the advisory committee on immunization practices—United States, 2017–2018 influenza season. MMWR Recomm. Rep. 2017, 66, 1–20. [Google Scholar] [CrossRef]
- Boda, B.; Benaoudia, S.; Huang, S.; Bonfante, R.; Wiszniewski, L.; Tseligka, E.D.; Tapparel, C.; Constant, S. Antiviral drug screening by assessing epithelial functions and innate immune responses in human 3D airway epithelium model. Antivir. Res. 2018, 156, 72–79. [Google Scholar] [CrossRef]
- Toots, M.; Yoon, J.J.; Cox, R.M.; Hart, M.; Sticher, Z.M.; Makhsous, N.; Plesker, R.; Barrena, A.H.; Reddy, P.G.; Mitchell, D.G.; et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019, 11, eaax5866. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H.; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef] [PubMed]
- DeVincenzo, J.; Tait, D.; Efthimiou, J.; Mori, J.; Kim, Y.I.; Thomas, E.; Wilson, L.; Harland, R.; Mathews, N.; Cockerill, S.; et al. A randomized, placebo-controlled, respiratory syncytial virus human challenge study of the antiviral efficacy, safety, and pharmacokinetics of RV521, an inhibitor of the RSV-F protein. Antimicrob. Agents Chemother. 2020, 64, e01884-19. [Google Scholar] [CrossRef] [PubMed]
- Memoli, M.J.; Czajkowski, L.; Reed, S.; Athota, R.; Bristol, T.; Proudfoot, K.; Fargis, S.; Stein, M.; Dunfee, R.L.; Shaw, P.A.; et al. Validation of the wild-type influenza A human challenge model H1N1pdMIST: An A(H1N1)pdm09 dose-finding investigational new drug study. Clin. Infect. Dis. 2015, 60, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Peretz, J.; Pekosz, A.; Lane, A.P.; Klein, S.L. Estrogenic compounds reduce influenza A virus replication in primary human nasal epithelial cells derived from female, but not male, donors. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 310, L415–L425. [Google Scholar] [CrossRef] [PubMed]
- Casimir, G.J.; Lefèvre, N.; Corazza, F.; Duchateau, J. Sex and inflammation in respiratory diseases: A clinical viewpoint. Biol. Sex. Differ. 2013, 4, 16. [Google Scholar] [CrossRef]
- Huang, C.G.; Lee, L.A.; Wu, Y.C.; Hsiao, M.J.; Horng, J.T.; Kuo, R.L.; Huang, C.H.; Lin, Y.C.; Tsao, K.C.; Chen, M.C.; et al. A pilot study on primary cultures of human respiratory tract epithelial cells to predict patients’ responses to H7N9 infection. Oncotarget 2018, 9, 14492–14508. [Google Scholar] [CrossRef]
- Honce, R.; Karlsson, E.A.; Wohlgemuth, N.; Estrada, L.D.; Meliopoulos, V.A.; Yao, J.; Schultz-Cherry, S. Obesity-related microenvironment promotes emergence of virulent influenza virus strains. MBio 2020, 11, e03341-19. [Google Scholar] [CrossRef]
- Zhang, L.; Bukreyev, A.; Thompson, C.I.; Watson, B.; Peeples, M.E.; Collins, P.L.; Pickles, R.J. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 2005, 79, 1113–1124. [Google Scholar] [CrossRef]
- Milewska, A.; Kula-Pacurar, A.; Wadas, J.; Suder, A.; Szczepanski, A.; Dabrowska, A.; Owczarek, K.; Marcello, A.; Ochman, M.; Stacel, T.; et al. Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Human Respiratory Epithelium. J. Virol. 2020, 94, e00957-20. [Google Scholar] [CrossRef]
- Griggs, T.F.; Bochkov, Y.A.; Basnet, S.; Pasic, T.R.; Brockman-Schneider, R.A.; Palmenberg, A.C.; Gern, J.E. Rhinovirus C targets ciliated airway epithelial cells. Respir. Res. 2017, 18, 84. [Google Scholar] [CrossRef]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.-D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 2004, 101, 4620–4624. [Google Scholar] [CrossRef] [PubMed]
- Warner, S.M.; Wiehler, S.; Michi, A.N.; Proud, D. Rhinovirus replication and innate immunity in highly differentiated human airway epithelial cells. Respir. Res. 2019, 20, 150. [Google Scholar] [CrossRef] [PubMed]
- Ehre, C.; Worthington, E.N.; Liesman, R.M.; Grubb, B.R.; Barbier, D.; O’Neal, W.K.; Sallenave, J.M.; Pickles, R.J.; Boucher, R.C. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc. Natl. Acad. Sci. USA 2012, 109, 16528–16533. [Google Scholar] [CrossRef] [PubMed]
- McAuley, J.L.; Corcilius, L.; Tan, H.X.; Payne, R.J.; McGuckin, M.A.; Brown, L.E. The cell surface mucin MUC1 limits the severity of influenza A virus infection. Mucosal Immunol. 2017, 10, 1581–1593. [Google Scholar] [CrossRef] [PubMed]
- Holly, M.K.; Diaz, K.; Smith, J.G. Defensins in viral infection and pathogenesis. Annu. Rev. Virol. 2017, 4, 369–391. [Google Scholar] [CrossRef] [PubMed]
- Doss, M.; White, M.R.; Tecle, T.; Gantz, D.; Crouch, E.C.; Jung, G.; Ruchala, P.; Waring, A.J.; Lehrer, R.I.; Hartshorn, K.L. Interactions of alpha-, beta-, and theta-defensins with influenza A virus and surfactant protein D. J. Immunol. 2009, 182, 7878–7887. [Google Scholar] [CrossRef]
- Kota, S.; Sabbah, A.; Chang, T.H.; Harnack, R.; Xiang, Y.; Meng, X.; Bose, S. Role of human beta-defensin-2 during tumor necrosis factor-alpha/NF-kappaB-mediated innate antiviral response against human respiratory syncytial virus. J. Biol. Chem. 2008, 283, 22417–22429. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, J.; Zhang, K.; Chu, H.; Liu, D.; Poon, V.K.M.; Chan, C.C.S.; Leung, H.C.; Fai, N.; Lin, Y.P.; et al. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016, 6, 22008. [Google Scholar] [CrossRef]
- Bertram, S.; Glowacka, I.; Müller, M.A.; Lavender, H.; Gnirss, K.; Nehlmeier, I.; Niemeyer, D.; He, Y.; Simmons, G.; Drosten, C.; et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011, 85, 13363–13372. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Lan, Q.; Su, S.; Wang, X.; Xu, W.; Liu, Z.; Zhu, Y.; Wang, Q.; Lu, L.; Jiang, S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Jaspers, I. Respiratory protease/antiprotease balance determines susceptibility to viral infection and can be modified by nutritional antioxidants. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L1189–L1201. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Ami, Y.; Tahara, M.; Kubota, T.; Anraku, M.; Abe, M.; Nakajima, N.; Sekizuka, T.; Shirato, K.; Suzaki, Y.; et al. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J. Virol. 2014, 88, 5608–5616. [Google Scholar] [CrossRef]
- Hatesuer, B.; Bertram, S.; Mehnert, N.; Bahgat, M.M.; Nelson, P.S.; Pöhlmann, S.; Schughart, K. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013, 9, e1003774. [Google Scholar] [CrossRef]
- Kühn, N.; Bergmann, S.; Kösterke, N.; Lambertz, R.L.O.; Keppner, A.; van den Brand, J.M.A.; Pöhlmann, S.; Weiß, S.; Hummler, E.; Hatesuer, B.; et al. The proteolytic activation of (H3N2) influenza A virus hemagglutinin is facilitated by different type II transmembrane serine proteases. J. Virol. 2016, 90, 4298–4307. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef]
- Zhou, Y.; Vedantham, P.; Lu, K.; Agudelo, J.; Carrion, R., Jr.; Nunneley, J.W.; Barnard, D.; Pöhlmann, S.; McKerrow, J.H.; Renslo, A.R.; et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015, 116, 76–84. [Google Scholar] [CrossRef]
- Zhou, N.; Pan, T.; Zhang, J.; Li, Q.; Zhang, X.; Bai, C.; Huang, F.; Peng, T.; Zhang, J.; Liu, C.; et al. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biol. Chem. 2016, 291, 9218–9232. [Google Scholar] [CrossRef]
- Shirato, K.; Kawase, M.; Matsuyama, S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 2018, 517, 9–15. [Google Scholar] [CrossRef]
- Xu, X.; Greenland, J.R.; Gotts, J.E.; Matthay, M.A.; Caughey, G.H. Cathepsin L helps to defend mice from infection with influenza, A. PLoS ONE 2016, 11, e0164501. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.D.; Ha, S.D.; Haeryfar, S.M.M.; Barr, S.D.; Kim, S.O. Cathepsin B plays a key role in optimal production of the influenza A virus. J. Virol. Antivir. Res. 2018, 2018, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Garten, W.; Braden, C.; Arendt, A.; Peitsch, C.; Baron, J.; Lu, Y.; Pawletko, K.; Hardes, K.; Steinmetzer, T.; Böttcher-Friebertshäuser, E. Influenza virus activating host proteases: Identification, localization and inhibitors as potential therapeutics. Eur. J. Cell Biol. 2015, 94, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Tarnow, C.; Engels, G.; Arendt, A.; Schwalm, F.; Sediri, H.; Preuss, A.; Nelson, P.S.; Garten, W.; Klenk, H.D.; Gabriel, G.; et al. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J. Virol. 2014, 88, 4744–4751. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, A.; Gravel, É.; Cloutier, A.; Marois, I.; Colombo, É.; Désilets, A.; Verreault, C.; Leduc, R.; Marsault, É.; Richter, M.V. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J. Virol. 2013, 87, 4237–4251. [Google Scholar] [CrossRef]
- Lambertz, R.L.O.; Gerhauser, I.; Nehlmeier, I.; Leist, S.R.; Kollmus, H.; Pöhlmann, S.; Schughart, K. Tmprss2 knock-out mice are resistant to H10 influenza A virus pathogenesis. J. Gen. Virol. 2019, 100, 1073–1078. [Google Scholar] [CrossRef]
- Rojas-Quintero, J.; Wang, X.; Tipper, J.; Burkett, P.R.; Zuñiga, J.; Ashtekar, A.R.; Polverino, F.; Rout, A.; Yambayev, I.; Hernández, C.; et al. Matrix metalloproteinase-9 deficiency protects mice from severe influenza A viral infection. JCI Insight 2018, 3, e99022. [Google Scholar] [CrossRef]
- Dabo, A.J.; Cummins, N.; Eden, E.; Geraghty, P. Matrix Metalloproteinase 9 exerts antiviral activity against respiratory syncytial virus. PLoS ONE 2015, 10, e0135970. [Google Scholar] [CrossRef]
- Kong, M.Y.F.; Whitley, R.J.; Peng, N.; Oster, R.; Schoeb, T.R.; Sullender, W.; Ambalavanan, N.; Clancy, J.P.; Gaggar, A.; Blalock, J.E. Matrix Metalloproteinase-9 mediates RSV infection in vitro and in vivo. Viruses 2015, 7, 4230–4253. [Google Scholar] [CrossRef]
- Dittmann, M.; Hoffmann, H.H.; Scull, M.A.; Gilmore, R.H.; Bell, K.L.; Ciancanelli, M.; Wilson, S.J.; Crotta, S.; Yu, Y.; Flatley, B.; et al. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell. 2015, 160, 631–643. [Google Scholar] [CrossRef]
- Wakabayashi, H.; Oda, H.; Yamauchi, K.; Abe, F. Lactoferrin for prevention of common viral infections. J. Infect. Chemother. 2014, 20, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Kesimer, M.; Scull, M.; Brighton, B.; DeMaria, G.; Burns, K.; O’Neal, W.; Pickles, R.J.; Sheehan, J.K. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: A possible role in innate defense. FASEB J. 2009, 23, 1858–1868. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [PubMed]
- Button, B.; Cai, L.H.; Ehre, C.; Kesimer, M.; Hill, D.B.; Sheehan, J.K.; Boucher, R.C.; Rubinstein, M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science 2012, 337, 937–941. [Google Scholar] [CrossRef]
- Song, Y.; Namkung, W.; Nielson, D.W.; Lee, J.-W.; Finkbeiner, W.E.; Verkman, A.S. Airway surface liquid depth measured in ex vivo fragments of pig and human trachea: Dependence on Na+ and Cl- channel function. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 297, L1131–L1140. [Google Scholar] [CrossRef]
- Jayaraman, S.; Song, Y.; Vetrivel, L.; Shankar, L.; Verkman, A.S. Noninvasive in vivo fluorescence measurement of airway-surface liquid depth, salt concentration, and pH. J. Clin. Investig. 2001, 107, 317–324. [Google Scholar] [CrossRef]
- Boucher, R.C. Airway surface dehydration in cystic fibrosis: Pathogenesis and therapy. Annu. Rev. Med. 2007, 58, 157–170. [Google Scholar] [CrossRef]
- Chen, E.Y.T.; Yang, N.; Quinton, P.M.; Chin, W.-C. A new role for bicarbonate in mucus formation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2010, 299, L542–L549. [Google Scholar] [CrossRef]
- Tang, X.X.; Ostedgaard, L.S.; Hoegger, M.J.; Moninger, T.O.; Karp, P.H.; McMenimen, J.D.; Choudhury, B.; Varki, A.; Stoltz, D.A.; Welsh, M.J. Acidic pH increases airway surface liquid viscosity in cystic fibrosis. J. Clin. Investig. 2016, 126, 879–891. [Google Scholar] [CrossRef]
- Fischer, H.; Widdicombe, J.H. Mechanisms of acid and base secretion by the airway epithelium. J. Membr. Biol. 2006, 211, 139–150. [Google Scholar] [CrossRef]
- Ermund, A.; Meiss, L.N.; Rodriguez-Pineiro, A.M.; Bähr, A.; Nilsson, H.E.; Trillo-Muyo, S.; Ridley, C.; Thornton, D.J.; Wine, J.J.; Hebert, H.; et al. The normal trachea is cleaned by MUC5B mucin bundles from the submucosal glands coated with the MUC5AC mucin. Biochem. Biophys. Res. Commun. 2017, 492, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Widdicombe, J.H.; Wine, J.J. Airway gland structure and function. Physiol. Rev. 2015, 95, 1241–1319. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Chen, G.; Subramani, D.B.; Wolf, M.; Gilmore, R.C.; Kato, T.; Radicioni, G.; Kesimer, M.; Chua, M.; Dang, H.; et al. Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am. J. Respir. Crit. Care Med. 2019, 199, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Amini, S.E.; Gouyer, V.; Portal, C.; Gottrand, F.; Desseyn, J.L. Muc5b is mainly expressed and sialylated in the nasal olfactory epithelium whereas Muc5ac is exclusively expressed and fucosylated in the nasal respiratory epithelium. Histochem. Cell Biol. 2019, 152, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.G.; Livraghi-Butrico, A.; Fletcher, A.A.; McElwee, M.M.; Evans, S.E.; Boerner, R.M.; Alexander, S.N.; Bellinghausen, L.K.; Song, A.S.; Petrova, Y.M.; et al. Muc5b is required for airway defence. Nature 2014, 505, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Hancock, L.A.; Hennessy, C.E.; Solomon, G.M.; Dobrinskikh, E.; Estrella, A.; Hara, N.; Hill, D.B.; Kissner, W.J.; Markovetz, M.R.; Grove Villalon, D.E.; et al. Muc5b overexpression causes mucociliary dysfunction and enhances lung fibrosis in mice. Nat. Commun. 2018, 9, 5363. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Y.; Qu, D.; Yu, J.; Yang, J. The possible pathogenesis of idiopathic pulmonary fibrosis considering MUC5B. Biomed. Res. Int. 2019, 2019, 9712464. [Google Scholar] [CrossRef]
- Ridley, C.; Thornton, D.J. Mucins: The frontline defence of the lung. Biochem. Soc. Trans. 2018, 46, 1099–1106. [Google Scholar] [CrossRef]
- Evans, C.M.; Raclawska, D.S.; Ttofali, F.; Liptzin, D.R.; Fletcher, A.A.; Harper, D.N.; McGing, M.A.; McElwee, M.M.; Williams, O.W.; Sanchez, E.; et al. The polymeric mucin Muc5ac is required for allergic airway hyperreactivity. Nat. Commun. 2015, 6, 6281. [Google Scholar] [CrossRef]
- Lachowicz-Scroggins, M.E.; Yuan, S.; Kerr, S.C.; Dunican, E.M.; Yu, M.; Carrington, S.D.; Fahy, J.V. Abnormalities in MUC5AC and MUC5B protein in airway mucus in asthma. Am. J. Respir. Crit. Care Med. 2016, 194, 1296–1299. [Google Scholar] [CrossRef]
- Bonser, L.R.; Erle, D.J. Airway mucus and asthma: The role of MUC5AC and MUC5B. J. Clin. Med. Res. 2017, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Peñia, M.T.; Aujla, P.K.; Zudaire, E.; Watson, A.M.; Przygodzki, R.; Zalzal, G.H.; Rose, M.C. Localization and expression of MUC5B and MUC7 mucins in pediatric sinus mucosa. Ann. Otol. Rhinol. Laryngol. 2007, 116, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Antón, A.; Debolós, C.; Garrido, M.; Roca-Ferrer, J.; Barranco, C.; Alobid, I.; Xaubet, A.; Picado, C.; Mullol, J. Mucin genes have different expression patterns in healthy and diseased upper airway mucosa. Clin. Exp. Allergy 2006, 36, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.J.; Song, K.S. Effect of MUC8 on airway inflammation: A friend or a foe? J. Clin. Med. Res. 2018, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.J.; Jung, M.S.; Ahn, D.W.; Choi, J.K.; Ock, M.S.; Kim, K.S.; Yoon, J.H.; Song, E.J.; Song, K.S. Silencing of MUC8 by siRNA increases P2Y₂-induced airway inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L495–L502. [Google Scholar] [CrossRef] [PubMed]
- van Putten, J.P.M.; Strijbis, K. Transmembrane mucins: Signaling receptors at the intersection of inflammation and cancer. J. Innate Immun. 2017, 9, 281–299. [Google Scholar] [CrossRef]
- Gipson, I.K.; Spurr-Michaud, S.; Tisdale, A.; Menon, B.B. Comparison of the transmembrane mucins MUC1 and MUC16 in epithelial barrier function. PLoS ONE 2014, 9, e100393. [Google Scholar] [CrossRef]
- Ueno, K.; Koga, T.; Kato, K.; Golenbock, D.T.; Gendler, S.J.; Kai, H.; Kim, K.C. MUC1 mucin is a negative regulator of toll-like receptor signaling. Am. J. Respir. Cell Mol. Biol. 2008, 38, 263–268. [Google Scholar] [CrossRef]
- Kato, K.; Lillehoj, E.P.; Lu, W.; Kim, K.C. MUC1: The first respiratory mucin with an anti-inflammatory function. J. Clin. Med. Res. 2017, 6, 110. [Google Scholar] [CrossRef]
- Mahanta, S.; Fessler, S.P.; Park, J.; Bamdad, C. A minimal fragment of MUC1 mediates growth of cancer cells. PLoS ONE 2008, 3, e2054. [Google Scholar] [CrossRef]
- Higuchi, T.; Orita, T.; Nakanishi, S.; Katsuya, K.; Watanabe, H.; Yamasaki, Y.; Waga, I.; Nanayama, T.; Yamamoto, Y.; Munger, W.; et al. Molecular cloning, genomic structure, and expression analysis of MUC20, a novel mucin protein, up-regulated in injured kidney. J. Biol. Chem. 2004, 279, 1968–1979. [Google Scholar] [CrossRef] [PubMed]
- Walters, R.W.; Pilewski, J.M.; Chiorini, J.A.; Zabner, J. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J. Biol. Chem. 2002, 277, 23709–23713. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P. Mucins: A biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta 2015, 1850, 236–252. [Google Scholar] [CrossRef] [PubMed]
- Priyadharshini, V.S.; Ramírez-Jiménez, F.; Molina-Macip, M.; Renteria-Rosales, C.; Santiago-Cruz, J.; Zarate-Segura, P.; Lara-Padilla, E.; Teran, L.M. Human Neutrophil Defensin-1, -3, and -4 are elevated in nasal aspirates from children with naturally occurring adenovirus infection. Can. Respir. J. 2018, 2018, 1038593. [Google Scholar] [CrossRef]
- Rohde, G.; Message, S.D.; Haas, J.J.; Kebadze, T.; Parker, H.; Laza-Stanca, V.; Khaitov, M.R.; Kon, O.M.; Stanciu, L.A.; Mallia, P.; et al. CXC chemokines and antimicrobial peptides in rhinovirus-induced experimental asthma exacerbations. Clin. Exp. Allergy 2014, 44, 930–939. [Google Scholar] [CrossRef]
- Gu, J.; Huang, Y. β-Defensin-2 is overexpressed in human vocal cord polyps. Eur. Arch. Otorhinolaryngol. 2017, 274, 901–907. [Google Scholar] [CrossRef]
- Taylor, K.; Clarke, D.J.; McCullough, B.; Chin, W.; Seo, E.; Yang, D.; Oppenheim, J.; Uhrin, D.; Govan, J.R.W.; Campopiano, D.J.; et al. Analysis and separation of residues important for the chemoattractant and antimicrobial activities of beta-defensin 3. J. Biol. Chem. 2008, 283, 6631–6639. [Google Scholar] [CrossRef]
- Funderburg, N.T.; Jadlowsky, J.K.; Lederman, M.M.; Feng, Z.; Weinberg, A.; Sieg, S.F. The Toll-like receptor 1/2 agonists Pam(3) CSK(4) and human β-defensin-3 differentially induce interleukin-10 and nuclear factor-κB signalling patterns in human monocytes. Immunology 2011, 134, 151–160. [Google Scholar] [CrossRef]
- Nagaoka, I.; Niyonsaba, F.; Tsutsumi-Ishii, Y.; Tamura, H.; Hirata, M. Evaluation of the effect of human beta-defensins on neutrophil apoptosis. Int. Immunol. 2008, 20, 543–553. [Google Scholar] [CrossRef]
- Semple, F.; Webb, S.; Li, H.-N.; Patel, H.B.; Perretti, M.; Jackson, I.J.; Gray, M.; Davidson, D.J.; Dorin, J.R. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur. J. Immunol. 2010, 40, 1073–1078. [Google Scholar] [CrossRef]
- Shen, Z.; Zhou, Y.; Qu, L.; Lei, H. ATP serves an anti-inflammatory role by enhancing β-defensin-2 response in acute pneumonia of rat. Biomed. Rep. 2017, 6, 649–653. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cui, D.; Lyu, J.; Li, H.; Lei, L.; Bian, T.; Li, L.; Yan, F. Human β-defensin 3 inhibits periodontitis development by suppressing inflammatory responses in macrophages. Mol. Immunol. 2017, 91, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Semple, F.; Dorin, J.R. β-Defensins: Multifunctional modulators of infection, inflammation and more? J. Innate Immun. 2012, 4, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Cole, A.M.; Selsted, M.E. θ-Defensins: Cyclic peptides with endless potential. J. Biol. Chem. 2012, 287, 27014–27019. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a005058. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Zhang, K.; Chen, J. Role of integrins in regulating proteases to mediate extracellular matrix remodeling. Cancer Microenviron. 2012, 5, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Knaapi, J.; Kiviranta, R.; Laine, J.; Kääpä, P.; Lukkarinen, H. Cathepsin K overexpression modifies lung development in newborn mice. Pediatr. Pulmonol. 2015, 50, 164–172. [Google Scholar] [CrossRef]
- Abboud, R.T.; Vimalanathan, S. Pathogenesis of COPD. Part, I. The role of protease-antiprotease imbalance in emphysema. Int. J. Tuberc. Lung Dis. 2008, 12, 361–367. [Google Scholar]
- Ashley, S.L.; Xia, M.; Murray, S.; O’Dwyer, D.N.; Grant, E.; White, E.S.; Flaherty, K.R.; Martinez, F.J.; Moore, B.B. Six-SOMAmer index relating to immune, protease and angiogenic functions predicts progression in IPF. PLoS ONE 2016, 11, e0159878. [Google Scholar] [CrossRef]
- Nikaido, T.; Tanino, Y.; Wang, X.; Sato, Y.; Togawa, R.; Kikuchi, M.; Misa, K.; Saito, K.; Fukuhara, N.; Kawamata, T.; et al. Serum decorin is a potential prognostic biomarker in patients with acute exacerbation of idiopathic pulmonary fibrosis. J. Thorac. Dis. 2018, 10, 5346–5358. [Google Scholar] [CrossRef]
- Kehlet, S.N.; Bager, C.L.; Willumsen, N.; Dasgupta, B.; Brodmerkel, C.; Curran, M.; Brix, S.; Leeming, D.J.; Karsdal, M.A. Cathepsin-S degraded decorin are elevated in fibrotic lung disorders—Development and biological validation of a new serum biomarker. BMC Pulm. Med. 2017, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Zou, C.; Ge, W.; Liu, Y.; Hu, B.; Wang, J.; Lin, B.; Li, Y.; Ma, E. A novel cathepsin L inhibitor prevents the progression of idiopathic pulmonary fibrosis. Bioorg. Chem. 2020, 94, 103417. [Google Scholar] [CrossRef] [PubMed]
- Weldon, S.; McNally, P.; McAuley, D.F.; Oglesby, I.K.; Wohlford-Lenane, C.L.; Bartlett, J.A.; Scott, C.J.; McElvaney, N.G.; Greene, C.M.; McCray, P.B., Jr.; et al. miR-31 dysregulation in cystic fibrosis airways contributes to increased pulmonary cathepsin S production. Am. J. Respir. Crit. Care Med. 2014, 190, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Small, D.M.; Brown, R.R.; Doherty, D.F.; Abladey, A.; Zhou-Suckow, Z.; Delaney, R.J.; Kerrigan, L.; Dougan, C.M.; Borensztajn, K.S.; Holsinger, L.; et al. Targeting of cathepsin S reduces cystic fibrosis-like lung disease. Eur. Respir. J. 2019, 53, 1801523. [Google Scholar] [CrossRef] [PubMed]
- Laguna, T.A.; Williams, C.B.; Nunez, M.G.; Welchlin-Bradford, C.; Moen, C.E.; Reilly, C.S.; Wendt, C.H. Biomarkers of inflammation in infants with cystic fibrosis. Respir. Res. 2018, 19, 6. [Google Scholar] [CrossRef]
- Nakajima, T.; Nakamura, H.; Owen, C.A.; Yoshida, S.; Tsuduki, K.; Chubachi, S.; Shirahata, T.; Mashimo, S.; Nakamura, M.; Takahashi, S.; et al. Plasma cathepsin S and cathepsin S/cystatin C ratios are potential biomarkers for COPD. Dis. Markers 2016, 2016, 4093870. [Google Scholar] [CrossRef]
- Zhou, P.P.; Zhang, W.Y.; Li, Z.F.; Chen, Y.R.; Kang, X.C.; Jiang, Y.X. Association between SNPs in the promoter region in cathepsin S and risk of asthma in Chinese Han population. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2070–2076. [Google Scholar]
- Craig, V.J.; Zhang, L.; Hagood, J.S.; Owen, C.A. Matrix metalloproteinases as therapeutic targets for idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2015, 53, 585–600. [Google Scholar] [CrossRef]
- Yamashita, C.M.; Radisky, D.C.; Aschner, Y.; Downey, G.P. The importance of matrix metalloproteinase-3 in respiratory disorders. Expert Rev. Respir. Med. 2014, 8, 411–421. [Google Scholar] [CrossRef]
- McKeown, S.; Richter, A.G.; O’Kane, C.; McAuley, D.F.; Thickett, D.R. MMP expression and abnormal lung permeability are important determinants of outcome in IPF. Eur. Respir. J. 2009, 33, 77–84. [Google Scholar] [CrossRef]
- Navratilova, Z.; Kolek, V.; Petrek, M. Matrix Metalloproteinases and their inhibitors in chronic obstructive pulmonary disease. Arch. Immunol. Ther. Exp. 2016, 64, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Grzela, K.; Litwiniuk, M.; Zagorska, W.; Grzela, T. Airway remodeling in chronic obstructive pulmonary disease and asthma: The role of Matrix Metalloproteinase-9. Arch. Immunol. Ther. Exp. 2016, 64, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Chokki, M.; Yamamura, S.; Eguchi, H.; Masegi, T.; Horiuchi, H.; Tanabe, H.; Kamimura, T.; Yasuoka, S. Human airway trypsin-like protease increases mucin gene expression in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2004, 30, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Zuo, K.; Qi, Y.; Yuan, C.; Jiang, L.; Xu, P.; Hu, J.; Huang, M.; Li, J. Specifically targeting cancer proliferation and metastasis processes: The development of matriptase inhibitors. Cancer Metastasis Rev. 2019, 38, 507–524. [Google Scholar] [CrossRef]
- Shi, G.P.; Villadangos, J.A.; Dranoff, G.; Small, C.; Gu, L.; Haley, K.J.; Riese, R.; Ploegh, H.L.; Chapman, H.A. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity 1999, 10, 197–206. [Google Scholar] [CrossRef]
- Driessen, C.; Bryant, R.A.; Lennon-Duménil, A.M.; Villadangos, J.A.; Bryant, P.W.; Shi, G.P.; Chapman, H.A.; Ploegh, H.L. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J. Cell Biol. 1999, 147, 775–790. [Google Scholar] [CrossRef]
- Nakagawa, T.; Roth, W.; Wong, P.; Nelson, A.; Farr, A.; Deussing, J.; Villadangos, J.A.; Ploegh, H.; Peters, C.; Rudensky, A.Y. Cathepsin L: Critical role in Ii degradation and CD4 T cell selection in the thymus. Science 1998, 280, 450–453. [Google Scholar] [CrossRef]
- Zhang, N.; Gao, P.; Yin, B.; Li, J.; Wu, T.; Kuang, Y.; Wu, W.; Li, J. Cathepsin L promotes secretory IgA response by participating in antigen presentation pathways during Mycoplasma Hyopneumoniae infection. PLoS ONE 2019, 14, e0215408. [Google Scholar] [CrossRef]
- Sevenich, L.; Hagemann, S.; Stoeckle, C.; Tolosa, E.; Peters, C.; Reinheckel, T. Expression of human cathepsin L or human cathepsin V in mouse thymus mediates positive selection of T helper cells in cathepsin L knock-out mice. Biochimie 2010, 92, 1674–1680. [Google Scholar] [CrossRef]
- Matsumoto, F.; Saitoh, S.I.; Fukui, R.; Kobayashi, T.; Tanimura, N.; Konno, K.; Kusumoto, Y.; Akashi-Takamura, S.; Miyake, K. Cathepsins are required for Toll-like receptor 9 responses. Biochem. Biophys. Res. Commun. 2008, 367, 693–699. [Google Scholar] [CrossRef]
- Patel, S.; Homaei, A.; El-Seedi, H.R.; Akhtar, N. Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed. Pharmacother. 2018, 105, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.G.S.; Batista, P.R.; Shida, C.S.; Robert, C.H.; Bisch, P.M.; Pascutti, P.G. How does heparin prevent the pH inactivation of cathepsin B? Allosteric mechanism elucidated by docking and molecular dynamics. BMC Genom. 2010, 11, S5. [Google Scholar] [CrossRef] [PubMed]
- Greenlee, K.J.; Werb, Z.; Kheradmand, F. Matrix metalloproteinases in lung: Multiple, multifarious, and multifaceted. Physiol. Rev. 2007, 87, 69–98. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, K.; de Rooij, J.D.; Suita, S.; Rottier, R.; Tibboel, D.; de Krijger, R.R. Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases during normal human pulmonary development. Histopathology 2005, 47, 410–419. [Google Scholar] [CrossRef]
- Hendrix, A.Y.; Kheradmand, F. The role of Matrix Metalloproteinases in development, repair, and destruction of the lungs. Prog. Mol. Biol. Transl. Sci. 2017, 148, 1–29. [Google Scholar]
- Brilha, S.; Chong, D.L.W.; Khawaja, A.A.; Ong, C.W.M.; Guppy, N.J.; Porter, J.C.; Friedland, J.S. Integrin α2β1 expression regulates Matrix Metalloproteinase-1-dependent bronchial epithelial repair in pulmonary tuberculosis. Front. Immunol. 2018, 9, 1348. [Google Scholar] [CrossRef]
- Herrera, I.; Cisneros, J.; Maldonado, M.; Ramírez, R.; Ortiz-Quintero, B.; Anso, E.; Chandel, N.S.; Selman, M.; Pardo, A. Matrix metalloproteinase (MMP)-1 induces lung alveolar epithelial cell migration and proliferation, protects from apoptosis, and represses mitochondrial oxygen consumption. J. Biol. Chem. 2013, 288, 25964–25975. [Google Scholar] [CrossRef]
- Blázquez-Prieto, J.; López-Alonso, I.; Amado-Rodríguez, L.; Huidobro, C.; González-López, A.; Kuebler, W.M.; Albaiceta, G.M. Impaired lung repair during neutropenia can be reverted by matrix metalloproteinase-9. Thorax 2018, 73, 321–330. [Google Scholar] [CrossRef]
- Howell, C.; Smith, J.R.; Shute, J.K. Targeting matrix metalloproteinase-13 in bronchial epithelial repair. Clin. Exp. Allergy 2018, 48, 1214–1221. [Google Scholar] [CrossRef]
- Rohani, M.G.; Parks, W.C. Matrix remodeling by MMPs during wound repair. Matrix Biol. 2015, 44, 113–121. [Google Scholar] [CrossRef]
- Fujita, Y.; Kadota, T.; Araya, J.; Ochiya, T.; Kuwano, K. Extracellular vesicles: New players in lung immunity. Am. J. Respir. Cell Mol. Biol. 2018, 58, 560–565. [Google Scholar] [CrossRef]
- Mueller, S.K.; Nocera, A.L.; Bleier, B.S. Exosome function in aerodigestive mucosa. Nanomedicine 2018, 14, 269–277. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Schageman, J.; Zeringer, E.; Li, M.; Barta, T.; Lea, K.; Gu, J.; Magdaleno, S.; Setterquist, R.; Vlassov, A.V. The complete exosome workflow solution: From isolation to characterization of RNA cargo. Biomed. Res. Int. 2013, 2013, 253957. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, A.; Ahmad, T.; Agrawal, A.; Ghosh, B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J. Allergy Clin. Immunol. 2013, 131, 1194–1203. [Google Scholar] [CrossRef] [PubMed]
- Bourdonnay, E.; Zasłona, Z.; Penke, L.R.K.; Speth, J.M.; Schneider, D.J.; Przybranowski, S.; Swanson, J.A.; Mancuso, P.; Freeman, C.M.; Curtis, J.L.; et al. Transcellular delivery of vesicular SOCS proteins from macrophages to epithelial cells blunts inflammatory signaling. J. Exp. Med. 2015, 212, 729–742. [Google Scholar] [CrossRef]
- Torregrosa Paredes, P.; Esser, J.; Admyre, C.; Nord, M.; Rahman, Q.K.; Lukic, A.; Rådmark, O.; Grönneberg, R.; Grunewald, J.; Eklund, A.; et al. Bronchoalveolar lavage fluid exosomes contribute to cytokine and leukotriene production in allergic asthma. Allergy 2012, 67, 911–919. [Google Scholar] [CrossRef]
- Nocera, A.L.; Miyake, M.M.; Seifert, P.; Han, X.; Bleier, B.S. Exosomes mediate interepithelial transfer of functional P-glycoprotein in chronic rhinosinusitis with nasal polyps. Laryngoscope 2017, 127, E295–E300. [Google Scholar] [CrossRef] [PubMed]
- Wahlund, C.J.E.; Eklund, A.; Grunewald, J.; Gabrielsson, S. Pulmonary extracellular vesicles as mediators of local and systemic inflammation. Front. Cell Dev. Biol. 2017, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Harit, D.; Subramani, D.B.; Arora, H.; Kumar, P.A.; Lai, S.K. Influenza-binding antibodies immobilise influenza viruses in fresh human airway mucus. Eur. Respir. J. 2017, 49, 1601709. [Google Scholar] [CrossRef]
- Kobayashi, K.; Suzukawa, M.; Watanabe, K.; Arakawa, S.; Igarashi, S.; Asari, I.; Hebisawa, A.; Matsui, H.; Nagai, H.; Nagase, T.; et al. Secretory IgA accumulated in the airspaces of idiopathic pulmonary fibrosis and promoted VEGF, TGF-β and IL-8 production by A549 cells. Clin. Exp. Immunol. 2020, 199, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Cereser, L.; De Carli, M.; d’Angelo, P.; Zanelli, E.; Zuiani, C.; Girometti, R. High-resolution computed tomography findings in humoral primary immunodeficiencies and correlation with pulmonary function tests. World J. Radiol. 2018, 10, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Charlson, E.S.; Bittinger, K.; Haas, A.R.; Fitzgerald, A.S.; Frank, I.; Yadav, A.; Bushman, F.D.; Collman, R.G. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011, 184, 957–963. [Google Scholar] [CrossRef] [PubMed]
- de Steenhuijsen Piters, W.A.A.; Sanders, E.A.M.; Bogaert, D. The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140294. [Google Scholar] [CrossRef]
- Ling, Z.; Liu, X.; Luo, Y.; Yuan, L.; Nelson, K.E.; Wang, Y.; Xiang, C.; Li, L. Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genom. 2013, 14, 390. [Google Scholar] [CrossRef]
- Charlson, E.S.; Chen, J.; Custers-Allen, R.; Bittinger, K.; Li, H.; Sinha, R.; Hwang, J.; Bushman, F.D.; Collman, R.G. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS ONE 2010, 5, e15216. [Google Scholar] [CrossRef]
- Yi, H.; Yong, D.; Lee, K.; Cho, Y.J.; Chun, J. Profiling bacterial community in upper respiratory tracts. BMC Infect. Dis. 2014, 14, 583. [Google Scholar] [CrossRef]
- Charlson, E.S.; Bittinger, K.; Chen, J.; Diamond, J.M.; Li, H.; Collman, R.G.; Bushman, F.D. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS ONE 2012, 7, e42786. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; Walker, N.; Scales, B.S.; Beck, J.M.; Martinez, F.J.; Curtis, J.L.; Lama, V.N.; Huffnagle, G.B. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS ONE 2014, 9, e97214. [Google Scholar] [CrossRef]
- Venkataraman, A.; Bassis, C.M.; Beck, J.M.; Young, V.B.; Curtis, J.L.; Huffnagle, G.B.; Schmidt, T.M. Application of a neutral community model to assess structuring of the human lung microbiome. MBio 2015, 6, e02284-14. [Google Scholar] [CrossRef]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 2015, 6, e00037. [Google Scholar] [CrossRef] [PubMed]
- Marsh, R.L.; Kaestli, M.; Chang, A.B.; Binks, M.J.; Pope, C.E.; Hoffman, L.R.; Smith-Vaughan, H.C. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 2016, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- de Steenhuijsen Piters, W.A.A.; Heinonen, S.; Hasrat, R.; Bunsow, E.; Smith, B.; Suarez-Arrabal, M.-C.; Chaussabel, D.; Cohen, D.M.; Sanders, E.A.M.; Ramilo, O.; et al. Nasopharyngeal Microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am. J. Respir. Crit. Care Med. 2016, 194, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Korten, I.; Mika, M.; Klenja, S.; Kieninger, E.; Mack, I.; Barbani, M.T.; Gorgievski, M.; Frey, U.; Hilty, M.; Latzin, P. Interactions of respiratory viruses and the nasal microbiota during the first year of life in healthy infants. mSphere 2016, 1, e00312-16. [Google Scholar] [CrossRef]
- Toivonen, L.; Camargo, C.A., Jr.; Gern, J.E.; Bochkov, Y.A.; Mansbach, J.M.; Piedra, P.A.; Hasegawa, K. Association between rhinovirus species and nasopharyngeal microbiota in infants with severe bronchiolitis. J. Allergy Clin. Immunol. 2019, 143, 1925–1928. [Google Scholar] [CrossRef]
- Wen, Z.; Xie, G.; Zhou, Q.; Qiu, C.; Li, J.; Hu, Q.; Dai, W.; Li, D.; Zheng, Y.; Wen, F. Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. Biomed. Res. Int. 2018, 2018, 6362716. [Google Scholar] [CrossRef]
- Edouard, S.; Million, M.; Bachar, D.; Dubourg, G.; Michelle, C.; Ninove, L.; Charrel, R.; Raoult, D. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1725–1733. [Google Scholar] [CrossRef]
- Poole, K.; Meder, D.; Simons, K.; Müller, D. The effect of raft lipid depletion on microvilli formation in MDCK cells, visualized by atomic force microscopy. FEBS Lett. 2004, 565, 53–58. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Liu, G.; Jia, Y.; Yang, J.; Shi, J.; Dong, J.; Wei, J.; Liu, X. Characterization of air-liquid interface culture of A549 alveolar epithelial cells. Braz. J. Med. Biol. Res. 2017, 51, e6950. [Google Scholar] [CrossRef]
- Joshi, S.; Kumar, S.; Bafna, S.; Rachagani, S.; Wagner, K.U.; Jain, M.; Batra, S.K. Genetically engineered mucin mouse models for inflammation and cancer. Cancer Metastasis Rev. 2015, 34, 593–609. [Google Scholar] [CrossRef]
- Zanin, M.; Baviskar, P.; Webster, R.; Webby, R. The interaction between respiratory pathogens and mucus. Cell Host Microbe 2016, 19, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Gagneux, P.; Cheriyan, M.; Hurtado-Ziola, N.; van der Linden, E.C.M.B.; Anderson, D.; McClure, H.; Varki, A.; Varki, N.M. Human-specific regulation of alpha 2-6-linked sialic acids. J. Biol. Chem. 2003, 278, 48245–48250. [Google Scholar] [CrossRef]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.-J.; Chun, S.-M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef] [PubMed]
- Jose, S.S.; De Zuani, M.; Tidu, F.; Hortová Kohoutková, M.; Pazzagli, L.; Forte, G.; Spaccapelo, R.; Zelante, T.; Frič, J. Comparison of two human organoid models of lung and intestinal inflammation reveals Toll-like receptor signalling activation and monocyte recruitment. Clin. Transl. Immunol. 2020, 9, e1131. [Google Scholar] [CrossRef] [PubMed]
- Stonebraker, J.R.; Wagner, D.; Lefensty, R.W.; Burns, K.; Gendler, S.J.; Bergelson, J.M.; Boucher, R.C.; O’Neal, W.K.; Pickles, R.J. Glycocalyx restricts adenoviral vector access to apical receptors expressed on respiratory epithelium in vitro and in vivo: Role for tethered mucins as barriers to lumenal infection. J. Virol. 2004, 78, 13755–13768. [Google Scholar] [CrossRef] [PubMed]
- Zanin, M.; Marathe, B.; Wong, S.-S.; Yoon, S.-W.; Collin, E.; Oshansky, C.; Jones, J.; Hause, B.; Webby, R. Pandemic Swine H1N1 Influenza Viruses with Almost Undetectable Neuraminidase Activity Are Not Transmitted via Aerosols in Ferrets and Are Inhibited by Human Mucus but Not Swine Mucus. J. Virol. 2015, 89, 5935–5948. [Google Scholar] [CrossRef] [PubMed]
- Vahey, M.D.; Fletcher, D.A. Influenza A virus surface proteins are organized to help penetrate host mucus. eLife 2019, 8, e43764. [Google Scholar] [CrossRef] [PubMed]
- Markovetz, M.R.; Subramani, D.B.; Kissner, W.J.; Morrison, C.B.; Garbarine, I.C.; Ghio, A.; Ramsey, K.A.; Arora, H.; Kumar, P.; Nix, D.B.; et al. Endotracheal tube mucus as a source of airway mucus for rheological study. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019, 317, L498–L509. [Google Scholar] [CrossRef]
- Schuster, B.S.; Suk, J.S.; Woodworth, G.F.; Hanes, J. Nanoparticle diffusion in respiratory mucus from humans without lung disease. Biomaterials 2013, 34, 3439–3446. [Google Scholar] [CrossRef]
- Yuan, S.; Hollinger, M.; Lachowicz-Scroggins, M.E.; Kerr, S.C.; Dunican, E.M.; Daniel, B.M.; Ghosh, S.; Erzurum, S.C.; Willard, B.; Hazen, S.L.; et al. Oxidation increases mucin polymer cross-links to stiffen airway mucus gels. Sci. Transl. Med. 2015, 7, 276ra27. [Google Scholar] [CrossRef]
- Innes, A.L.; Carrington, S.D.; Thornton, D.J.; Kirkham, S.; Rousseau, K.; Dougherty, R.H.; Raymond, W.W.; Caughey, G.H.; Muller, S.J.; Fahy, J.V. Ex vivo sputum analysis reveals impairment of protease-dependent mucus degradation by plasma proteins in acute asthma. Am. J. Respir. Crit. Care Med. 2009, 180, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.B.; Vasquez, P.A.; Mellnik, J.; McKinley, S.A.; Vose, A.; Mu, F.; Henderson, A.G.; Donaldson, S.H.; Alexis, N.E.; Boucher, R.C.; et al. A biophysical basis for mucus solids concentration as a candidate biomarker for airways disease. PLoS ONE 2014, 9, e87681. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.L.; Lai, S.K. Tuning barrier properties of biological hydrogels. ACS Appl. Bio Mater. 2020, 3, 2875–2890. [Google Scholar] [CrossRef]
- Frickmann, H.; Jungblut, S.; Hirche, T.O.; Groß, U.; Kuhns, M.; Zautner, A.E. Spectrum of viral infections in patients with cystic fibrosis. Eur. J. Microbiol. Immunol. 2012, 2, 161–175. [Google Scholar] [CrossRef]
- Boucher, R.C. On the pathogenesis of acute exacerbations of mucoobstructive lung diseases. Ann. Am. Thorac. Soc. 2015, 12 (Suppl. 2), S160–S163. [Google Scholar]
- Duncan, G.A.; Jung, J.; Joseph, A.; Thaxton, A.L.; West, N.E.; Boyle, M.P.; Hanes, J.; Suk, J.S. Microstructural alterations of sputum in cystic fibrosis lung disease. JCI Insight 2016, 1, e88198. [Google Scholar] [CrossRef]
- Anderson, W.H.; Coakley, R.D.; Button, B.; Henderson, A.G.; Zeman, K.L.; Alexis, N.E.; Peden, D.B.; Lazarowski, E.R.; Davis, C.W.; Bailey, S.; et al. The relationship of mucus concentration (hydration) to mucus osmotic pressure and transport in chronic bronchitis. Am. J. Respir. Crit. Care Med. 2015, 192, 182–190. [Google Scholar] [CrossRef]
- Kesimer, M.; Ford, A.A.; Ceppe, A.; Radicioni, G.; Cao, R.; Davis, C.W.; Doerschuk, C.M.; Alexis, N.E.; Anderson, W.H.; Henderson, A.G.; et al. Airway mucin concentration as a marker of chronic bronchitis. N. Engl. J. Med. 2017, 377, 911–922. [Google Scholar] [CrossRef]
- Essaidi-Laziosi, M.; Brito, F.; Benaoudia, S.; Royston, L.; Cagno, V.; Fernandes-Rocha, M.; Piuz, I.; Zdobnov, E.; Huang, S.; Constant, S.; et al. Propagation of respiratory viruses in human airway epithelia reveals persistent virus-specific signatures. J. Allergy Clin. Immunol. 2018, 141, 2074–2084. [Google Scholar] [CrossRef]
- Lai, S.K.; Wang, Y.Y.; Wirtz, D.; Hanes, J. Micro- and macrorheology of mucus. Adv. Drug Deliv. Rev. 2009, 61, 86–100. [Google Scholar] [CrossRef]
- Bansil, R.; Turner, B.S. The biology of mucus: Composition, synthesis and organization. Adv. Drug Deliv. Rev. 2018, 124, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Duncan, G.A.; Jung, J.; Hanes, J.; Suk, J.S. The mucus barrier to inhaled gene therapy. Mol. Ther. 2016, 24, 2043–2053. [Google Scholar] [CrossRef] [PubMed]
- Huck, B.C.; Hartwig, O.; Biehl, A.; Schwarzkopf, K.; Wagner, C.; Loretz, B.; Murgia, X.; Lehr, C.-M. Macro- and microrheological properties of mucus surrogates in comparison to native intestinal and pulmonary mucus. Biomacromolecules 2019, 20, 3504–3512. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Ensign, L.M.; Boylan, N.J.; Schön, A.; Gong, X.; Yang, J.C.; Lamb, N.W.; Cai, S.; Yu, T.; Freire, E.; et al. Impact of surface Polyethylene Glycol (PEG) density on biodegradable nanoparticle transport in mucus ex vivo and distribution in vivo. ACS Nano 2015, 9, 9217–9227. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, K.R.; Reznikov, L.R. Strategies for measuring airway mucus and mucins. Respir. Res. 2019, 20, 261. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.A.; Rushton, Z.L.; Ehre, C. Mucin agarose gel electrophoresis: Western blotting for high-molecular-weight glycoproteins. J. Vis. Exp. 2016, 112, 54153. [Google Scholar] [CrossRef]
- Abdullah, L.H.; Wolber, C.; Kesimer, M.; Sheehan, J.K.; Davis, C.W. Studying mucin secretion from human bronchial epithelial cell primary cultures. Methods Mol. Biol. 2012, 842, 259–277. [Google Scholar]
- Sedaghat, M.H.; Shahmardan, M.M.; Norouzi, M.; Heydari, M. Effect of Cilia beat frequency on muco-ciliary clearance. J. Biomed. Phys. Eng. 2016, 6, 265–278. [Google Scholar]
- Smith, D.J.; Gaffney, E.A.; Blake, J.R. A viscoelastic traction layer model of muco-ciliary transport. Bull. Math. Biol. 2007, 69, 289–327. [Google Scholar] [CrossRef][Green Version]
- Sears, P.R.; Yin, W.N.; Ostrowski, L.E. Continuous mucociliary transport by primary human airway epithelial cells in vitro. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 309, L99–L108. [Google Scholar] [CrossRef]
- Smith, C.; Radhakrishnan, P.; DoHyang Lee, D.; Hessel, E.M.; Williamson, R.; O’Callaghan, C. Rhinovirus infection of human ciliated respiratory epithelial cultures. In Proceedings of the 10.1 Respiratory Infections, European Respiratory Society International Congress, London, UK, 3–7 September 2016; p. 2608. [Google Scholar]
- Feriani, L.; Juenet, M.; Fowler, C.J.; Bruot, N.; Chioccioli, M.; Holland, S.M.; Bryant, C.E.; Cicuta, P. Assessing the Collective Dynamics of Motile Cilia in Cultures of Human Airway Cells by Multiscale DDM. Biophys. J. 2017, 113, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, G.; Pifferi, M.; Vozzi, G. Automated software for analysis of ciliary beat frequency and metachronal wave orientation in primary ciliary dyskinesia. Eur. Arch. Otorhinolaryngol. 2010, 267, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Bustamante-Marin, X.M.; Ostrowski, L.E. Cilia and Mucociliary Clearance. Cold Spring Harb. Perspect. Biol. 2017, 9, a028241. [Google Scholar] [CrossRef] [PubMed]
- Button, B.; Picher, M.; Boucher, R. Differential effects of cyclic and constant stress on ATP release and mucociliary transport by human airway epithelia. J. Physiol. 2007, 580, 577–592. [Google Scholar] [CrossRef]
- Duncan, G.A.; Kim, N.; Colon-Cortes, Y.; Rodriguez, J.; Mazur, M.; Birket, S.E.; Rowe, S.M.; West, N.E.; Livraghi-Butrico, A.; Boucher, R.C.; et al. An adeno-associated viral vector capable of penetrating the mucus barrier to inhaled gene therapy. Mol. Ther. Methods Clin. Dev. 2018, 9, 296–304. [Google Scholar] [CrossRef]
- Liu, S.L.; Wang, Z.G.; Xie, H.Y.; Liu, A.A.; Lamb, D.C.; Pang, D.W. Single-virus tracking: From imaging methodologies to virological applications. Chem. Rev. 2020, 120, 1936–1979. [Google Scholar] [CrossRef]
- Liu, S.L.; Zhang, Z.L.; Tian, Z.Q.; Zhao, H.S.; Liu, H.; Sun, E.Z.; Xiao, G.F.; Zhang, W.; Wang, H.Z.; Pang, D.W. Effectively and efficiently dissecting the infection of influenza virus by quantum-dot-based single-particle tracking. ACS Nano 2012, 6, 141–150. [Google Scholar] [CrossRef]
- Zhang, L.J.; Wang, S.; Xia, L.; Lv, C.; Tang, H.W.; Liang, Z.; Xiao, G.; Pang, D.W. Lipid-specific labeling of enveloped viruses with quantum dots for single-virus tracking. MBio 2020, 11. [Google Scholar] [CrossRef]
- Griffiths, C.D.; Bilawchuk, L.M.; McDonough, J.E.; Jamieson, K.C.; Elawar, F.; Cen, Y.; Duan, W.; Lin, C.; Song, H.; Casanova, J.-L.; et al. IGF1R is an entry receptor for respiratory syncytial virus. Nature 2020, 583, 615–619. [Google Scholar] [CrossRef]
- Cohen, M.; Zhang, X.-Q.; Senaati, H.P.; Chen, H.W.; Varki, N.M.; Schooley, R.T.; Gagneux, P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol. J. 2013, 10, 321. [Google Scholar] [CrossRef]
- Schuster, B.S.; Kim, A.J.; Kays, J.C.; Kanzawa, M.M.; Guggino, W.B.; Boyle, M.P.; Rowe, S.M.; Muzyczka, N.; Suk, J.S.; Hanes, J. Overcoming the cystic fibrosis sputum barrier to leading adeno-associated virus gene therapy vectors. Mol. Ther. 2014, 22, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shan, X.; Patel, U.; Huang, X.; Lu, J.; Li, J.; Tao, N. Label-free imaging, detection, and mass measurement of single viruses by surface plasmon resonance. Proc. Natl. Acad. Sci. USA 2010, 107, 16028–16032. [Google Scholar] [CrossRef] [PubMed]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, Y.; Goda, T.; Matsumoto, A.; Takeuchi, H.; Yamaoka, S.; Miyahara, Y. Direct and label-free influenza virus detection based on multisite binding to sialic acid receptors. Biosens. Bioelectron. 2017, 92, 234–240. [Google Scholar] [CrossRef]
- Joyner, K.; Song, D.; Hawkins, R.F.; Silcott, R.D.; Duncan, G.A. A rational approach to form disulfide linked mucin hydrogels. Soft Matter 2019, 15, 9632–9639. [Google Scholar] [CrossRef]
- Yan, H.; Hjorth, M.; Winkeljann, B.; Dobryden, I.; Lieleg, O.; Crouzier, T. Glyco-modification of mucin hydrogels to investigate their immune activity. ACS Appl. Mater. Interfaces 2020, 12, 19324–19336. [Google Scholar] [CrossRef]
- Werlang, C.; Cárcarmo-Oyarce, G.; Ribbeck, K. Engineering mucus to study and influence the microbiome. Nat. Rev. Mater. 2019, 4, 134–145. [Google Scholar] [CrossRef]
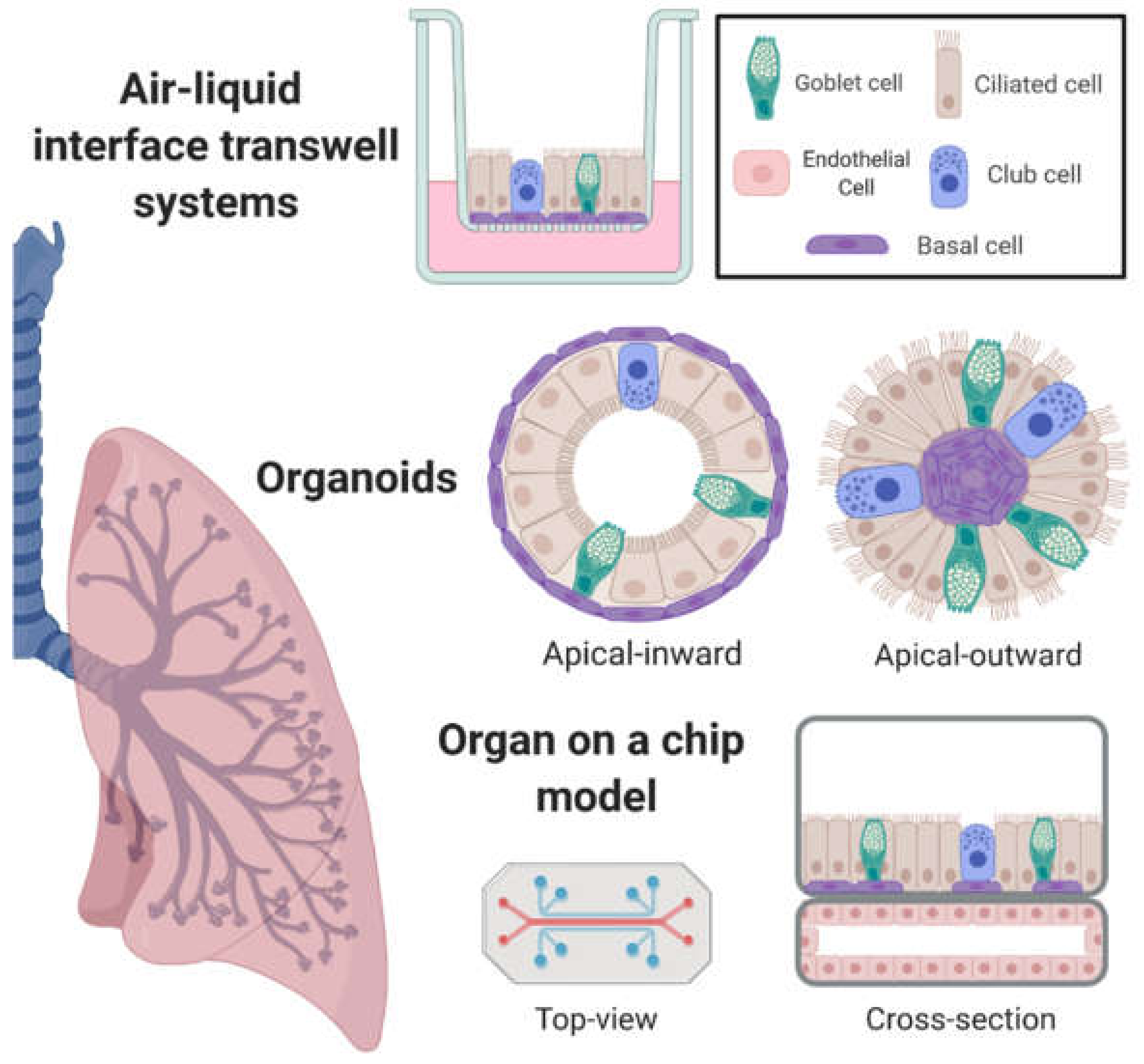
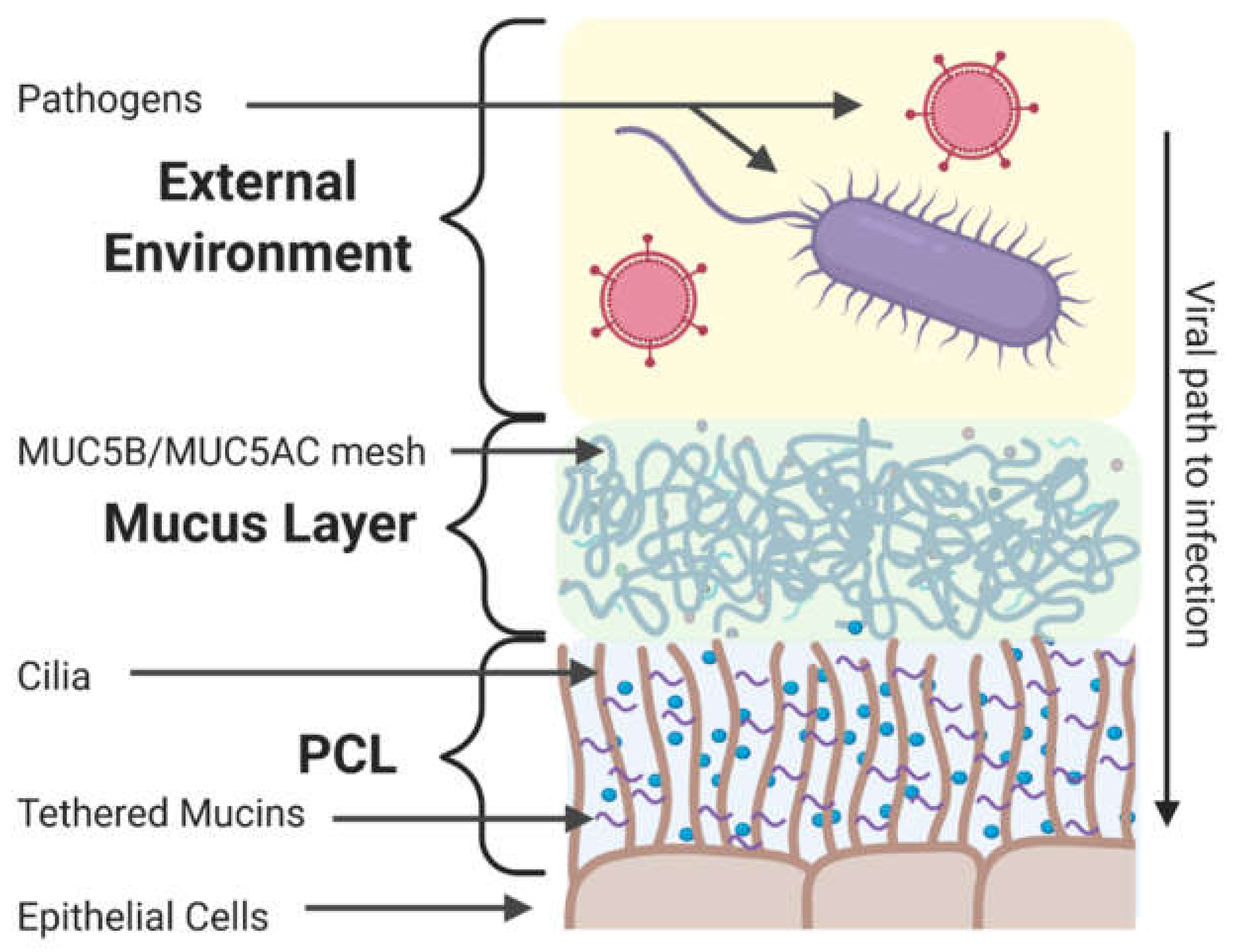
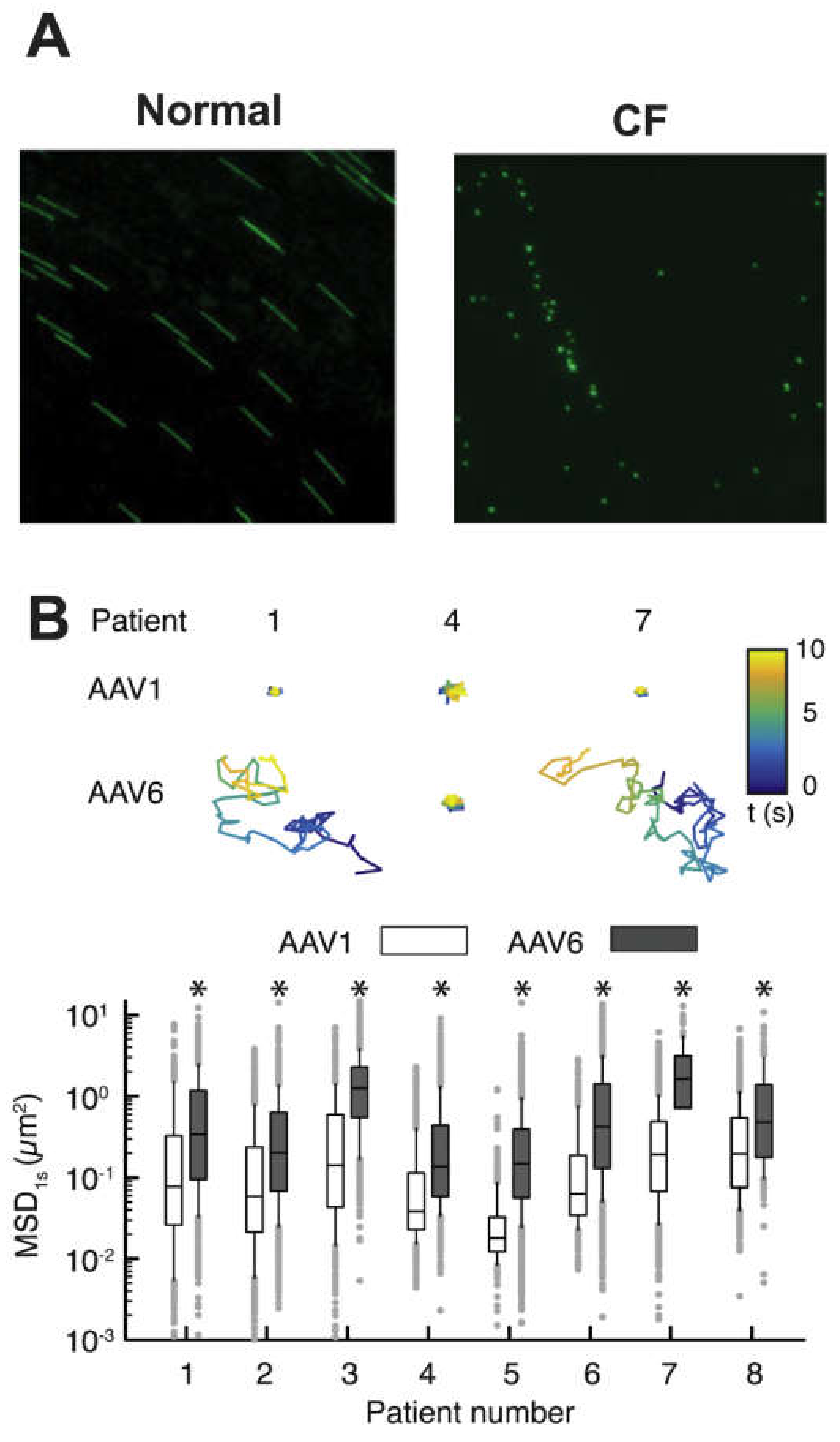
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iverson, E.; Kaler, L.; Agostino, E.L.; Song, D.; Duncan, G.A.; Scull, M.A. Leveraging 3D Model Systems to Understand Viral Interactions with the Respiratory Mucosa. Viruses 2020, 12, 1425. https://doi.org/10.3390/v12121425
Iverson E, Kaler L, Agostino EL, Song D, Duncan GA, Scull MA. Leveraging 3D Model Systems to Understand Viral Interactions with the Respiratory Mucosa. Viruses. 2020; 12(12):1425. https://doi.org/10.3390/v12121425
Chicago/Turabian StyleIverson, Ethan, Logan Kaler, Eva L. Agostino, Daniel Song, Gregg A. Duncan, and Margaret A. Scull. 2020. "Leveraging 3D Model Systems to Understand Viral Interactions with the Respiratory Mucosa" Viruses 12, no. 12: 1425. https://doi.org/10.3390/v12121425
APA StyleIverson, E., Kaler, L., Agostino, E. L., Song, D., Duncan, G. A., & Scull, M. A. (2020). Leveraging 3D Model Systems to Understand Viral Interactions with the Respiratory Mucosa. Viruses, 12(12), 1425. https://doi.org/10.3390/v12121425







